Abstract
Background
Several laboratories have shown that cells with a memory B-cell phenotype can have the same clonotype as multiple myeloma tumor cells.
Design and Methods
The aim of this study was to determine whether some memory B cells have the same genetic alterations as their corresponding multiple myeloma malignant plasma cells. The methodology included sorting multiple myeloma or memory B cells into RNA stabilizing medium for generation of subset-specific polymerase chain reaction complementary DNA libraries from one or 100 cells.
Results
Cells with the phenotype of tumor plasma cells (CD38++CD19−CD45−/+CD56−/+/++) or memory B cells (CD38−/CD19+/CD27+) were isolated by flow activated cell sorting. In samples from all four patients with multiple myeloma and from two of the three with monoclonal gammopathy of undetermined significance, we identified memory B cells expressing multiple myeloma-specific oncogenes (FGFR3; IGH-MMSET; CCND1 high) dysregulated by an IGH translocation in the respective tumor plasma cells. By contrast, in seven patients with multiple myeloma, each of whom had tumor plasma cells with a K-RAS61 mutation, a total of 32,400 memory B cells were analyzed using a sensitive allele-specific, competitive blocker polymerase chain reaction assay, but no K-RAS mutations were identified.
Conclusions
The increased expression of a specific “early” oncogene of multiple myeloma (monoclonal gammopathy of undetermined significance) in some memory B cells suggests that dysregulation of the oncogene occurs in a precursor B-cell that can generate memory B cells and transformed plasma cells. However, if memory B cells lack “late” oncogene (K-RAS) mutations but express the “early” oncogene, they cannot be involved in maintaining the multiple myeloma tumor, but presumably represent a clonotypic remnant that is only partially transformed.
Keywords: multiple myeloma, monoclonal gammopathy of undetermined significance, memory B cells, K-RAS, clonotype
Introduction
Multiple myeloma (MM) is a mostly incurable B-cell malignancy characterized by uncontrolled growth and accumulation of MM plasma cells in the bone marrow.1 A number of laboratories have identified clonotypic cells (i.e., cells having the same somatically mutated immunoglobulin genes as the MM cells) with a pre-plasma cell phenotype in peripheral blood, lymph nodes and bone marrow.2–8 Although some studies support the idea of a MM “stem-cell” or tumor-propagating cell present in a preplasmacytic compartment,9–13 other studies failed to confirm this possibility.14–16
MM oncogenesis is a multistep process, which usually is preceded by a pre-malignant tumor called monoclonal gammopathy of undetermined significance (MGUS).17,18 Two early pathways of MM oncogenesis, both of which are shared by MGUS and MM, have been proposed: a hyperdiploid pathway that is associated with multiple trisomies involving eight chromosomes, and a non-hyper-diploid pathway that is typically characterized by the presence of primary immunoglobulin heavy chain (IGH) translocations involving mostly the genes for cyclin D1 (CCND1) [t(11;14)] or FGFR3/MMSET [t(4;14)].19,20 Most hyperdiploid tumors have a CCND1 low phenotype, with the levels of ectopic and bi-allelic CCND1 expression being consistently lower than those in tumors with a t(11;14) translocation that have a CCND1 high phenotype.21–23
The prevalence of activating mutations of the K- and N-RAS oncogenes is about 30–40% in MM, with some studies showing a somewhat higher prevalence of N-RAS mutations and other studies a higher prevalence of K-RAS mutations.24–28 The prevalence of RAS mutations is much higher in malignant MM - independently of stage - than in pre-malignant MGUS. Significantly, however, although N-RAS mutations are somewhat less prevalent in MGUS than in MM, no K-RAS mutations have been identified in MGUS. Therefore, unlike hyperdiploidy and the CCND1 low phenotype, or primary IGH translocations, K-RAS mutations must be a relatively late event in pathogenesis.19,20
MM tumor plasma cells harboring a primary IGH translocation, multiple trisomies, or a RAS mutation should also have these genetic alterations in a potential MM “stem cell”, which is ultimately responsible for proliferation of the MM tumor. Here we demonstrate that we can identify “early” oncogenic events in some memory B cells but not “late” K-RAS61 mutations, suggesting that these cells are not tumor stem cells but rather premalignant, incompletely transformed remnants that coexist with the malignant cells but are not directly involved in tumor maintenance and proliferation.
Design and Methods
Patients and samples
Bone marrow aspirates and peripheral blood samples from 120 patients with MM, and three MGUS patients with a CCND1 high phenotype, were collected from our biobank of diagnostic material from clinical trials. In addition, peripheral blood samples from three healthy individuals were used as controls. All samples were obtained after informed consent and the study was approved by the Ethical Committee of Copenhagen County and was performed in accordance with the Helsinki Declaration. Bone marrow mononuclear cells, peripheral blood mononuclear cells and RNA were isolated as reported elsewhere.7,28,29
Flow cytometry and single-cell sorting
Bone marrow and peripheral blood mononuclear cells were stained with three or four colors with the monoclonal antibodies CD19-FITC, CD27-FITC/PE, CD38-FITC/APC, CD45-PERCP, CD56-PE [Becton Dickinson Immunocytometry Systems (BDIS), San Jose, CA, USA], CD19-Cy5 (Dako, Glostrup, Denmark). IgG1-FITC, IgG1-PE, IgG1-PerCP and IgG1-APC (BDIS) were used as negative controls. Fluorescence activated cell sorting (FACS) of memory B cells or plasma cells, using a FACS Vantage (BDIS) in counter mode, was performed to separate cells directly to polymerase chain reaction (PCR) tubes.7,29,30
Oncogene expression levels in purified plasma cells
We have previously described an approach to examine the gene expression profile in an immunophenotypically defined population of plasma cells.28,31–33 Briefly, single or 100 plasma cells/memory B cells were FACS-sorted directly to PCR tubes: global reverse transcriptase (RT)-PCR was performed to generate a PCR cDNA archive. The levels of expression of CCND1, FGFR3 and MMSET, normalized to the level of expression of β-actin, were determined by real-time RT-PCR for each archived cDNA. For quantitative reactions (qRT-PCR), the amount of cDNA was adjusted to give a Ct of 16–18 for β-actin.
