Dop functions as a depupylase in the prokaryotic ubiquitin-like modification pathway
Bacteria modify proteins with a prokaryotic ubiquitin-like protein (Pup). Here the enzyme that removes Pup in a process analogous to deubiquitination has been identified.
Keywords: Dop, depupylation, pupylation, proteasome, Mpa
Abstract
Post-translational modification of proteins with prokaryotic ubiquitin-like protein (Pup) is the bacterial equivalent of ubiquitination in eukaryotes. Mycobacterial pupylation is a two-step process in which the carboxy-terminal glutamine of Pup is first deamidated by Dop (deamidase of Pup) before ligation of the generated γ-carboxylate to substrate lysines by the Pup ligase PafA. In this study, we identify a new feature of the pupylation system by demonstrating that Dop also acts as a depupylase in the Pup proteasome system in vivo and in vitro. Dop removes Pup from substrates by specific cleavage of the isopeptide bond. Depupylation can be enhanced by the unfolding activity of the mycobacterial proteasomal ATPase Mpa.
Introduction
In eukaryotes, post-translational modification of proteins with ubiquitin has been linked to important cellular processes, such as protein degradation, signal transduction and differentiation (Kerscher et al, 2006). Only recently it was discovered that certain bacteria, among them Mycobacterium tuberculosis (Mtb), have a related modification pathway termed ‘pupylation' (Pearce et al, 2008; Burns et al, 2009). Although ubiquitination is carried out by the sequential action of three enzymes (E1, E2 and E3; Kerscher et al, 2006), in the pupylation process, prokaryotic ubiquitin-like protein (Pup) is ligated to target proteins by the Pup ligase PafA (Striebel et al, 2009). PafA forms an isopeptide bond between the γ-carboxylate of Pup's carboxy-terminal glutamate and the ɛ-amino group of a lysine side chain of the target protein (Sutter et al, 2010). In mycobacteria, Pup is encoded with a C-terminal glutamine residue necessitating a preparation step before ligation that converts this glutamine to a glutamate. The deamidation of the glutamine residue is carried out by Dop (deamidase of Pup; Striebel et al, 2009)—an enzyme homologous to the Pup ligase PafA and to the γ-glutamyl cysteine ligase family (Iyer et al, 2008). Although, in Mtb, pupylation has been linked to proteasomal degradation of target proteins (Pearce et al, 2008; Wang et al, 2009; Striebel et al, 2010), the role of the Pup modification pathway in actinobacteria that do not have proteasomal subunit genes (for example, corynebacteria) is less clear. It is possible that pupylation has additional roles similar to non-degradative ubiquitination in eukaryotes.
Ubiquitination in eukaryotes is reversible due to the existence of deubiquitinases acting on the isopeptide linkage between ubiquitin and target lysines (Reyes-Turcu et al, 2009). Deubiquitinases oppose the ubiquitination process and thus provide a crucial regulative element in the ubiquitin modification pathway in addition to ensuring recycling of ubiquitin at the proteasome (Finley, 2009).
To investigate the potential existence of enzymatic activities able to reverse the modification of proteins with Pup, we assessed the depupylating activity of mycobacterial and corynebacterial lysates, demonstrating that they can depupylate an exogenously added Pup-modified proteasomal substrate. We identify the depupylating enzyme as Dop, the enzyme that also prepares Pup for ligation in mycobacteria. Furthermore, we provide evidence that substrate unfolding by the mycobacterial proteasomal ATPase Mpa can enhance depupylation in vitro.
