BRG1 helps RNA polymerase II to overcome a nucleosomal barrier during elongation, in vivo
This report shows that a positioned nucleosome in the body of a transcription unit impairs RNAPII progression, provokes RNAPII accumulation upstream of the positioned nucleosomes and reduces transcription in vivo. BRG1, the enzymatic motor of the SWI–SNF chromatin-remodelling complex, facilitates nucleosomal traversal by RNAPII.
Abstract
RNA polymerase II (RNAPII) transcribes genes in a chromatin context. We have designed a system to investigate the role of chromatin remodelling during elongation in vivo, which involves inserting a strong nucleosome-positioning sequence between a promoter and a reporter gene. Our data indicate that a nucleosome positioned in the body of a transcription unit impairs RNAPII progression, provokes RNAPII accumulation upstream to the positioned nucleosome and reduces transcription. By using this system, we show that BRG1, the enzymatic motor of the SWI–SNF chromatin-remodelling complex, is recruited to the positioned nucleosome in a transcription elongation-dependent manner and facilitates traversal of the nucleosome by RNAPII.
Introduction
The basic building block of chromatin is the nucleosome, which consists of about 150 bp of DNA wrapped around a histone octamer containing two units of each of the histones H2A, H2B, H3 and H4 (Luger et al, 1997). Enzymatic machines that travel along the DNA have to deal with its compact nucleoprotein organization. Biochemical studies in vitro have demonstrated that nucleosomes form a high barrier for transcribing RNA polymerase II (RNAPII; Izban & Luse, 1991; Bondarenko et al, 2006; Kulaeva et al, 2007; Hodges et al, 2009). Thus, nucleosomes increase the chance that RNAPII will pause at intrinsic transient pause sites at which RNAPII would eventually arrest, even in naked DNA. Furthermore, some of these in vitro studies have shown that RNAPII elongation provokes the displacement of an H2A–H2B dimer which generates a hexasome, but does not change the position of the nucleosome on the DNA. In vivo experiments have shown a negative correlation between RNAPII-dependent transcription and density of histones, especially H2A and H2B, in the body of the gene (Thiriet & Hayes, 2005). However, direct in vivo evidence of RNAPII blocking due to a nucleosome barrier at the transcribed region of a gene has not been reported.
ATP-dependent chromatin-remodelling machines are able to destabilize the interactions between DNA and histone octamers (for a review, see Clapier & Cairns, 2009). Although the role of these enzymes in chromatin remodelling at promoters and regulatory regions has been extensively documented, their role during elongation remains poorly understood. Workman and colleagues demonstrated that a purified RSC complex, one of the two switch–sucrose non-fermentable (SWI–SNF)-related complexes from yeast, facilitates RNAPII elongation on a mononucleosome template in vitro (Carey et al, 2006). However, it is unclear whether SWI–SNF is required to help RNAPII overcome a nucleosomal barrier at the body of the genes, in vivo. To explore this issue, we have developed a system based on autonomously replicating episomal plasmid, to investigate the role of a high nucleosomal barrier in transcription elongation. We show that strong nucleosome-positioning sequences impair transcriptional elongation and provoke an accumulation of elongating polymerase upstream to the nucleosomal barrier. Brahma-related gene 1 (BRG1), the ATPase subunit of the human SWI–SNF complex, helps RNAPII to overcome this nucleosomal barrier.