IGH-MMSET reverse transcriptase polymerase chain reaction
Nested IGH-MMSET RT-PCR was performed as described by Malgeri et al.34 The identity of all PCR products was confirmed by direct sequencing. The frequency of cells containing IGH-MMSET hybrid transcripts was estimated by limiting dilution, using the equation f = ∑ki/[N-∑(ki x ci/2)] (where f = frequency of the target cell, ki = the number of positive wells at the i’th dilution, ci = number of cells per well at dilution i and N = the total number of cells in all the wells at all dilutions).31
Blimp-1 and XBP-1 polymerase chain reaction
Blimp-1 and XBP-1 PCR was performed on global RT-PCR amplified products from one or 100 sorted cells using the following primers: Blimp-1, forward: 5-AGATTTAA-GCCCCTTGCCACA-3, reverse: 5-TTCAACGTCTTCTTAGGGTCTCTTG-3. XBP-1, forward: 5-TCCAGTTTTAGGTCCTTTAGTTTGC-3, reverse: 5-ATACATGAGGGA-AAGAGCCCC-3. The identity of the PCR products was determined by sequencing as previously described.32
RAS mutation analysis by direct sequencing of purified plasma cell lysate
In a previous study we identified MM patients having K-RAS61 mutations by direct sequencing of a plasma cell-lysate generated from 100 FACS-sorted immunophenotypic aberrant plasma cells (CD38+/CD45−/i/CD19−/CD56++).28
Allele-specific competitive blocker polymerase chain reaction for detection of K-RAS codon 61 mutations in purified plasma cell lysate
We previously designed an allele-specific competitive blocker (ACB)-PCR generating preferential amplification of the allele carrying K-RAS61 (CAA to CAC and CAA to CTA) mutations.28 The method used a mutant-specific primer in combination with a non-extendable blocker primer with preferential binding for the wild-type allele. ACB-PCR was performed on a lysate generated from 100 FACS-purified memory B cells or plasma cells with an aberrant immunophenotype.
Results
Selection of bone marrow plasma cells and peripheral blood memory B cells
Normal plasma cells were identified as CD38++/CD19+/CD45+/CD56− and MM plasma cells as CD38++ and CD19−and/or CD45− and/or CD56+. The MGUS patients analyzed often had mixtures of normal and aberrant plasma cells (MM immunophenotype) in the bone marrow, but only immunophenotypically aberrant plasma cells were FACS-sorted for further analyses, as described previously.28 Memory B cells with the CD38−/CD19+/CD27+ immunophenotype were selected for FACS-sorting in all cases (healthy individuals, MGUS and MM), as described previously.7
Selected memory B cells are Blimp-1 and XBP-1 negative
MM plasma cells and memory B cells were separated based on their pattern of expression of CD antigens. However, the immunophenotype of the malignant plasma cells could be a reflection of the environment (bone marrow versus peripheral blood). Thus, the observed cells in the CD38−/CD19+/CD27+ FACS-sorted fraction with a dysregulated oncogene could be MM plasma cells with a yet unidentified aberrant immunophenotype, or simply contaminating circulating myeloma plasma cells as a result of the sorting procedure. To exclude the above possibilities we analyzed all FACS-sorted cells for expression of Blimp-1, a transcriptional repressor important for the final differentiation of B cells into plasma cells, which is expressed in all plasma cells, but absent in memory B cells;35,36 and XBP-1, a second transcription factor that is unique to plasma cell differentiation.37 The Blimp-1 and XBP-1 PCR assays could reproducibly detect a single FACS-sorted MM plasma cell among 100 memory B cells FACS-sorted from a healthy individual (data not shown). In all cases the FACS-sorted CD38−/CD19+/CD27+ cells (≤ 100 per tube) were negative for Blimp-1 and XBP-1, whereas all FACS-sorted CD38++/CD19+/CD45i/CD56− and CD38++/CD19−/CD45−/i/CD56−/+/++ plasma cells were positive (Figure 1), indicating that an aberrant surface phenotype or contamination had no impact on the results.
Figure 1.
Blimp-1 and XBP-1 expression. Global RT-PCR products from plasma cells and memory B cells were analyzed by RT-PCR for expression of the transcription factors XBP-1 and Blimp-1. Results are shown for three myeloma patients and three healthy individuals, with 100 sorted cells per tube in each case. Lanes 1–3, oncogene-negative (healthy) memory B cells; lanes 4–6, oncogene-positive (myeloma) memory B cells; lanes 7–9, MM plasma cells; lanes 10–12, healthy plasma cells.
Healthy memory B cells do not express CCND1 or FGFR3
Hematopoietic cells do not express CCND1 or FGFR3 and a complete absence of both CCND1 and FGFR3 expression was verified analyzing a total of 3600 healthy memory B cells isolated from the peripheral blood of three individuals, and 700 healthy plasma cells from seven individuals (Table 1 and Figures 2 and 3).33,38 Thus, the expression of CCND1 or FGFR3 in memory B cells, as found in their autologous MM plasma cells, would be a myeloma-specific event.
Table 1.
Expression of dysregulated oncogenes identified in tumor plasma cells sometimes detectable in the memory B-cell compartment.
Figure 2.
