Role of the RNA/DNA kinase Grc3 in transcription termination by RNA polymerase I
GRC3 of S.cerevisiae encodes a polynucleotide kinase that is required for efficient termination of transcription by RNA polymerase I. This study proposes that GRC3 acts to maintain the 5′-phosphate end of the downstream Rnt1 cleavage product thus allowing Rat1 to efficiently degrade this RNA and so “torpedo” RNA polymerase I.
Keywords: Grc3, kinase, Pol I, transcription termination
Abstract
Transcription termination by RNA polymerase I in Saccharomyces cerevisiae is mediated by a ‘torpedo' mechanism: co-transcriptional RNA cleavage by Rnt1 at the ribosomal DNA 3′-region generates a 5′-end that is recognized by the 5′–3′ exonuclease Rat1; this degrades the downstream transcript and eventually causes termination. In this study, we identify Grc3 as a new factor involved in this process. We demonstrate that GRC3, an essential gene of previously unknown function, encodes a polynucleotide kinase that is required for efficient termination by RNA polymerase I. We propose that it controls the phosphorylation status of the downstream Rnt1 cleavage product and thereby regulates its accessibility to the torpedo Rat1.
Introduction
The enzyme RNA polymerase (Pol) I is dedicated to the synthesis of ribosomal RNA (rRNA). Despite using distinct transcription systems, Pol I and Pol II show close parallels in the process of transcription termination (El Hage et al, 2008; Kawauchi et al, 2008), as they both terminate by a ‘torpedo' mechanism (Kim et al, 2004; West et al, 2004). On ribosomal DNA (rDNA), co-transcriptional cleavage of the transcript by Rnt1 releases the 35S pre-rRNA, but also generates a free 5′-end in the downstream Pol I-associated 3′-transcript. This acts as a substrate for the 5′–3′ exonuclease Rat1 that degrades the 3′-transcript. This process is associated with destabilization of the transcription complex and consequent termination (El Hage et al, 2008; Kawauchi et al, 2008).
Eukaryotic Clp1 is a component of the messenger RNA (mRNA) cleavage and polyadenylation machinery (Minvielle-Sebastia et al, 1997; de Vries et al, 2000). Human Clp1 (hClp1) has been identified as a RNA kinase and phosphorylates the 5′-end of both synthetic short interfering RNAs and transfer RNA (tRNA) 3′-exons during tRNA splicing (Weitzer & Martinez, 2007). A potential enzymatic role for hClp1 in Pol II transcription termination has also been proposed, in which it acts to maintain a 5′-phosphate on the downstream cleavage product generated by mRNA 3′-end processing. This might provide Rat1 (Xrn2 in mammals) with a favourable substrate for exonucleolytic degradation and consequent Pol II termination (Weitzer & Martinez, 2007).
Although the human protein is an RNA kinase, no such activity has been identified in Saccharomyces cerevisiae Clp1 (Noble et al, 2007; Ramirez et al, 2008). A polynucleotide kinase-active Clp1 has been characterized in Archaea, suggesting the ancestral Clp1 possessed enzymatic activity (Jain & Shuman, 2009). It is possible that this activity was lost during yeast evolution, as the tRNA ligase Trl1 has intrinsic kinase activity.
We looked for other potential RNA kinases in yeast that are functionally related to Clp1. From this work, we identified Grc3 by bioinformatic analysis (Supplementary Fig S1 online).
GRC3 is an essential gene in S. cerevisiae and its transcription is cell cycle regulated (El-Moghazy et al, 2000), but its function is unknown. A genome-wide study has shown that Grc3 is associated with rRNA processing, and in particular with the removal of internal transcribed sequence 2 (ITS2; Peng et al, 2003). Grc3 was also observed, together with ribosomal proteins, in a protein fraction that was isolated by affinity purification of Rai1 (Sydorskyy et al, 2003), which co-purifies with and enhances Rat1 exoribonuclease activity (Xue et al, 2000; Xiang et al, 2009). Interestingly, pyrophosphatase activity has also been identified in Schizosaccharomyces pombe Rai1 (Xiang et al, 2009).