Results
Description of the reporter system
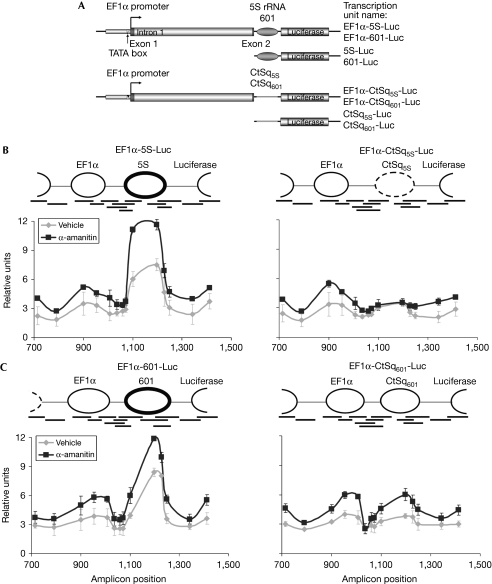
A system for in vivo study of the involvement of chromatin remodelling during transcription elongation was generated using pREP4-Luc episomal reporter vector. pREP4 contains Epstein–Barr virus replication origin and encodes nuclear antigen 1 (Langle-Rouault et al, 1998). This episomal vector has been shown previously to form a correct nucleosomal chromatin structure on transfection (van der Vlag et al, 2000). Our experimental design involved the insertion of a sequence that strongly positions a nucleosome between a promoter and a reporter luciferase gene (Fig 1A). Two non-related nucleosome-positioning sequences were used: a fragment of the 5S ribosomal RNA gene from Lytechinus variegatus (FitzGerald & Simpson, 1985) and the 601 sequence (Lowary & Widom, 1998). Both of these sequences have been extensively characterized, and have strong nucleosome-positioning abilities. As a control sequence, we used a fragment of bacterial ampicillin resistance gene of the same length as the nucleosome sequences (CtSq5S and CtSq601), 195 and 147 bp, respectively. Sequences were placed downstream to an approximately 1.2-kb translation elongation factor 1α (EF1α) gene fragment including the promoter, exon 1, intron 1 and 9bp of exon 2 of the gene (Fig 1A; Uetsuki et al, 1989). Finally, the Luciferase gene was placed downstream to the nucleosome positioning and control sequences. Therefore, four transcriptional units were generated, termed EF1α-5S-Luc, EF1α-CtSq5S-Luc, EF1α-601-Luc and EF1α-CtSq601-Luc. The messenger RNA generated from these transcriptional units included exon 1 and nine bases of exon 2 of EF1α, the positioning or control sequences and the luciferase sequence (data not shown). Constructs without the EF1α fragment were also generated as controls (Fig 1A).
Figure 1.
A system to study the role of nucleosome-positioning during transcription elongation, in vivo. (A) Schematic representation of the transcription units inserted into the pREP4 plasmids. (B,C) Line graphs of nucleosomal positioning at the indicated transcription units in vivo. Constructs containing the indicated transcription units were transfected into 293T cells. Twenty-four hours after transfection cells were treated with 2 μg/ml α-amanitin (see supplementary Fig S2 online) or non-treated, for an additional 24 h before chromatin isolation. The mapping of nucleosomal positions by MNase and qPCR is described in the supplementary information online. The position of nucleosomes deduced from the data are represented as solid line ovals. Black lines under representation denote the locations of amplicons for qPCR analysis. Numbers in the x-axis correspond to nucleotide positions with respect to the transcription start point. Data are means of three independent experiments and error bars represent ±s.d. values. EF1α, elongation factor 1α; qPCR, quantitative PCR.
First, we confirmed in vivo positioning of the nucleosomes at the 5S and 601 episomal constructs. Cells were fixed with formaldehyde and nuclei were obtained. After micrococcal nuclease (MNase) digestion, mononucleosomal DNA was isolated (supplementary Fig S1 online) and subjected to quantitative PCR (qPCR), using primer pairs that cover 700 bp around the positioning or the control sequences. The results were normalized against those obtained using MNase-digested genomic DNA. There was a strong decrease in MNase sensitivity at the 5S and 601 sequences, indicating the presence of a strongly positioned nucleosome (Fig 1B,C). Some nucleosome enrichment was also observed upstream and downstream from the 5S and 601 sequences, suggesting the presence of less stably positioned nucleosomes. As expected, nucleosome enrichment was low or absent at control sequences. Transcription inhibition with α-amanitin increased nucleosome occupancy, indicating that transcription is accompanied by nucleosome remodelling. As a control, we checked that α-amanitin blocks transcription and strongly decreases occupancy of RNAPII from the 5S and 601 elements of the EF1α-5S-Luc and EF1α-601-Luc transcription units (supplementary Fig S2A,B,C online).