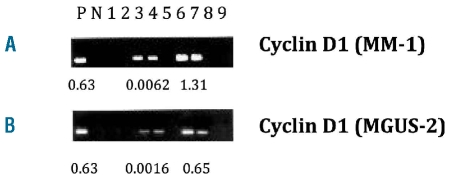
Oncogene analyses of plasma cells and memory B cells: (A) from a CCND1-positive MM patient (MM-1) and (B) from a CCND1-positive MGUS patient (MGUS-2). Global RT-PCR products were screened for CCND1 expression using real-time PCR, with 100 sorted cells per tube in each case. CCND1-positive or negative real-time PCR products were run on a gel and bands corresponding to CCND1 were excised from the gel and sequenced. The oncogene/β-actin ratio determined by real-time PCR is shown below each positive case. P, positive control; N, negative control (H2O); lanes 1 and 2, healthy memory B cells; lanes 3 and 4, oncogene-positive memory B cells; lane 5, RT control; lanes 6 and 7, myeloma plasma cells; lanes 8 and 9, healthy plasma cells.
Figure 3.
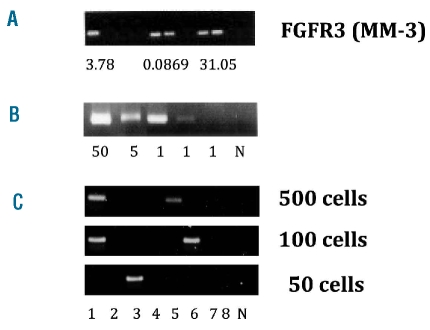
Identification of IGH-MMSET hybrid transcripts in memory B cells. (A) The FGFR3-positive MM patient (MM-3) was analyzed as described in Figure 2. (B) The CD38++/CD19−/CD56++ plasma cells were flow-sorted directly to PCR tubes in numbers of 50, 5 and 1 cell/PCR tube followed by nested RT-PCR for IGH-MMSET hybrid transcripts. (C) RT-PCR analysis of IGH-MMSET hybrid transcripts in sorted CD19+/CD38−/CD27+ memory B cells. Memory B cells were flow sorted in numbers of 500, 100 and 50 cells/PCR tube, with eight PCR tubes for each cell number followed by RT-PCR analysis of IGH-MMSET hybrid transcripts. N = negative control (H2O).
Two patterns of CCND1 expression in multiple myeloma or monoclonal gammopathy of undetermined significance
Previously, we identified a high level expression of oncogenes involved in IGH translocations (FGFR3/MMSET, CCND1), or expression of hybrid transcripts associated with translocation t(4;14) (IGH-MMSET) in FACS-purified MM plasma cells.33,38 There are two distinct groups of CCND1-positive MM: the t(11;14) group, all showing high expression of CCND1, and a group with low to intermediate CCND1 expression associated with hyperdiploidy, including 11q trisomy.22 To identify which MM patients belonged to the two different CCND1 groups for further studies we determined the level of CCND1 expression in FACS-purified plasma cells from 120 MM patients (Online Supplementary Figure S1). As in previous studies, which included only about half of these patients, we identified 21(17.5%) MM patients with plasma cells showing high CCND1 expression (CCND1/β-actin >0.1) indicating a t(11;14), and 30 (25%) patients with low CCND1 expression (CCND1/β-actin >0.001 and <0.1).21,22,28,33,39 By contrast, the CCND1/β-actin was less than 0.0001 (but mostly <0.000001) in the other 69 cases of MM, and also in plasma cells and memory B cells from healthy individuals (Table 1, Figure 2, and data not shown).
CCND1 expression in memory B cells from patients with multiple myeloma or monoclonal gammopathy of undetermined significance with t(11;14)
Two patients with MM and three with MGUS with a CCND1 high phenotype were selected for further analyses (Table 1, MM 1-2 and MGUS 1-3).33 Memory B cells from the two MM and three MGUS patients were FACS-purified, followed by global RT-PCR amplifications of the RNA pools from one or 100 cells (Table 1). The expression of CCND1 was analyzed using quantitative real-time PCR assays (reproducibility shown in Online Supplementary Figure S2). From patient MM-1, for whom 4% of memory B cells had been previously identified as clonotypic,40,41 a single memory B cell was FACS-sorted into each of 120 PCR tubes, with an amplified cDNA product (a cDNA archive) generated in 96 tubes. Analysis of CCND1 expression (Figure 2A, Table 1) revealed 5/98 positive wells, indicating that memory B cells had a similar frequency of CCND1+ cells (5%) as clonotypic cells (4%). Memory B cells from patient MM-2 were sorted into nine tubes, each containing 100 sorted cells, and CCND1 expression was detected in five of these nine tubes (Table 1). Therefore, less than 1% of the memory B cells from patient MM-2 expressed CCND1, consistent with the results of a previous study in which we showed that less than 1% of the memory B cells were clonotypic in this patient.41 We also identified CCND1+ memory B cells in two out of the three CCND1+ MGUS cases, by analyzing 1500, 900, and 800 (negative case) memory B cells available for analyses (Table 1 and Figure 2B).
We observed a lower level of oncogene expression – in the range of 94 to 710 times lower – in memory B cells than in autologous MM plasma cells (Table 1). This could be due to several factors. Firstly, the expected ratio of oncogene-positive cells to normal cells is nearly 100% when sorting plasma cells, but usually 0 or 1 of 100 when sorting memory B cells (except for patient MM-1, Table 1). Secondly, as the dysregulation of the oncogenes is believed to be under the control of an IGH enhancer, oncogene expression could be further induced and increased upon plasma cell differentiation.