These molecular connections led us to investigate a possible role for Grc3 in Pol I transcription termination. In particular, we considered the possibility that the phosphorylation status of the 5′-end generated by Rnt1 cleavage on rRNA is fine-tuned by RNA kinase and phosphatase activities. Rat1 is closely related to the exonuclease Xrn1 (Kenna et al, 1993), which preferentially hydrolyses substrates with a 5′-monophosphate end (Stevens, 1980). This end is normally generated by the RNase III-like activity of Rnt1 (Gan et al, 2008) but other modifying enzymes could also be involved.
In this study, we show that Grc3 is a polynucleotide kinase, is present on rDNA and that its inactivation reduces the efficiency of termination by Pol I. We propose that Grc3 kinase activity is required to maintain the phosphorylated status of the downstream Rnt1 cleavage product, which in turn allows the torpedo activity of Rat1 to efficiently terminate Pol I transcription.
Results
Grc3 is a polynucleotide kinase
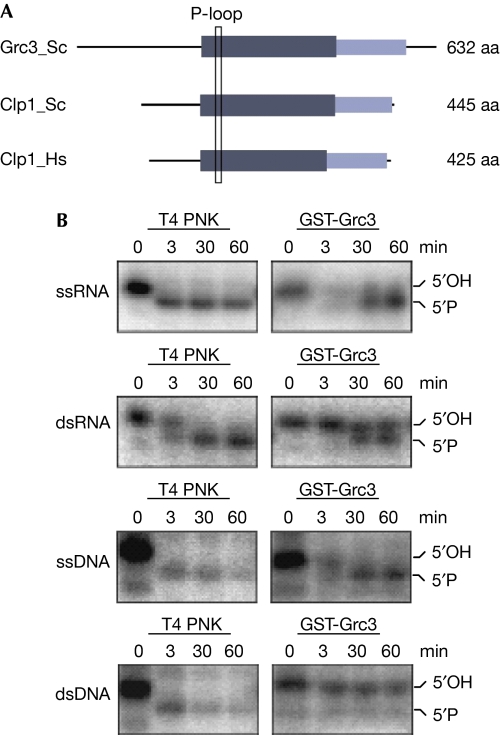
In S. cerevisiae, no polynucleotide kinase activity has been identified except for tRNA ligase Trl1. Although hClp1 is an RNA kinase, yeast Clp1 lacks kinase activity (J. Martinez, unpublished results; Noble et al, 2007; Ramirez et al, 2008). Performing National Center for Biotechnology Information–protein basic local alignment search tool (NCBI–BLASTP) searches within the NCBI non-redundant database, we identified two protein families related to Clp1 orthologues (Supplementary Fig S1 online): a group of hypothetical proteins from Eubacteria and Archaebacteria and a eukaryotic protein family including S. cerevisiae Grc3. These proteins share a carboxy-terminal domain of unknown function and a central nucleotide-binding domain containing a highly conserved structural element of the NTPase fold, the P-loop/Walker A box (Fig 1A).
Figure 1.
Grc3 is a polynucleotide kinase. (A) Domain architecture of Clp1 and Grc3. The Clp1 domain architecture is derived from Noble et al (2007) and was transferred to Grc3 using an alignment. The central kinase domain is depicted in dark grey, the carboxy-terminal domain in light grey. Location of the P-loop/Walker A box is indicated with a rectangle. (B) Kinase assays of glutathione-S-transferase (GST)-tagged Grc3. Purified Grc3 was incubated with the indicated substrates (single- or double-stranded RNA or DNA) for the indicated time. Grc3 is active on RNA or single-stranded DNA substrates. T4 polynucleotide kinase (PNK) was assayed in parallel as a control. Phosphorylation was monitored by electrophoresis. aa, amino acid; ds, double-stranded; Hs, Homo sapiens; Sc, Saccharomyces cerevisiae; ss, single-stranded.
To determine whether yeast Grc3 has kinase activity, we expressed and purified the glutathione-S-transferase-tagged protein and performed an in vitro kinase assay using radiolabelled single- or double-stranded RNA or DNA as substrates (Fig 1B). As a positive control, we tested T4 polynucleotide kinase in parallel. Phosphorylation was detected as a mobility shift on a polyacrylamide denaturing gel. As shown in Fig 1B, Grc3 displayed polynucleotide kinase activity on both single- and double-stranded RNA and on single-stranded DNA alone, but not double-stranded DNA alone. Therefore, these results support our bioinformatic analysis and confirm that Grc3 is a polynucleotide kinase.