Results
Actinobacteria depupylate a Pup-tagged substrate
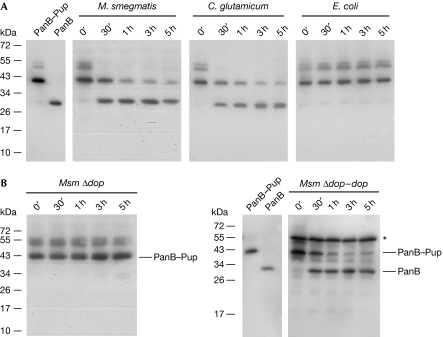
Pup is a small protein modifier conjugated to substrate lysine residues through an isopeptide bond (Pearce et al, 2008; Burns et al, 2009). To test for an activity that can reverse the modification of target proteins, we first conjugated purified His6-tagged proteasomal substrate PanB (ketopantoate hydroxymethyltransferase; Pearce et al, 2006) to His6-tagged Pup in vitro by using recombinantly produced Pup ligase PafA and Pup-GGE (a Pup Q64E variant). The PanB–Pup conjugate was then incubated with mycobacterial or corynebacterial lysates and samples were drawn at the indicated time points (Fig 1A). The samples were analysed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) followed by anti-His6 detection of the PanB and PanB–Pup bands to check for depupylation activity. In both Mycobacterium smegmatis (Msm) and Corynebacterium glutamicum (Cglu) lysate, the intensity of the PanB–Pup band decreases over the course of 5 h while concomitantly the PanB band appears. By contrast, the PanB–Pup band is stable in lysate of Escherichia coli, which does not harbour the Pup proteasome system, demonstrating that the activity is specific for actinobacterial lysates. Degradation of the exogenously added substrate is not observed, indicating that in the prepared lysates, the depupylation takes place on a faster time scale than degradation.
Figure 1.
Actinobacterial lysates exhibit depupylating activity that is due to Dop. (A) PanB–Pup (0.3 μM), a substrate modified with Pup using the ligase PafA, was added to cell extracts of Mycobacterium smegmatis (Msm), Corynebacterium glutamicum or Escherichia coli, and depupylation activity was monitored by anti-His6 immunoblot. (B) In Msm Δdop lysate, PanB–Pup is stable over the course of the experiment. Depupylation is restored in the complemented strain Msm Δdop–dop. Purified PanB–Pup and PanB are shown for comparison. The asterisk denotes expressed Dop-His6. Dop, deamidase of Pup; Pup, prokaryotic ubiquitin-like protein.
Depupylase activity is carried out by Dop
We previously identified Dop as the enzyme that deamidates mycobacterial Pup at its C-terminal glutamine residue to render it competent for ligation to substrate lysines by the Pup-ligase PafA (Striebel et al, 2009). The deamidation of Pup's terminal glutamine by Dop shows mechanistic similarities to reactions carried out by isopeptidases (see later). Therefore, we investigated whether lysate produced from a dop-deficient Msm strain (Msm Δdop) could remove Pup from His6-tagged PanB–Pup. Noticeably, PanB–Pup is stable in the dop deletion strain, whereas complementation of Msm Δdop with dop restores depupylation (Fig 1B). This demonstrates that Dop accounts for the observed depupylating activity.
In agreement with a function of Dop as depupylase, our recent characterization of Msm Δdop showed that the deletion strain produces a greater increase in pupylated substrates than the respective parent strain when expressing Pup-GGE (Imkamp et al, 2010). This was detectable as an increase not only in total intensity but also in amount of bands.
Dop cleaves the isopeptide bond of Pup-tagged substrates
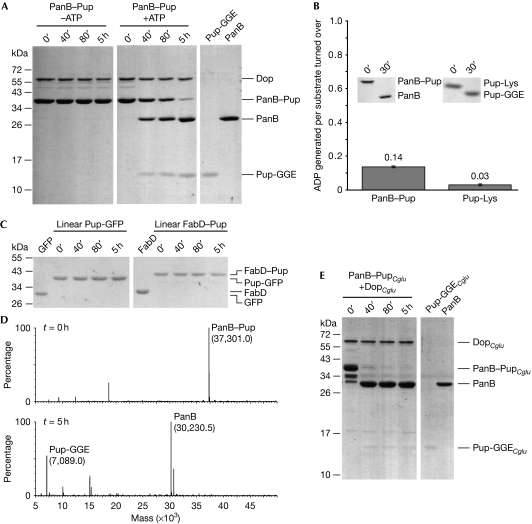
Next we analysed the depupylase activity of Dop in vitro. We had previously shown that deamidation of the C-terminal glutamine of Pup by Dop requires ATP as a cofactor (Striebel et al, 2009). To observe depupylation of PanB–Pup, we mixed PanB–Pup with Dop (Fig 2A). In the presence of ATP, but not in its absence, we observed a decrease in the PanB–Pup band over time that correlates with the appearance of the PanB band. Consistent with this, a Dop variant with a point mutation in the predicted ATP-binding motif (DopE10A; Imkamp et al, 2010) does not exhibit depupylating activity (supplementary Fig S1 online). Another previously identified proteasomal substrate, FabD–Pup (malonyl-CoA transacylase; Pearce et al, 2006), was also depupylated by Dop (supplementary Fig S2 online).