Expression of FGFR3 and MMSET hybrid transcripts in memory B cells
Two patients (MM-3 and MM-4) had MM tumors with t(4;14) translocations, as determined by ectopic and enhanced expression of FGFR3, and also MMSET hybrid transcripts. For both patients, we demonstrated increased expression of FGFR3 in about 0.1 to 1% of purified memory B cells (Table 1 and Figure 3A), consistent with the presence of less than 1% clonotypic memory B cells in these patients, as determined previously.41 For patient MM-3, there was enough material to enable us to perform a limiting dilution experiment to verify the presence and frequency of IGH-MMSET hybrid transcript-positive cells in the memory B-cell pool. IGH-MMSET hybrid transcripts were detected in single flow-sorted plasma cells (Figure 3B). IGH-MMSET hybrid transcripts were detected in 2/8, 2/8 and 1/8 PCR analyses with 500, 100, 50 cells/PCR tube, respectively (Figure 3C). Limiting dilution statistical analysis of the results shown in Figure 3C gave an estimated frequency of 0.1% IGH-MMSET-positive memory B cells, similar to the frequency of FGFR3-expressing memory B cells in this patient.
CCND1 low memory B cells were present in only two of nine patients with CCND1 low multiple myeloma
Memory B cells were FACS-sorted from nine MM patients belonging to the CCND1 low group (Table 1, MM 5 - 13). In two of these MM patients, CCND1+ memory B cells were identified in 2/12 and 5/96 PCR tubes with 100 cells/tube (Table 1, patients MM-9 and MM-13). However, in the other seven of nine patients with CCND1 low MM, we did not detect CCND1 expression in any of the purified memory B cell pools.
Activating K-RAS61 mutations are absent in memory B cells
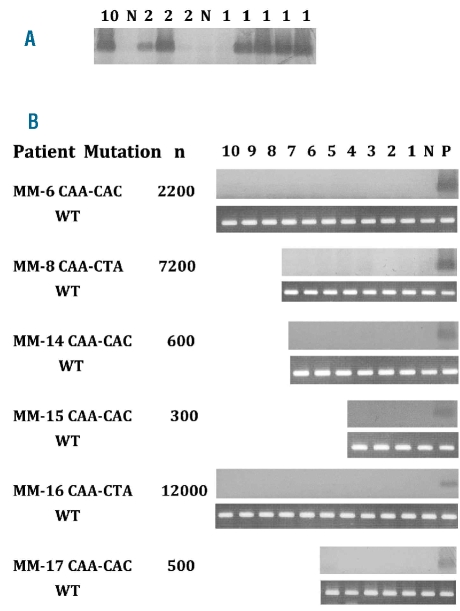
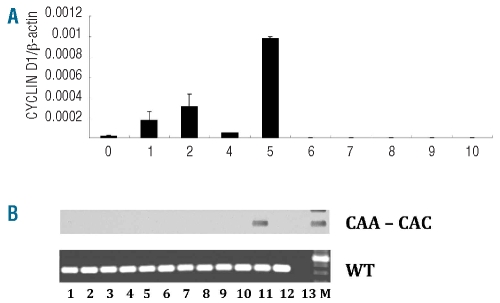
FACS-sorted MM plasma cells (CD38++/CD19−/CD45−/i/ CD56−/+/++) were analyzed previously for the presence of mutations in the N-RAS and K-RAS genes using RT-PCR followed by direct sequencing, enabling the identification of patients who had aberrant MM plasma cells with a RAS mutation at the time of diagnosis.28 From seven MM patients with a known CAC or CTA K-RAS61 mutation in their MM plasma cells, CD38−/CD19+/CD27+ memory cells from peripheral blood mononuclear cells were FACS-sorted to produce 100 cells/PCR tube, then subjected to ACB-PCR for detection of K-RAS61 mutations. As a positive control for the ACB-PCR method, a single RAS-mutated MM cell was FACS-sorted into 100 non-mutated healthy B cells in each experiment (Figure 4A), confirming that it was possible to efficiently detect a single cell containing the K-RAS61 mutation in the presence of 100 cells without the mutation. Different numbers of CD38−/CD19+/CD27+ cells were available from the obtained blood samples, ranging from 300 to 12,000 cells, so that a total of 32,400 CD38−/CD19+/CD27+ cells were analyzed (Figures 4B and 5B). However, all 32,400 memory B cells analyzed from seven different MM patients with RAS mutations in their plasma cells were found to be negative for K-RAS61 mutations in the memory B-cell populations (Figures 4B and 5B).
Figure 4.
Screening memory B cells for K-RAS61 mutations. (A) Sensitivity of ACB-PCR when analyzing FACS-sorted plasma cells. FACS-sorting of 1, 2 or 10 MM plasma cells from a patient with a K-RAS61 mutation to PCR tubes containing 100 FACS-sorted CD19+ B cells from a healthy individual. The results are only shown for the CAA to CAC mutation, but similar results was obtained for the CAA to CTA mutation. In four out of five PCR tubes with a single FACS-sorted K-RAS61+ MM plasma cell among 100 healthy B cells, a K-RAS61+ MM plasma cell was detected. (B) From six patients with a K-RAS61 mutation, 100 CD19+/CD38−/CD27+ memory B cells were FACS sorted for each ACB-PCR analysis. For each of the MM patients the K-RAS61 mutation type is shown and the total number of CD19+/CD38−/CD27+ memory B cells available for analysis is given (n). P = positive control, 100 FACS-sorted plasma cells from a patient with a K-RAS61 mutation. N = negative control, 100 FACS-sorted CD19+ B cells from a healthy donor. WT = wild-type, an analysis for wild-type RAS was performed for each FACS-sorted memory B-cell population as a positive control.
Figure 5.