A positioned nucleosome impairs RNAPII progression
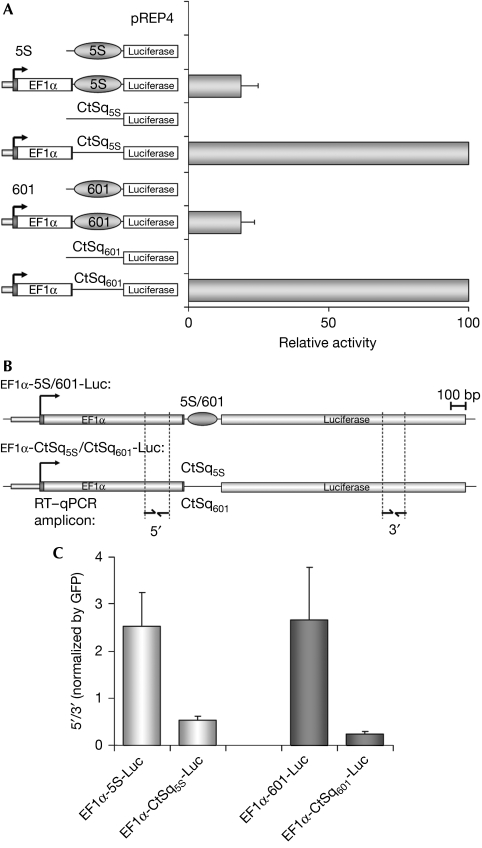
We then analysed whether 5S and 601 nucleosomal positioning sequences affect expression of the luciferase reporter gene. Constructs containing nucleosome-positioning sequences showed significantly lower (4–5-fold) luciferase activity levels than control constructs (Fig 2A). Nucleosomes have been shown to represent a strong barrier for RNAPII traversal in vitro (for a review, see Kulaeva et al, 2007). Therefore, a possible explanation of our result was that the decrease in luciferase activity was a consequence of a defect in transcription elongation. We reasoned that, if transcription elongation was impaired by the positioned nucleosome, truncated transcripts containing sequences upstream to the positioning sequences should accumulate. An accumulation of transcripts of the EF1α-5S-Luc and EF1α-601-Luc transcription units containing sequences upstream to the positioning sequences was observed (Fig 2B,C), suggesting that the positioned nucleosome had impaired RNAPII progression.
Figure 2.
Nucleosome-positioning impairs transcription. (A) Constructs containing the indicated transcription units were transfected into 293T cells. Forty-eight hours after transfection, cells were collected and Luc activity was measured. Data are expressed as a percentage of mean relative activity from three independent experiments. Error bars represent ±s.d. values. (B) Schematic representation of the indicated transcription units with the locations of amplicons for RT–qPCR analysis. (C) Constructs containing the indicated transcription units were transfected into 293T cells. Forty-eight hours after transfection RNA expression was analysed by using RT–qPCR with primers specific for the 5′ and 3′ amplicons. Data are means of three independent experiments and error bars represent ±s.d. values. EF1α, elongation factor 1α;RT–qPCR, reverse transcriptase–quantitative PCR.