Figure 2.
Dop specifically cleaves the isopeptide bond between Pup and the substrate lysine residue. (A) PanB–Pup (3 μM)—a substrate modified with Pup using the ligase PafA—is depupylated by Dop (0.5 μM) only in the presence of ATP as analysed by SDS–PAGE and Coomassie staining (first two panels). The expected amounts of Pup (3 μM) and PanB (3 μM) after complete cleavage are shown in the right panel. (B) Depupylation activity of Dop does not require stoichiometric ATP turnover. PanB–Pup (20 μM) or Pup-Lys (20 μM) were incubated in the presence of DopCglu (10 or 4 μM) and ATP (50 μM). Nucleotides were analysed after completion of the depupylation reaction (see inset). Error bars represent s.d. values from three experiments. (C) Pup-GFP (3 μM) and FabD–Pup (1.5 μM), in which Pup is amino-terminally fused to the substrates, are not depupylated by Dop (0.5 μM). (D) Depupylation reaction as described in (A) analysed by ESI-MS at t=0 h (upper panel) and t=5 h (lower panel). The theoretical masses are: 37,300.7 (PanB–Pup), 30,229.3 (PanB) and 7,089.4 (Pup-GGE). (E) PanB–PupCglu is depupylated by Corynebacterium glutamicum Dop (DopCglu). Assay conditions were as described under (A). C. glutamicum proteins are labelled with the subscript Cglu. Dop, deamidase of Pup; ESI-MS, electrospray ionization-mass spectrometry; GFP, green fluorescent protein; Pup, prokaryotic ubiquitin-like protein; SDS–PAGE, sodium dodecyl sulphate–polyacrylamide gel electrophoresis.
Analysis of nucleotides present after depupylation of 20 μM PanB–Pup or 20 μM Pup-Lys—a model substrate in which Pup has been ligated to the ɛ-amino group of free lysine—by corynebacterial Dop, showed the production of only 2.7 and 0.5 μM ADP, respectively. This is equivalent to 0.14 and 0.03 ADP produced per substrate turned over (Fig 2B). No generation of AMP was observed. In comparison, the ligase PafA was shown to produce 1.1 ADP per Pup ligated (Striebel et al, 2009). This clearly argues against stoichiometric turnover of ATP during depupylation and is in agreement with the finding that deamidation by Dop also does not require ATP turnover.
Linear fusions of Pup to substrates, in which Pup is amino-terminally fused to either FabD or GFP (Fig 2C) by way of a regular peptide bond rather than an isopeptide bond, were not cleaved by Dop. This suggests that the Dop depupylase activity is specific for the isopeptide bond. To confirm that Pup is cleaved at the isopeptide bond, aliquots of a time course carried out with Dop acting on PanB–Pup were analysed by using electrospray mass spectrometry (Fig 2D). The observed masses correspond to the molecular weights expected for unmodified PanB and for free Pup-GGE. This provides direct proof that the isopeptide bond between the substrate lysine and Pup is cleaved by Dop and no additional residues remain on PanB after the reaction.
Although corynebacteria do not harbour proteasomal subunit genes, they do have genes for Pup, the ligase PafA, Dop and ARC (ATPase forming ring-shaped complexes; the corynebacterial homologue of Mpa). Interestingly, in these organisms Pup is sometimes encoded as the deamidated form (Pup-GGE), as in Cglu. In agreement with the observed depupylation activity of Cglu lysate, corynebacterial PupCglu coupled to mycobacterial PanB is removed from PanB after incubation with corynebacterial DopCglu, suggesting that in corynebacteria Dop also acts as a depupylase, counteracting the PafA ligase (Fig 2E). Notably, DopCglu was heterologously expressed in E. coli and thus purified from a source that does not show depupylase activity (see Fig 1A) and that does not contain any enzyme related to the Pup proteasome pathway. Therefore, one can conclude that DopCglu accounts for the observed depupylation activity. As with mycobacterial Dop, linearly fused Pup is not removed by DopCglu (supplementary Fig S3A online).