Simultaneous screening for CCND1 expression and K-RAS61 mutations. A cell lysate obtained from 100 FACS-sorted memory B cells from patient MM-13 was split into two aliquots, one for global RT-PCR followed by real-time PCR for CCND1 and one for ACB-PCR detection of K-RAS61 mutations. (A) In 5/96 PCR tubes CCND1+ memory B cells were identified, the measured CCND1 level is shown (n. 1–5). N. 6–10 show five representative negative CCND1 measurements. (B) Each memory B-cell lysate was analyzed for the KRAS61 CAA to CAC mutation present in the patients’ MM plasma cells. As a control for the presence of cDNA, each cell lysate was analyzed for the presence of wild-type (WT) RAS. Lanes 1–5, CCND1-positive memory B cells (see Online Supplementary Figure 2A). Lanes 6–10, CCND1-negative memory B cells. Lane 11, positive control, 100 FACS-sorted plasma cells from patient MM-13 with a K-RAS61 mutation. Lane 12, negative control, 100 FACS-sorted CD19+ B cells from a healthy donor. Lane 13, negative control (H2O). M = molecular size marker. RAS mutations were absent from all memory B cells analyzed.
The presence of CCND1 low memory B cells lacking K-RAS61 mutations
To directly compare CCND1 expression and KRAS61 mutations in the same pools of memory B cells, we FACS-sorted memory B cells from patient MM-13 who had MM plasma cells with both CCND1 low expression and a K-RAS61 mutation (Table 1). A total of 9600 memory B cells were FACS-sorted into 96 PCR tubes with 100 cells/tube (Table 1, MM-13), and the cell lysate was split into two aliquots, one for global RT-PCR followed by real-time PCR for CCND1 and one for ACB-PCR detection of K-RAS61 mutations. Memory B cells expressing CCND1 were identified in 5/96 PCR tubes (Figure 5A, Table 1). However, RAS mutations were absent in all memory B cells FACS-sorted from patient MM-13, including the tubes containing memory B cells that expressed CCND1 (Figure 5B).
Discussion
All MM tumors analyzed have stable V(D)J rearrangements with an extensive accumulation of somatic mutations in the complementarity determining regions, and an absence of intraclonal somatic mutation.42,43 These findings support the idea that MM cells are derived from germinal center or post-germinal center B cells. A normal germinal center B cell that has undergone repeated cycles of somatic hypermutation and antigen selection can differentiate into either a memory B cell or a terminally differentiated, long-lived plasma cell that has undergone productive IgH switch recombination, and is typically found in the bone marrow.44 A number of laboratories have identified clonotypic CD19+ cells in the peripheral blood that share identical V(D)J sequences with the MM tumor cells in the same patient.2–8 Some clonotypic CD19+ cells are pre-switch, expressing IgM, whereas others have switched to the same IgH isotype as the CD138+ MM tumor cells. We previously identified a CD19+/CD27+/CD38− subset of clonotypic cells in the peripheral blood which fulfills the immunophenotypic characteristics of a memory B cell.7,40,44
Recently, it has been shown that most – if not all – MM tumors are generated from premalignant MGUS tumors.17,18 Primary IGH translocations, which are shared by MGUS and MM tumors, mostly have breakpoints that fall within or near IGH switch regions.45,46 It is, therefore, thought that these translocations, which are present in about 40% of MM tumors, occur as a result of errors in IgH switch recombination, which takes place mostly in germinal center or post-germinal center B cells. Since hyperdiploidy and cyclin D dysregulation, an apparently unifying event in the pathogenesis of MM, are present in both MGUS and MM, it seems likely that these early oncogenic events may also occur in germinal center or post-germinal center B cells, although this needs to be documented.20,21
It is possible that either fully or incompletely transformed cells can generate progeny with memory B cell or plasmablast/plasma cell phenotypes. Hence, there are several possible explanations for the presence of clonotypic B cells in the peripheral blood of MM patients. First, the clonotypic B cells could be incompletely transformed memory B cells that have undergone only some of the very early oncogenic events, and perhaps these cells have some proliferative or survival advantage compared to their normal counterparts. Second, the clonotypic B cells could be the MM “stem/tumor-propagating cell” that has a critical role in the maintenance of the MM tumor, and includes the full spectrum of oncogenic changes as the MM cells with a plasma cell phenotype. Third, the clonotypic B cells could be phenotypic variants of the MM tumor cells that have lost some plasma cell markers and gained some B-cell markers, possibly related to the loss of contact with the bone marrow microenvironment. In principle, any of these three types of clonotypic B cells could generate a clonotypic tumor in an immunodeficient xenogenic model. However, the first type of clonotypic B cell would differ from the other two types by sharing early but not late oncogenic events with the CD38+ MM tumor cells.
Our study was designed to isolate the CD38−CD19+CD27+ memory B cells from the peripheral blood of patients with MM and MGUS and sort them into pools containing from one up to 100 cells, from which a global PCR cDNA archive was made. Our results suggest that sorted cells were not significantly contaminated by MM tumor cells. First, we were unable to detect Blimp-1 or XBP-1 transcripts using an assay that could detect one normal plasma cell or one MM aberrant plasma cell in a pool of 100 purified memory B cells. Secondly, we could not detect CCND1 expression in the purified memory B-cell population isolated from seven of nine MM patients in whom the tumor cells had a CCND1 low phenotype. Third, we could not detect K-RAS61 mutant transcripts in the purified memory B-cell populations isolated from all seven MM patients whose MM tumor cells contained the K-RAS61 mutation. The lack of significant contamination of the memory B-cell populations most likely reflects the fact that there were very few CD38+ MM tumor cells in the peripheral blood.1,47 In addition, the FACS sorting procedure would be expected to exclude CD38+CD19− cells from the purified CD38−CD19+CD27+ memory B-cell population more effectively than immunomagnetic selection of CD19+ cells.