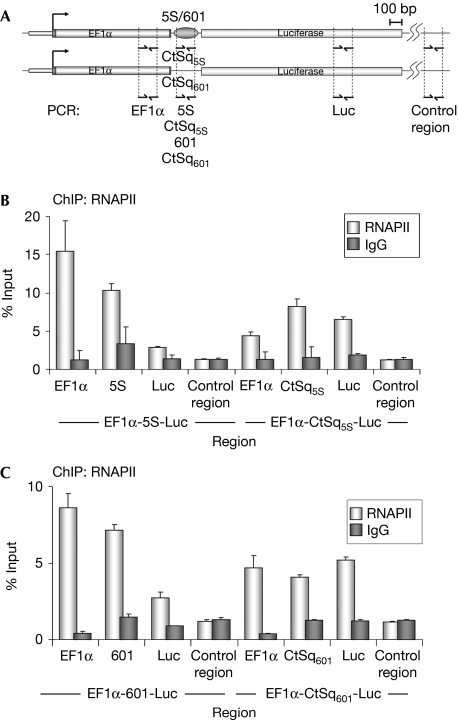
Next, in chromatin immunoprecipitation (ChIP) experiments, we analysed RNAPII occupancy along the transcription units. RNAPII was evenly distributed along the transcription units that lacked nucleosome-positioning sequences (Fig 3). However, when transcription units containing nucleosome-positioning sequences were analysed, a significant accumulation of RNAPII was observed upstream to these sequences. Concomitantly, a reduction in RNAPII occupancy was observed in the luciferase region (Fig 3B,C). Similar results were obtained using an antibody that specifically recognizes the elongating form of RNAPII (data not shown). Transcription units with and without positioned nucleosomes have very different profiles of RNAPII occupancy. This strongly suggests that RNAPII elongation is being blocked, in vivo, by the nucleosomal barrier.
Figure 3.
Nucleosome-positioning sequences cause RNAPII accumulation upstream from the positioned nucleosome. (A) Schematic representation of transcription units with the locations of amplicons for ChIP analysis. (B,C) Constructs containing the indicated transcription units were transfected into 293T cells. Forty-eight hours after transfection, cells were collected and used for ChIP experiments with RNAPII antibody or rabbit IgG as a control. The precipitated DNA fragments were subjected to qPCR quantification with primers for the indicated construct regions. Data (percentage of input) are the mean of at least n=6 qPCR reactions from three independent experiments. Error bars represent ±s.d. values. ChIP, chromatin immunoprecipitation; EF1α, elongation factor 1α; RNAPII, RNA polymerase II; qPCR, quantitative PCR.
BRG1 facilitates nucleosome traversal by RNAPII
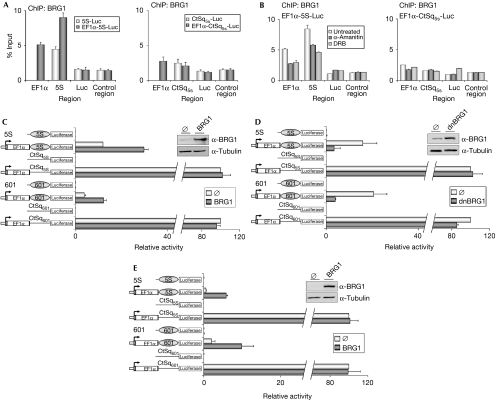
We used the system described above to investigate the role of BRG1—one of the ATPases of the mammalian SWI–SNF complexes—in chromatin remodelling during elongation. First, we determined by using ChIP analysis whether endogenous BRG1 is recruited to the transcribed region within the EF1α-5S-Luc and EF1α-CtSq5S-Luc transcription units (Fig 4A). A high level of BRG1 enrichment was observed at the 5S and EF1α intron regions of the EF1α-5S-Luc transcription unit, but not at the EF1α-CtSq5S-Luc transcription unit (Fig 4A). Furthermore, recruitment of BRG1 was dependent on the presence of the EF1α promoter and was significantly impaired in the presence of α-amanitin, which blocks both transcription initiation and elongation (Bushnell et al, 2002), or in the presence of 5,6-dichlorobenzimidazole 1-β-D-ribofuranoside, which blocks elongation (Chodosh et al, 1989; Fig 4B), indicating that BRG1 recruitment requires transcription. These data suggest that BRG1 escorts RNAPII during elongation and is stabilized at regions with a high density of positioned nucleosomes.
Figure 4.