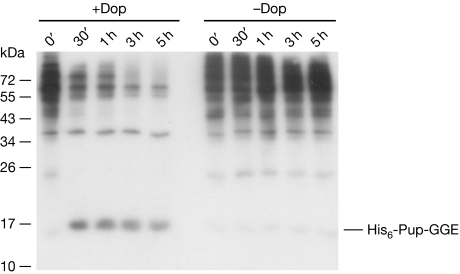
To investigate the substrate specificity of Dop in depupylation, we carried out depupylase reactions using the affinity-purified ‘pupylome' of Msm as a substrate pool. A decrease in the intensity of the ladder of pupylated substrates is observed, which is absent in the reaction without Dop (Fig 3). This indicates that Dop has broad substrate specificity and might act as the main depupylase in mycobacteria.
Figure 3.
Dop has a broad substrate specificity with respect to pupylated proteins. Pupylated proteins (300 μg) isolated from Msm Δdop expressing amino-terminally His6-tagged Pup-GGE were incubated with ATP (5 mM) in the presence or absence of Dop (1 μM). Samples were withdrawn at the time points indicated and analysed by an anti-Pup immunoblot. Dop, deamidase of Pup; Pup, prokaryotic ubiquitin-like protein.
The proteasomal ATPase Mpa can enhance depupylation
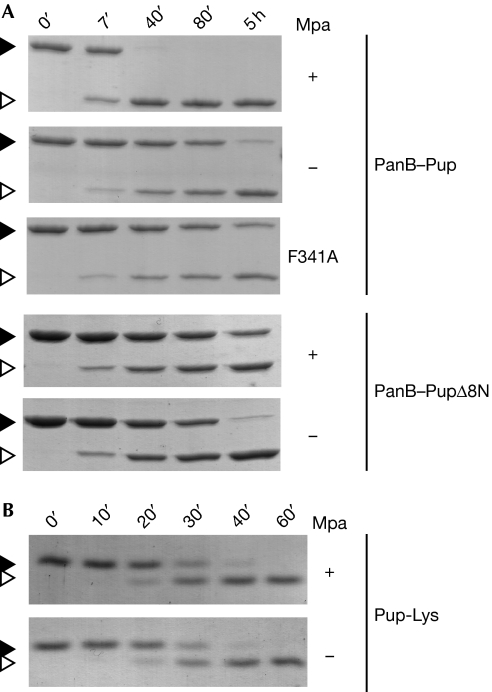
Intriguingly, corynebacteria maintain a proteasomal ATPase gene despite the absence of the proteasomal subunit genes. This led us to hypothesize that Mpa might have a role in assisting other, non-degradative features of the Pup-tagging system. To test this with respect to depupylation, we carried out PanB–Pup depupylation assays in the absence or presence of the ATPase Mpa (Fig 4A). Remarkably, Mpa enhances the depupylation activity of mycobacterial Dop significantly. Similar results are obtained with depupylation reactions using DopCglu (supplementary Fig S3B online). We previously showed that the mycobacterial ATPase Mpa alone can unfold a pupylated model substrate (Striebel et al, 2010). We thus speculated that Mpa might unfold PanB–Pup, and thereby increase the accessibility of the isopeptide bond for interaction with the depupylase Dop.
Figure 4.
Mpa enhances the depupylation of proteasomal substrates in vitro. (A) The depupylation of PanB–Pup (3 μM) by Dop (0.5 μM) is accelerated by Mpa (0.2 μM) in the presence of ATP as analysed by SDS–PAGE and Coomassie staining. By contrast, the depupylation of PanB–PupΔ8N (3 μM; Pup is truncated by the first eight amino acids) is not accelerated by the presence of Mpa and a translocation-defective Mpa variant (F341A) cannot enhance depupylation of PanB–Pup. Black arrowhead, PanB–Pup; white arrowhead, PanB. (B) The presence of Mpa (0.2 μM) does not affect the rate of Pup-Lys depupylation. Reaction described in (A) using Pup modified with lysine (Pup-Lys; 10 μM) as substrate. Black arrowhead, Pup-Lys; white arrowhead, Pup. Dop, deamidase of Pup; Mpa; mycobacterial proteasomal ATPase; Pup, prokaryotic ubiquitin-like protein; SDS–PAGE, sodium dodecyl sulphate–polyacrylamide gel electrophoresis.