Grc3 ChIPs on rRNA encoding and terminator regions
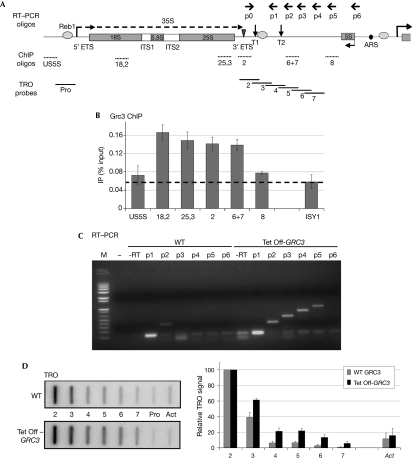
Genome-wide studies suggest that Grc3 is involved in rRNA processing (Peng et al, 2003), which implies that it is present in the nucleolus. To determine whether this protein is recruited onto rDNA, we prepared a 3 × haemagglutinin (3HA–GRC3)-tagged strain and performed chromatin immunoprecipitation (ChIP) analysis across the Pol I transcription unit. As a control, we also probed the coding region of ISY1, a Pol II-transcribed gene. ChIP signals were detected above background across the rDNA gene (Fig 2A,B; regions 18,2 and 25,3), as well as over the terminator region (2 and 6+7). No significant enrichment was detected upstream to the transcription start site or further downstream in the 35S 3′-regions (US5S and 8). ChIP signals were relatively low but reproducible, consistent with a transient association of Grc3 with chromatin as an enzyme involved in rRNA processing. However, the presence of Grc3 over the terminator region prompted us to test further whether Grc3 is involved in 35S 3′-processing and/or Pol I transcription termination.
Figure 2.
Grc3 depletion results in defective transcription termination by RNA polymerase I. (A) Schematic of a Saccharomyces cerevisiae rDNA repeat. In addition to the sequence encoding 18S, 5.8S and 25S rRNA (grey rectangles), the Pol I transcription unit includes ETSs and ITSs; the 35S primary transcript is shown as a dashed arrow. Grey ovals denote binding sites for Reb1, the triangle denotes the Rnt1 cleavage site and vertical arrows indicate T1 and T2 terminator elements. 5S rDNA, transcribed by Pol III in the opposite orientation, and ARS are shown. Primers used for RT–PCR are shown above, chromatin immunoprecipitation (ChIP) oligonuleotides and TRO probes below. (B) ChIP analysis of 3HA-tagged Grc3 along rDNA. Specific enrichment (above dashed line) is visible over the rRNA encoding and terminator sequence. Pol II-transcribed ISY1 is shown as a control. An average of two independent experiments is shown, error bars indicate s.d. values. (C) RT–PCR analysis of transcripts downstream to the Rnt1 cleavage site in WT or Grc3-depleted (Tet Off-GRC3) cells. Reverse transcription was primed with oligonuleotides p1–p6, PCR with the common forward primer p0 and the indicated reverse primers. Grc3 depletion results in stabilization of the Pol I transcripts over the 35S 3′-region. Oligonucleotide p1 and no reverse transcriptase (−RT) or no cDNA were used as a template in control PCR reactions. M, molecular weight marker (Invitrogen 1 kb Plus). (D) TRO analysis of Pol I transcripts over the 35S 3′-region in WT or Tet Off-GRC3 cells. Grc3 depletion results in a Pol I termination defect. Quantification of the signals is shown in the right-hand panel. Background signal (Pro) was subtracted to each probe value. Data were then normalized towards probe 2, set to 100%. The Pol II-transcribed actin gene (Act) is shown as a control. The average of three independent experiments is shown, error bars indicate s.d. values. ARS, autonomously replicating sequence; ETS, external transcribed sequence; HA, haemagglutinin; ITS, internal transcribed sequence; RT–PCR, reverse transcriptase PCR; rDNA, ribosomal DNA; rRNA, ribosomal RNA; TRO, transcriptional run on; WT, wild type.