As summarized in the Results section, we detected ectopic FGFR3 expression in the memory B-cell population of both MM patients who had a t(4;14) translocation, and were also able to confirm this result in one of these two patients by identifying MMSET hybrid transcripts that represent direct products of the t(4;14) translocation, a result reported previously for a single MM patient.14 We also detected CCND1 expression in peripheral blood memory B-cell populations derived from both MM patients and two of three MGUS patients with a CCND1 D1 high phenotype which is associated with t(11;14) in their MM or MGUS tumor cells. It is possible that the third MGUS patient had memory B cells with CCND1 expression, but that these were not present in the 800 memory B cells that we were able to analyze. Significantly, for the four cases of MM above, the fraction of memory B cells expressing a translocation-associated oncogene product was the same as the fraction of clonotypic memory B cells, as reported previously for the same patients.41 We were unable to detect CCND1 expression in the memory B cells derived from seven of nine MM patients with a CCND1 low phenotype, which is almost invariably associated with hyperdiploidy. Importantly, in all cases, the dysregulated oncogene identified in memory B cells was the same as that found in the corresponding MM tumor.
In contrast to the identification of oncogene products dysregulated by translocations in the memory B cells from all four MM patients and two of three MGUS patients, we were unable to identify K-RAS61 mutations in the memory B cells of all seven patients with K-RAS61 mutations in their CD38+ MM tumor cells. Moreover, in one patient who had a MM tumor with a CCND1 low phenotype and a K-RAS61 mutation, we could not identify the K-RAS61 mutation in several pools of memory B cells that expressed CCND1.
Although there is little information about oncogenic changes in clonotypic B cells, it is important to mention a prior study that appears to be in disagreement with our results. Zojer et al.47 identified a few percent of purified CD19+ peripheral blood B cells containing both early (RB1 deletion, trisomy 11) and late (P53 deletion) karyotypic abnormalities that are shared with CD138+ MM tumor cells. However, the authors estimated that the immunobead purification of CD19+ cells averaged about 95% so that the low fraction of cells with karyotypic abnormalities might represent contaminating CD138+ MM tumor cells. In fact, this possibility seems to be supported by their finding that the rare karyotypically abnormal cells in the CD19-selected population expressed the same high levels of cytoplasmic immunoglobulin as found in CD138+ MM tumor cells. In addition, their late event (P53 deletion) is associated with advanced tumors that have a poor prognosis, and may be more likely to be found in the peripheral blood.1,20
There are two conflicting albeit potentially reconcilable views about the phenotype of a MM “stem/tumor-propagating” cell. On the one hand, Epstein et al. have shown that CD138+ cells can undergo substantial proliferation and several rounds of serial transplantation in ectopic bone in SCID-hu or SCID-rab immunodeficient models, whereas CD38− or CD138− cells do not proliferate significantly in this model.15,16 On the other hand, Matsui, Pilarski and others have shown that CD38−CD19+CD27+ cells - but not CD38+ or CD138+ cells - can form in vitro clones, or in vivo tumors that give rise to CD138+ cells in immunodeficient NOD/SCID mice.10,11,13 So, how do our results fit into this unresolved issue? Given that others have identified clonotypic B cells that express IgM,2,3 it is not surprising that we have found that some memory B cells can share early oncogenic events with MM tumor cells but not K-RAS61 mutations. Although we found no evidence for a MM “stem/tumor-propagating” cell with a memory B-cell phenotype in peripheral blood, it is possible that this cell might be found mainly in another location, or exist at too low a level in peripheral blood to be detected by our assays.
In conclusion, our results strongly suggest the existence of clonotypic memory B cells in the peripheral blood that have “early” (ectopic translocation products) but not “late” (K-RAS61 mutations) oncogenic events. Memory B cells with this genotype must represent pre-malignant, incompletely transformed remnants that could have some proliferative advantage compared to normal memory B cells. It remains to be determined whether clonotypic B cells that can be cloned in vitro or can form tumors in NOD/SCID mice are truly MM “stem/tumor-propagating” cells or phenotypically aberrant MM tumor cells that share essentially all early and late oncogenic changes with MM tumor cells.
Acknowledgment
the authors thank Kirsten Nikolajsen for technical assistance with FACS sorting.
Footnotes
Funding: this project was supported in part by the Danish Cancer Society, grant number DP 00006 to TR and DP 22-00-0314 and DP 2002-04 to HEJ. HEJ was supported by a senior grant from the Multiple Myeloma Research Foundation 2003-4 and EU 6th FP to MSC-NET (LSHC-CT-2006-037602) and MK was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
The online version of this article has a Supplementary Appendix
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Malpas JS, Bergsagel DE, Kyle RA, Anderson KC. Myeloma: Biology and Management. Third ed. Philadelphia: Saunders; 2004. [Google Scholar]