As we previously demonstrated, a Pup variant lacking the eight N-terminal residues (PupΔ8N) can still bind to Mpa, but the substrate modified with truncated Pup (PanB–PupΔ8N) can no longer be unfolded, which is in agreement with a role of the N-terminal segment in the initiation of unfolding (Striebel et al, 2010). In this study, we show that depupylation of PanB–PupΔ8N is not enhanced by Mpa but is inhibited slightly by its presence (Fig 4A), suggesting that unfolding by Mpa is the mechanism by which depupylation of this substrate is enhanced. Consistent with this, a translocation-deficient variant of Mpa (Mpa F341A) cannot enhance depupylation (Fig 4A). For another proteasomal substrate of Mtb, FabD–Pup (supplementary Fig S2 online), we also observe enhancement of the depupylation rate in the presence of Mpa, demonstrating that the observed effect is not restricted to PanB as a substrate. However, the depupylation of a pupylated substrate with a completely accessible isopeptide linkage to Pup should be unaffected by Mpa. Concordantly, the model substrate Pup-Lys (Sutter et al, 2010) is depupylated at the same rate whether Mpa is present or not (Fig 4B).
Dop and PafA adjust the pupylation state in vitro
To assess whether ligation of Pup to a substrate protein and depupylation of the same substrate occur in similar time windows, we carried out pupylation and depupylation reactions at similar concentrations of PafA or Dop and PanB or PanB–Pup, respectively (supplementary Fig S4A online). The appearance of the PanB–Pup product in the ligation reaction shows a similar time course as the disappearance of the PanB–Pup band in the depupylation reaction. This suggests that the two processes contribute in a competitive way to the adjustment of the pupylation level of substrate proteins in the cell (Fig 5B). When the respective opposing enzyme activity was added to either the ligation or the depupylation reaction (supplementary Fig S4B online), the overall ligation or depupylation was slowed and both reactions ultimately reach a similar equilibrium ratio of PanB to PanB–Pup.
Figure 5.
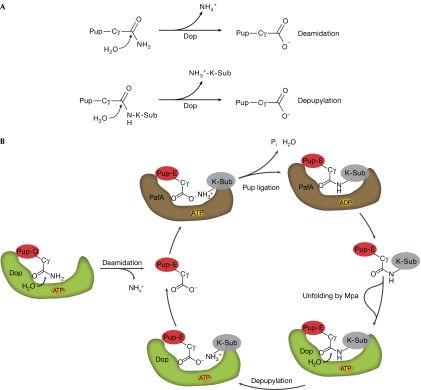
The Pup proteasome system is regulated by opposing activities. (A) Deamidation and depupylation performed by Dop are chemically similar reactions whereby water attacks the γ-carboxyl of Pup, leading to the release of ammonia in the case of deamidation or of a substrate lysine in the case of depupylation. (B) Schematic showing the proposed cycle of pupylation and depupylation in mycobacteria. Pup is shown as a red oval with indicated glutamine or glutamate C-terminus, and the substrate protein is depicted in grey carrying the label ‘K-Sub' to highlight the linkage through a lysine residue. Dop and PafA are depicted as green and brown shapes, respectively. Dop, deamidase of Pup; Pup, prokaryotic ubiquitin-like protein.
Discussion
Until recently, the use of small protein modifiers, such as ubiquitin, was thought to be a feature of eukaryotic cells. Since the discovery of pupylation in actinobacteria (Pearce et al, 2008; Burns et al, 2009) and the observation of conjugates between small archaeal modifier proteins and substrate lysine residues in archaea (Humbard et al, 2010), it is clear that the post-translational modification repertoire in prokaryotes also includes macromolecular tags. A key feature of most post-translational modification mechanisms is reversibility to allow for a highly dynamic system that can respond quickly. In eukaroytes, ubiquitination is mediated by hundreds of ubiquitin ligases and is counterbalanced by deubiquitinating enzymes (Wilkinson, 2009). Although the prokaryotic pupylation pathway seems to represent a more reductionist version of eukaryotic ubiquitination, an opposing activity to tagging of target proteins with Pup would seem an important element as a means of regulating pupylation levels.