Grc3 depletion stabilizes 3′-ETS Pol I transcripts
We first determined whether the levels of 3′-external transcribed sequence (ETS) transcripts were influenced by Grc3 using a strain in which GRC3 is under the control of a regulatable promoter (Tet Off). The addition of doxycycline severely affected cell growth and GRC3 mRNA levels dropped to about 20% of the wild-type (WT) level after 5 h (data not shown). We extracted total RNA from WT or Tet Off-GRC3 cells, performed reverse transcription with oligonucleotides p1–p6 (Fig 2A) and PCR with the same oligonucleotides and a common forward primer p0, annealing downstream to the Rnt1 cleavage site. With this method we detected transcripts produced by Pol I downstream to the main terminator element T1. On rDNA, a small proportion of polymerases read through this signal, mostly terminating at the downstream T2 element (Lang et al, 1994; El Hage et al, 2008). In the WT strain, reverse transcriptase PCR (RT–PCR) signal was detectable with oligonucleotide p1 (upstream to T2), very weak with p2 and not present at all further downstream (Fig 2C; left). By contrast, on depletion of Grc3, 3′-extended transcripts were visible (oligonucleotides p2–p5), up to the 5S gene (Fig 2C; right). We conclude that Grc3 is involved in Pol I transcription termination or in stabilization of 3′-transcripts that extend beyond the normal termination site.
Grc3 depletion causes defective Pol I termination
Although RT–PCR measures steady-state RNA levels, transcriptional run on (TRO) analysis monitors nascent transcription and, thus, provides a map of the active polymerase density indicating where transcription termination occurs. Using TRO analysis, we further compared WT and Tet Off-GRC3 strains, using probes covering the rDNA terminator region (outlined in Fig 2A). As a control, we also used a probe upstream to the Pol I transcriptional start site (Pro), providing background signal, and over the actin gene (Act), transcribed by Pol II. All data were quantified relative to probe 2, overlapping the T1 terminator, as previously described (Jones et al, 2007). In WT, we obtained the expected termination profile with a strong signal over probe 2, reduced signal over probe 3 and near to background signal downstream (Fig 2D). On depletion of Grc3, we observed a proportionally higher signal over the downstream probes (3–7), indicating a termination defect. We conclude that not only RNA stability, but also Pol I termination itself is affected by the loss of Grc3.
Pol I termination requires Grc3 kinase activity
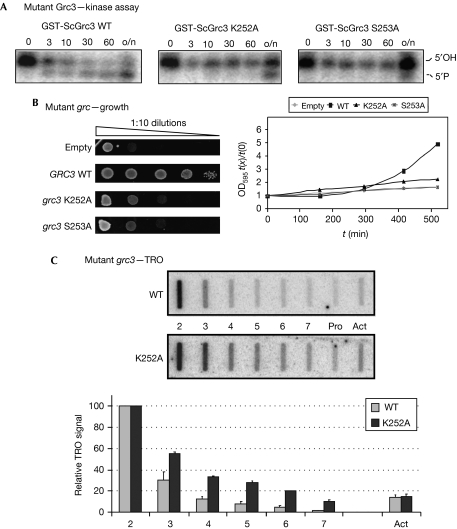
Next, we aimed to determine whether the effect of Grc3 on Pol I termination is mediated by its polynucleotide kinase activity. We mutated specific conserved residues in the Walker A box of Grc3, based on the sequence homology with hClp1. In particular, we produced K252A and S253A mutants because the corresponding mutations abolish the kinase activity in hClp1 (Weitzer & Martinez, 2007).
We assessed the kinase activity of the mutants using purified glutathione-S-transferase-tagged Grc3 (WT, K252A or S253A) with a mixture of single- and double-stranded RNA substrate in the kinase assay, as before. The WT protein rapidly phosphorylated the RNA (Fig 3A), whereas the mutant K252A showed partial activity only after overnight incubation, and S253A was almost completely inactive.
Figure 3.