- 2.Bakkus MH, Van Riet I, Van Camp B, Thielemans K. Evidence that the clonogenic cell in multiple myeloma originates from a pre-switched but somatically mutated B cell. Br J Haematol. 1994;87(1):68–74. doi: 10.1111/j.1365-2141.1994.tb04872.x. [DOI] [PubMed] [Google Scholar]
- 3.Billadeau D, Ahmann G, Greipp P, Van Ness B. The bone marrow of multiple myeloma patients contains B cell populations at different stages of differentiation that are clonally related to the malignant plasma cell. J Exp Med. 1993;178(3):1023–31. doi: 10.1084/jem.178.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caligaris-Cappio F, Bergui L, Gregoretti MG, Gaidano G, Gaboli M, Schena M, et al. Role of bone marrow stromal cells in the growth of human multiple myeloma. Blood. 1991;77(12):2688–93. [PubMed] [Google Scholar]
- 5.Chen BJ, Epstein J. Circulating clonal lymphocytes in myeloma constitute a minor subpopulation of B cells. Blood. 1996;87(5):1972–6. [PubMed] [Google Scholar]
- 6.Corradini P, Boccadoro M, Voena C, Pileri A. Evidence for a bone marrow B cell transcribing malignant plasma cell VDJ joined to C mu sequence in immunoglobulin (IgG)- and IgA-secreting multiple myelomas. J Exp Med. 1993;178(3):1091–6. doi: 10.1084/jem.178.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen T, Lodahl M, Hancke S, Johnsen HE. In multiple myeloma clonotypic CD38−/CD19+/CD27+ memory B cells recirculate through bone marrow, peripheral blood and lymph nodes. Leuk Lymphoma. 2004;45(7):1413–7. doi: 10.1080/10428190410001655157. [DOI] [PubMed] [Google Scholar]
- 8.Szczepek AJ, Seeberger K, Wizniak J, Mant MJ, Belch AR, Pilarski LM. A high frequency of circulating B cells share clonotypic Ig heavy-chain VDJ rearrangements with autologous bone marrow plasma cells in multiple myeloma, as measured by single-cell and in situ reverse transcriptase-polymerase chain reaction. Blood. 1998;92(8):2844–55. [PubMed] [Google Scholar]
- 9.Huff CA, Matsui W. Multiple myeloma cancer stem cells. J Clin Oncol. 2008;26(17):2895–900. doi: 10.1200/JCO.2007.15.8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kukreja A, Hutchinson A, Dhodapkar K, Mazumder A, Vesole D, Angitapalli R, et al. Enhancement of clonogenicity of human multiple myeloma by dendritic cells. J Exp Med. 2006;203(8):1859–65. doi: 10.1084/jem.20052136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsui W, Wang Q, Barber JP, Brennan S, Smith BD, Borrello I, et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 2008;68(1):190–7. doi: 10.1158/0008-5472.CAN-07-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pilarski LM, Hipperson G, Seeberger K, Pruski E, Coupland RW, Belch AR. Myeloma progenitors in the blood of patients with aggressive or minimal disease: engraftment and self-renewal of primary human myeloma in the bone marrow of NOD SCID mice. Blood. 2000;95(3):1056–65. [PubMed] [Google Scholar]
- 13.Pilarski LM, Seeberger K, Coupland RW, Eshpeter A, Keats JJ, Taylor BJ, et al. Leukemic B cells clonally identical to myeloma plasma cells are myelomagenic in NOD/SCID mice. Exp Hematol. 2002;30(3):221–8. doi: 10.1016/s0301-472x(01)00788-3. [DOI] [PubMed] [Google Scholar]
- 14.Guikema JE, Vellenga E, Bakkus MH, Bos NA. Myeloma clonotypic B cells are hampered in their ability to undergo B-cell differentiation in vitro. Br J Haematol. 2002;119(1):54–61. doi: 10.1046/j.1365-2141.2002.03789.x. [DOI] [PubMed] [Google Scholar]
- 15.Yaccoby S, Epstein J. The proliferative potential of myeloma plasma cells manifest in the SCID-hu host. Blood. 1999;94(10):3576–82. [PubMed] [Google Scholar]
- 16.Yata K, Yaccoby S. The SCID-rab model: a novel in vivo system for primary human myeloma demonstrating growth of CD138-expressing malignant cells. Leukemia. 2004;18(11):1891–7. doi: 10.1038/sj.leu.2403513. [DOI] [PubMed] [Google Scholar]
- 17.Landgren O, Kyle RA, Pfeiffer RM, Katzmann JA, Caporaso NE, Hayes RB, et al. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood. 2009;113(22):5412–7. doi: 10.1182/blood-2008-12-194241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiss BM, Abadie J, Verma P, Howard RS, Kuehl WM. A monoclonal gammopathy precedes multiple myeloma in most patients. Blood. 2009;113(22):5418–22. doi: 10.1182/blood-2008-12-195008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chng WJ, Glebov O, Bergsagel PL, Kuehl WM. Genetic events in the pathogenesis of multiple myeloma. Best Pract Res Clin Haematol. 2007;20(4):571–96. doi: 10.1016/j.beha.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23(12):2210–21. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergsagel PL, Kuehl WM, Zhan F, Sawyer J, Barlogie B, Shaughnessy J., Jr Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood. 2005;106(1):296–303. doi: 10.1182/blood-2005-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Specht K, Haralambieva E, Bink K, Kremer M, Mandl-Weber S, Koch I, et al. Different mechanisms of cyclin D1 overexpression in multiple myeloma revealed by fluorescence in situ hybridization and quantitative analysis of mRNA levels. Blood. 2004;104(4):1120–6. doi: 10.1182/blood-2003-11-3837. [DOI] [PubMed] [Google Scholar]
- 23.Zhan F, Barlogie B, Arzoumanian V, Huang Y, Williams DR, Hollmig K, et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood. 2007;109(4):1692–700. doi: 10.1182/blood-2006-07-037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bezieau S, Devilder MC, Avet-Loiseau H, Mellerin MP, Puthier D, Pennarun E, et al. High incidence of N and K-Ras activating mutations in multiple myeloma and primary plasma cell leukemia at diagnosis. Hum Mutat. 2001;18(3):212–24. doi: 10.1002/humu.1177. [DOI] [PubMed] [Google Scholar]