In our study, we demonstrate that Dop acts as an opposing activity to the PafA ligase, by efficiently depupylating Pup-modified proteins (Fig 5B). Reversibility greatly increases the functional and regulatory potential of pupylation, not only with respect to the Pup proteasome pathway, but also for use of pupylation as a degradation-independent modifier.
The fact that we identified Dop, the enzyme that renders Pup ligation competent in mycobacteria, as the depupylase in the Pup-modification pathway seems at the same time surprising and consequential. Mycobacteria encode a Pup protein that has a glutamine as its C-terminal residue, necessitating the action of Dop as a deamidase to generate the γ-carboxylate that can be coupled to substrate lysine residues by PafA (Striebel et al, 2009; Sutter et al, 2010). Dop thus acts as an enzyme that is ‘on pathway' for the pupylation of target proteins in mycobacteria. That the same enzyme can act on the final product of this pathway, the Pup-coupled substrate, to reverse the tagging process, is interesting. Possibly, in mycobacteria there is more evolutionary pressure to keep the level of pupylated substrates tightly controlled than in those bacteria that encode Pup with a C-terminal glutamate. Mechanistically, it is not surprising that Dop has depupylase activity, as the chemical steps and intermediates involved in either deamidation of the glutamine residue of Pup or in the cleavage of an isopeptide bond are similar (Fig 5A).
The depupylase activity of Dop also explains why some bacteria that encode a Pup with a C-terminal glutamate maintain Dop in their genome, even though they do not need it for deamidation (Striebel et al, 2009). The function of Dop as the opposing activity to pupylation makes it a crucial element of any Pup-modification pathway.
Our experimental results also shed light on the question why ARC, the homologue of the proteasomal ATPase Mpa, is maintained in organisms that contain all elements of the pupylation pathway but lack the proteasomal genes. Our findings (Fig 4A) suggest that the ATPase-dependent pulling on the Pup tag initiates unfolding and renders the isopeptide bond more accessible, thereby enhancing the depupylation reaction. This might be particularly important when the pupylated substrate is part of a larger functional complex. It might be possible that in certain cases Pup cannot be removed without Mpa activity, resulting in a constant ‘on' state of the modification. This would provide strong evolutionary pressure to maintain the proteasomal ATPase.
One crucial question is whether depupylation by Dop promotes or rescues substrates from degradation by Mpa and the proteasome. In vitro, using Mpa in combination with an open-gate variant of the proteasome, Pup is not recycled but degraded along with the substrate (Striebel et al, 2010). Furthermore, the N-terminal segment of Pup was shown to provide an unfolding initiation site for Mpa in vitro (Striebel et al, 2010) and in vivo (Burns et al, 2010), indicating a mechanism in which Pup is a two-part degron providing tethering and the unfolding initiation site. However, a combination of in vivo and in vitro approaches will ultimately be necessary to address this question.
Methods
Cloning of constructs and purification of proteins. For cloning of constructs and purification of proteins refer to supplementary information online. For Mpa, the concentrations refer to the hexamer, for all others to the monomer. Proteins used in this study are M. tuberculosis proteins unless stated otherwise.
Production of Pup-modified substrates. Pup-modified substrates were generated and purified as described previously (Striebel et al, 2010). For details see supplementary information online.
Depupylation assay in vitro. The Pup-modified substrate (3 μM) or substrate linearly fused to Pup (3 μM Pup-GFP or 1.5 μM FabD–Pup) was incubated with Dop (0.5 μM) in 50 mM Tris (pH 7.5), 150 mM NaCl, 20 mM MgCl2, 10% glycerol (v/v), 1 mM dithiothreitol (buffer R) supplemented with 5 mM ATP at 30°C. Depupylation of Pup-Lys (10 μM) was carried out at 23°C. Depupylation reactions containing Mpa (0.2 μM) were supplemented with 30 mM phosphocreatine and 0.4 U/ml creatine phosphokinase. The reactions were terminated at the indicated time points with SDS sample buffer and analysed by SDS–PAGE and Coomassie staining. For electrospray ionization-mass spectrometry, samples were withdrawn and frozen in liquid nitrogen until analysis.