Kinase-inactive Grc3 mutants are defective in cell growth and RNA polymerase I termination. (A) Kinase assays of WT and mutant Grc3. Purified GST-tagged Grc3 (WT, K252A or S253A) was incubated with a 2:1 mixture of single- and double-stranded RNA for the indicated time (in min; o/n, overnight). Substrate phosphorylation was monitored by electrophoresis. Grc3 kinase activity is strongly delayed in the K252A and absent in the S253A mutant. (B) Growth phenotype of cells expressing WT or mutant Grc3 (K252A or S253A) from a centromeric plasmid. Endogenous GRC3 transcription was repressed in glucose. Cell proliferation is severely affected by Grc3 mutation. Cells transformed with an empty plasmid are shown as a control. Left panel: serial dilutions drop plate. Right panel: growth curves in liquid culture. (C) TRO analysis of cells expressing WT or K252A mutant Grc3. The kinase-inactive Grc3 mutant is defective in Pol I termination. Probes and quantification as in Fig 2D. GST, glutathione-S-transferase; Pol I, RNA polymerase I; TRO, transcriptional run on; WT, wild type.
To test these mutants in vivo, we replaced the endogenous GRC3 promoter with the regulatable GAL1 promoter, repressed its transcription in glucose and expressed Grc3, WT or mutant forms, from a centromeric plasmid. As shown in Fig 3B, control cells transformed with the empty plasmid showed scarce growth, both on plate and in liquid culture as expected, because GRC3 is essential. Also transformation with WT Grc3 produced viable cells with nearly normal growth phenotype. Both K252A and S253A mutation strongly affected cell growth, the latter more severely, confirming that we had mutated critical residues in the Grc3 active site.
Next, we tested Pol I transcription termination of the K252A mutant by TRO (Fig 3C). The S253A mutant was too severely growth-retarded to allow further analysis. However, quantitative analysis of K252A produced a Pol I termination profile similar to that obtained after Grc3 depletion. Thus, a clear termination defect (higher polymerase density over probes 3–7) was obtained, strongly suggesting that the kinase activity of Grc3 is required for efficient termination. We conclude that Grc3 polynucleotide kinase activity has an important role in determining efficient termination by Pol I.
Discussion
We have shown that Grc3, an essential yeast protein of previously unknown function, is a polynucleotide kinase with similarities to Clp1. Furthermore, this enzyme seems to have a physiological role in Pol I transcription termination, as kinase-inactive Grc3 mutants produce increased read-through transcription in the rDNA 3′-ETS.
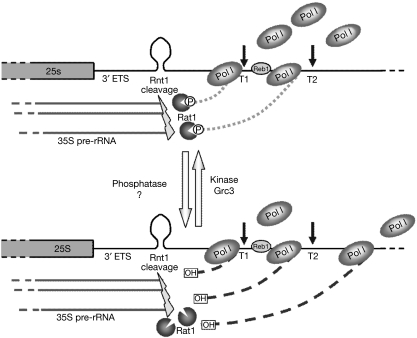
How is a polynucleotide kinase involved in the termination process? Pol I transcription termination involves co-transcriptional RNA cleavage by Rnt1, followed by degradation of the downstream cleavage product by the 5′–3′ exonuclease Rat1. This ‘torpedoes' Pol I and eventually causes termination by releasing it from the DNA template (El Hage et al, 2008; Kawauchi et al, 2008). The phosphorylation status of the downstream Rnt1 cleavage product might have a critical role in its recognition by Rat1. Substrates with 5′-monophosphate ends are strongly preferred to non-phosphorylated 5′-ends by the 5′–3′ exonuclease Xrn1 (Stevens, 1980). It is likely that Rat1, closely related to Xrn1, has similar specificity. We propose that Grc3 acts to maintain the phosphorylated status of the downstream RNA after Rnt1 cleavage, possibly by counteracting phosphatase activity (Fig 4). This equilibrium might regulate the kinetic and overall efficiency of transcription termination by Pol I. In support of this model, Grc3 has been observed to immunoprecipitate with Rai1, the Rat1-activating partner (Sydorskyy et al, 2003). Previous results have connected Grc3 with rRNA processing (Peng et al, 2003). Interestingly, Rat1 is also the RNA exonuclease involved in 5′-trimming of rRNA-processing intermediates (Kufel et al, 1999). Therefore, our studies predict a functional coupling between these two activities.
Figure 4.