- 25.Chng WJ, Gonzalez-Paz N, Price-Troska T, Jacobus S, Rajkumar SV, Oken MM, et al. Clinical and biological significance of RAS mutations in multiple myeloma. Leukemia. 2008;22(12):2280–4. doi: 10.1038/leu.2008.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corradini P, Ladetto M, Voena C, Palumbo A, Inghirami G, Knowles DM, et al. Mutational activation of N- and K-ras oncogenes in plasma cell dyscrasias. Blood. 1993;81(10):2708–13. [PubMed] [Google Scholar]
- 27.Liu P, Leong T, Quam L, Billadeau D, Kay NE, Greipp P, et al. Activating mutations of N- and K-ras in multiple myeloma show different clinical associations: analysis of the Eastern Cooperative Oncology Group Phase III Trial. Blood. 1996;88(7):2699–706. [PubMed] [Google Scholar]
- 28.Rasmussen T, Kuehl M, Lodahl M, Johnsen HE, Dahl IM. Possible roles for activating RAS mutations in the MGUS to MM transition and in the intramedullary to extramedullary transition in some plasma cell tumors. Blood. 2005;105(1):317–23. doi: 10.1182/blood-2004-03-0833. [DOI] [PubMed] [Google Scholar]
- 29.Rasmussen T. The presence of circulating clonal CD19+ cells in multiple myeloma. Leuk Lymphoma. 2001;42(6):1359–66. doi: 10.3109/10428190109097764. [DOI] [PubMed] [Google Scholar]
- 30.Rasmussen T, Jensen L, Honore L, Johnsen HE. Frequency and kinetics of polyclonal and clonal B cells in the peripheral blood of patients being treated for multiple myeloma. Blood. 2000;96(13):4357–9. [PubMed] [Google Scholar]
- 31.Molesh DA, Hall JM. Quantitative analysis of CD34+ stem cells using RT-PCR on whole cells. PCR Methods Appl. 1994;3(5):278–84. doi: 10.1101/gr.3.5.278. [DOI] [PubMed] [Google Scholar]
- 32.Rasmussen T, Jensen L, Honore L, Andersen H, Johnsen HE. Circulating clonal cells in multiple myeloma do not express CD34 mRNA, as measured by single-cell and real-time RT-PCR assays. Br J Haematol. 1999;107(4):818–24. doi: 10.1046/j.1365-2141.1999.01770.x. [DOI] [PubMed] [Google Scholar]
- 33.Rasmussen T, Theilgaard-Monch K, Hudlebusch HR, Lodahl M, Johnsen HE, Dahl IM. Occurrence of dysregulated oncogenes in primary plasma cells representing consecutive stages of myeloma pathogenesis: indications for different disease entities. Br J Haematol. 2003;123(2):253–62. doi: 10.1046/j.1365-2141.2003.04577.x. [DOI] [PubMed] [Google Scholar]
- 34.Malgeri U, Baldini L, Perfetti V, Fabris S, Vignarelli MC, Colombo G, et al. Detection of t(4;14)(p16.3;q32) chromosomal translocation in multiple myeloma by reverse transcription-polymerase chain reaction analysis of IGH-MMSET fusion transcripts. Cancer Res. 2000;60(15):4058–61. [PubMed] [Google Scholar]
- 35.Angelin-Duclos C, Cattoretti G, Lin KI, Calame K. Commitment of B lymphocytes to a plasma cell fate is associated with Blimp-1 expression in vivo. J Immunol. 2000;165(10):5462–71. doi: 10.4049/jimmunol.165.10.5462. [DOI] [PubMed] [Google Scholar]
- 36.Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77(2):297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 37.Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412(6844):300–7. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 38.Rasmussen T, Knudsen LM, Johnsen HE. Frequency and prognostic relevance of cyclin D1 dysregulation in multiple myeloma. Eur J Haematol. 2001;67(5–6):296–301. doi: 10.1034/j.1600-0609.2001.00559.x. [DOI] [PubMed] [Google Scholar]
- 39.Soverini S, Cavo M, Cellini C, Terragna C, Zamagni E, Ruggeri D, et al. Cyclin D1 overexpression is a favorable prognostic variable for newly diagnosed multiple myeloma patients treated with high-dose chemotherapy and single or double autologous transplantation. Blood. 2003;102(5):1588–94. doi: 10.1182/blood-2002-12-3789. [DOI] [PubMed] [Google Scholar]
- 40.Rasmussen T, Jensen L, Johnsen HE. The clonal hierachy in multiple myeloma. Acta Oncol. 2000;39(7):765–70. doi: 10.1080/028418600750063479. [DOI] [PubMed] [Google Scholar]
- 41.Rasmussen T, Jensen L, Johnsen HE. The CD19 compartment in myeloma includes a population of clonal cells persistent after high-dose treatment. Leuk Lymphoma. 2002;43(5):1075–7. doi: 10.1080/10428190290021524. [DOI] [PubMed] [Google Scholar]
- 42.Bakkus MH, Heirman C, Van Riet I, Van Camp B, Thielemans K. Evidence that multiple myeloma Ig heavy chain VDJ genes contain somatic mutations but show no intraclonal variation. Blood. 1992;80(9):2326–35. [PubMed] [Google Scholar]
- 43.Vescio RA, Cao J, Hong CH, Lee JC, Wu CH, Der Danielian M, et al. Myeloma Ig heavy chain V region sequences reveal prior antigenic selection and marked somatic mutation but no intraclonal diversity. J Immunol. 1995;155(5):2487–97. [PubMed] [Google Scholar]
- 44.Agematsu K, Hokibara S, Nagumo H, Komiyama A. CD27: a memory B-cell marker. Immunol Today. 2000;21(5):204–6. doi: 10.1016/s0167-5699(00)01605-4. [DOI] [PubMed] [Google Scholar]
- 45.Bergsagel PL, Kuehl WM. Chromosome translocations in multiple myeloma. Oncogene. 2001;20(40):5611–22. doi: 10.1038/sj.onc.1204641. [DOI] [PubMed] [Google Scholar]
- 46.Gabrea A, Martelli ML, Qi Y, Roschke A, Barlogie B, Shaughnessy JD, Jr, et al. Secondary genomic rearrangements involving immunoglobulin or MYC loci show similar prevalences in hyperdiploid and nonhyperdiploid myeloma tumors. Genes Chromosomes Cancer. 2008;47(7):573–90. doi: 10.1002/gcc.20563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zojer N, Schuster-Kolbe J, Assmann I, Ackermann J, Strasser K, Hubl W, et al. Chromosomal aberrations are shared by malignant plasma cells and a small fraction of circulating CD19+ cells in patients with myeloma and monoclonal gammopathy of undetermined significance. Br J Haematol. 2002;117(4):852–9. doi: 10.1046/j.1365-2141.2002.03529.x. [DOI] [PubMed] [Google Scholar]