Depupylation assay in cell extracts. Msm SMR5 (Sander et al, 1995), Msm Δdop and Msm Δdop–dop (Imkamp et al, 2010) were grown to an OD600 of 1.5 at 37°C in 7H9 liquid broth supplemented with 0.025% Tween 80. C. glutamicum (DSM 20300) was grown in medium 53 (DSMZ, Braunschweig, Germany) at 30°C, E. coli DH5α was grown in Luria–Bertani medium at 37°C. C. glutamicum and E. coli were grown to an OD600 of ∼2.0. Cell extracts (4.5 mg/ml) were prepared as described previously (Imkamp et al, 2010). Purified His6-Pup-tagged PanB-His6 (0.3 μM), dithiothreitol (1 mM), MgCl2 (20 mM), ATP (5 mM) and ATP regeneration system (creatin phosphate/creatin phosphokinase) were incubated at 23°C for 5–10 min. The reaction was started by adding cell extract and was incubated at 30°C. Samples were withdrawn at the time points indicated and analysed by immunoblot using a His6 antibody (Acris). For detection, a mouse IgG alkaline phosphatase antibody produced in goat (Sigma) was used with the CDP Star substrate (Roche).
Analysis of nucleotides in the depupylation reaction. DopCglu (10 or 4 μM) was incubated with PanB–Pup (20 μM) or Pup-Lys (20 μM), respectively, in the presence of ATP (50 μM) for 30 min at 30°C. Nucleotides were then analysed and [ADP] calculated as described previously (Striebel et al, 2009). As control reactions, DopCglu or the substrate were incubated with ATP. The fraction f of ADP generated per substrate turned over was calculated as f=([ADP]depupylation−[ADP]controls)/[substrate].
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank the Functional Genomics Center Zurich (FGCZ) for mass spectrometry. This work was supported by the Swiss National Science Foundation (SNF), the National Center for Excellence in Research (NCCR) Structural Biology programme of the SNF, an Eidgenössische Technische Hochschule research grant and a Kekulé fellowship by the ‘Fonds der Chemischen Industrie' to F.S.
Footnotes
The authors declare that they have no conflict of interest.
References
- Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CE III (2009) Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem 284: 3069–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KE, Pearce MJ, Darwin KH (2010) Prokaryotic ubiquitin-like protein provides a two-part degron to Mycobacterium proteasome substrates. J Bacteriol 192: 2933–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D (2009) Recognition and processing of ubiquitin–protein conjugates by the proteasome. Annu Rev Biochem 78: 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbard MA, Miranda HV, Lim JM, Krause DJ, Pritz JR, Zhou G, Chen S, Wells L, Maupin-Furlow JA (2010) Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature 463: 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkamp F, Rosenberger T, Striebel F, Keller PM, Amstutz B, Sander P, Weber-Ban E (2010) Deletion of dop in Mycobacterium smegmatis abolishes pupylation of protein substrates in vivo. Mol Microbiol 75: 744–754 [DOI] [PubMed] [Google Scholar]
- Iyer LM, Burroughs AM, Aravind L (2008) Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination. Biol Direct 3: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Pearce MJ, Arora P, Festa RA, Butler-Wu SM, Gokhale RS, Darwin KH (2006) Identification of substrates of the Mycobacterium tuberculosis proteasome. EMBO J 25: 5423–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH (2008) Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 322: 1104–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 78: 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander P, Meier A, Böttger EC (1995) rpsL+: a dominant selectable marker for gene replacement in mycobacteria. Mol Microbiol 16: 991–1000 [DOI] [PubMed] [Google Scholar]
- Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E (2009) Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol 16: 647–651 [DOI] [PubMed] [Google Scholar]
- Striebel F, Hunkeler M, Summer H, Weber-Ban E (2010) The mycobacterial Mpa-proteasome unfolds and degrades pupylated substrates by engaging Pup′s N-terminus. EMBO J 29: 1262–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter M, Damberger FF, Imkamp F, Allain FH, Weber-Ban E (2010) Prokaryotic ubiquitin-like protein (Pup) is coupled to substrates via the side chain of its C-terminal glutamate. J Am Chem Soc 132: 5610–5612 [DOI] [PubMed] [Google Scholar]
- Wang T, Li H, Lin G, Tang C, Li D, Nathan C, Darwin KH, Li H (2009) Structural insights on the Mycobacterium tuberculosis proteasomal ATPase Mpa. Structure 17: 1377–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KD (2009) DUBs at a glance. J Cell Sci 122: 2325–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.