Model of Grc3 kinase activity involvement in the process of transcription termination by Pol I. Rnt1 co-transcriptionally cleaves the transcript RNA at a stem loop structure in the 3′-ETS. The phosphorylation status of the Rnt1 cleavage 3′-product is controlled by the equilibrium between a putative phosphatase activity and the RNA kinase Grc3. The phosphorylated 5′-end (top) is recognized by the 5′–3′ exonuclease Rat1 that ‘torpedoes' Pol I and thus promotes transcription termination. When Grc3 is absent or inactive, the equilibrium is shifted and the 3′-transcripts present a 5′-hydroxyl end (bottom). This is suboptimal substrate for the ‘torpedo' Rat1. As a consequence, the 3′-transcripts are stabilized and Pol I termination is impaired. ETS, external transcribed sequence; Pol I, RNA polymerase I; rRNA, ribosomal RNA.
Methods
Clp1/Grc3 protein family collection. hClp1 (NP_006822.1) was used as a query for NCBI–BLASTP searches within the NCBI non-redundant database (Altschul et al, 1997). To expand the protein family, significant hits (E-values <1 × 104) were used as queries in further searches. Selected proteins were aligned using MUSCLE (Edgar, 2004).
Expression and purification of Grc3. The open reading frame of Grc3 (WT, K252A or S253A) was cloned into pDEST20 using the Gateway technique (Invitrogen) and expressed in sf9 insect cells. After collection, cells were lysed in 100 mM NaCl, 50 mM Tris–HCl (pH 8), 5 mM MgCl2, 0.1 mM 4-(2-aminoethyl) benzenesulphonyl fluoride hydrochloride, 1 mM dithiothreitol and sonicated. Purification was performed using glutathione Sepharose 4B (GE Healthcare). Protein was eluted with 20 mM glutathione, concentrated on Vivaspin 500 columns (Sartorius) and dialysed against 30 mM 4-(2-hydroxyethyl)-1-piperazineethanesulphonic acid (pH 7.4), 5 mM MgCl2, 100 mM KCl, 10% glycerol and 0.1 mM 4-(2-aminoethyl) benzenesulphonyl fluoride hydrochloride.
In vitro kinase assay. Kinase assays were performed as described previously (Weitzer & Martinez, 2007) in the presence of 5 mM ATP at 30°C. Substrates were 21-nucleotides long and either single-stranded or annealed to a complementary oligonucleotide to create blunt double strands. The RNA substrates were 3′-end-labelled with 32P pCp using T4 RNA Ligase (Amersham) and then dephosphorylated with alkaline phosphatase (Roche) to obtain 5′, 3′-hydroxylated RNA substrates. The DNA substrates were labelled with 32P cordycepin and recombinant terminal deoxynucleotidyl transferase (Promega). Phosphorylation was monitored on 15% polyacrylamide/urea gel.
Yeast strains and plasmids. Strains used were pGAL-3HA-GRC3 (MAT-a; ade2-1; can1-100; his3-11,15; leu2-3,112; trp1-1; ura3-1; PGAL1-3HA-GRC3 KANMX6) and untagged W303-1a (MAT-a; ade2-1; can1-100; his3-11,15; leu2-3,112; trp1-1; ura3-1) (Fig 2B) and WT R1158 (MAT-a; his3-1; leu2-0; met15-0; URA∷CMV-tTA) and Tet Off-GRC3 (MAT-a; his3-1; leu2-0; met15-0; URA∷CMV-tTA; kanR-tetO7-TATA-GRC3) (Fig 2C,D). GRC3 was repressed by treating the cells with 10 μg/ml doxycycline for 5 h. pGAL-3HA-GRC3 strain was transformed with a modified pCM252 (lacking the Tet-O box repeats) containing GRC3 open reading frame (WT or mutant) cloned at the StuI site (Fig 3). Standard media and growth conditions were used.
ChIP. The ChIP analysis was performed as described previously (Kawauchi et al, 2008), using Anti-HA Clone F7 antibody (Santa Cruz Biotechnology). Background signal from the untagged strain was subtracted to 3HA-GRC3 signal at each position. Oligonucleotide sequences have been described previously (Kawauchi et al, 2008).
RT–PCR. RT–PCR was performed with Superscript III RT (Invitrogen) on 400 ng RNA priming the reaction with a mixture of oligonucleotides p1–p6. A total of 28 cycles of PCR were performed with the communal oligonulceotide p0 and each of the p1–p6 oligonucleotides. The PCRs with oligonucleotide p1 and no RT were used as a negative control. Primer sequences have been described previously (Kawauchi et al, 2008).
TRO. The TRO analysis and probes have been described previously (Kawauchi et al, 2008). Five micrograms of each single-stranded DNA probe was immobilized on Hybond-N membrane (Amersham). Cells collected in log-phase were permeabilized with sarkosyl and then incubated in transcription buffer containing α-32P UTP for 5 min to label nascent transcripts. Extracted RNA was then partly hydrolysed and hybridized to the membrane.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank F. Lampert and S. Westermann at the Research Institute of Molecular Pathology, Vienna. This study was supported by a Wellcome Trust Programme Grant to N.J.P. and by Genomic Research in Austria and Fonds zur Förderung der wissenschaftlichen Forschung (grant P20502-B11) to K.H. and J.M.
Footnotes
The authors declare that they have no conflict of interest.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W (2000) Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J 19: 5895–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, Koper M, Kufel J, Tollervey D (2008) Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev 22: 1069–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Moghazy AN et al. (2000) Functional analysis of six novel ORFs on the left arm of chromosome XII in Saccharomyces cerevisiae reveals two essential genes, one of which is under cell-cycle control. Yeast 16: 277–288 [DOI] [PubMed] [Google Scholar]
- Gan J, Shaw G, Tropea JE, Waugh DS, Court DL, Ji X (2008) A stepwise model for double-stranded RNA processing by ribonuclease III. Mol Microbiol 67: 143–154 [DOI] [PubMed] [Google Scholar]
- Jain R, Shuman S (2009) Characterization of a thermostable archaeal polynucleotide kinase homologous to human Clp1. RNA 15: 923–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HS, Kawauchi J, Braglia P, Alen CM, Kent NA, Proudfoot NJ (2007) RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat Struct Mol Biol 14: 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ (2008) Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev 22: 1082–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna M, Stevens A, McCammon M, Douglas MG (1993) An essential yeast gene with homology to the exonuclease-encoding XRN1/KEM1 gene also encodes a protein with exoribonuclease activity. Mol Cell Biol 13: 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S (2004) The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432: 517–522 [DOI] [PubMed] [Google Scholar]
- Kufel J, Dichtl B, Tollervey D (1999) Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 5: 909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang WH, Morrow BE, Ju Q, Warner JR, Reeder RH (1994) A model for transcription termination by RNA polymerase I. Cell 79: 527–534 [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Preker PJ, Wiederkehr T, Strahm Y, Keller W (1997) The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc Natl Acad Sci USA 94: 7897–7902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble CG, Beuth B, Taylor IA (2007) Structure of a nucleotide-bound Clp1–Pcf11 polyadenylation factor. Nucleic Acids Res 35: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng WT et al. (2003) A panoramic view of yeast noncoding RNA processing. Cell 113: 919–933 [DOI] [PubMed] [Google Scholar]
- Ramirez A, Shuman S, Schwer B (2008) Human RNA 5′-kinase (hClp1) can function as a tRNA splicing enzyme in vivo. RNA 14: 1737–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A (1980) Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5′-mononucleotides by a 5′ leads to 3′ mode of hydrolysis. J Biol Chem 255: 3080–3085 [PubMed] [Google Scholar]
- Sydorskyy Y, Dilworth DJ, Yi EC, Goodlett DR, Wozniak RW, Aitchison JD (2003) Intersection of the Kap123p-mediated nuclear import and ribosome export pathways. Mol Cell Biol 23: 2042–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzer S, Martinez J (2007) The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature 447: 222–226 [DOI] [PubMed] [Google Scholar]
- West S, Gromak N, Proudfoot NJ (2004) Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432: 522–525 [DOI] [PubMed] [Google Scholar]
- Xiang S, Cooper-Morgan A, Jiao X, Kiledjian M, Manley JL, Tong L (2009) Structure and function of the 5′ → 3′ exoribonuclease Rat1 and its activating partner Rai1. Nature 458: 784–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Bai X, Lee I, Kallstrom G, Ho J, Brown J, Stevens A, Johnson AW (2000) Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol Cell Biol 20: 4006–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.