BRG1 promotes transcription through a nucleosomal barrier. (A) Constructs containing the indicated transcription units were transfected into 293T cells. At 48 h after transfection, BRG1 occupancy at the indicated construct regions (see Fig 3A) was analysed by ChIP. (B) ChIP analysis of BRG1 in 293T cells untreated or treated with 2 μg/ml α-amanitin (α-amanitin) or 50 μM DRB (see supplementary Fig S4 online) for 24 h. (A,B) Data (percentage of input) are means from at least n=6 qPCR reactions from three independent experiments. (C–E) Constructs containing the indicated transcription units were co-transfected with empty vector (∅) or pSV-BRG1 (BRG1) in (C) 293T cells or in (E) C33A cells and empty vector (∅) or pTS-dnBRG1 (dnBRG1) in 293T cells (D). At 48 h after transfection, cells were collected and Luc activity was measured. Data are expressed as a percentage of mean relative activity from three independent experiments. Error bars represent ±s.d. values. BRG1 and dnBRG1 expression was analysed by western blot analysis with BRG1 antibody (left panels in C, D and E). BRG1, Brahma-related gene 1; ChIP, chromatin immunoprecipitation; dnBRG1, dominant-negative Brahma-related gene 1; DRB, 5,6-dichlorobenzimidazole 1-β-D-ribofuranoside.
The involvement of BRG1 in transcription elongation was explored by testing the effect of overexpressing BRG1 or a dominant-negative BRG1 form (dnBRG1), containing a mutation in the ATP-binding site domain on luciferase activity (de La Serna et al, 2000). First, we confirmed by PCR that expression of BRG1 or dnBRG1 did not affect the number of copies of the episomal vectors (data not shown). Second, we verified that BRG1 did not alter initiation of transcription from the EF1α promoter in our system (supplementary Fig S3 online). Consistent with the lack of effect on transcription initiation, overexpression of wild-type BRG1 or dnBRG1 did not affect the luciferase activity levels reached by control sequence vectors (Fig 4C,D). However, BRG1 enhanced the EF1α-dependent luciferase activity observed with 5S and 601 nucleosome-positioning constructs. By contrast, a dnBRG1 form impaired the EF1α-dependent luciferase activity of episomal vectors containing 5S and 601 sequences (Fig 4C,D). Together, these results suggest that BRG1 is involved in overcoming the 5S and 601 nucleosomal barriers during transcription elongation. Next, we assayed the role of BRG1 in C33A cells, a cervical carcinoma cell line that contains very small amounts of BRG1 and non-detectable amounts of the BRG1 paralogue human BRAHMA (Muchardt & Yaniv, 1993; Dunaief et al, 1994). In these cells, BRG1 strongly enhanced the expression of EF1α-5S-Luc (14-fold) and the EF1α-601-Luc (ninefold) transcription units, but not of transcription units, containing control sequences (Fig 4E).
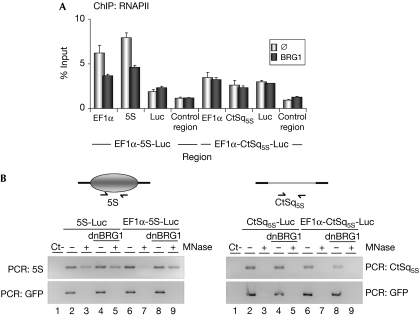
Next, we wondered how BRG1 affects RNAPII distribution in our system. Expression of BRG1 in C33A cells provoked a significant reduction in the level of RNAPII accumulated upstream to or at the nucleosome-positioning sequence in the EF1α-5S-Luc transcription unit (Fig 5A). These data suggest that BRG1 helps elongating RNAPII to overcome the barrier created by the positioned nucleosome. Finally, we showed that expression of dnBRG1 in 293T cells strongly reduced MNase accessibility of the positioned nucleosome at the EF1α-5S-Luc transcription unit (Fig 5B, left panel, compare lanes 6–7 and 8–9), suggesting that the SWI–SNF complex is responsible for the remodelling of this nucleosome during elongation. Consistent with the fact that BRG1 is recruited in a transcription-dependent manner, expression of dnBRG1 had no effect on the 5S-Luc transcription unit that lacks the EF1α promoter (Fig 5B, left panel, compare lanes 2–3 and 4–5).
Figure 5.
BRG1 helps RNAPII to overcome a nucleosomal barrier. (A) Constructs containing the indicated transcription units were co-transfected with empty vector (∅) or pSV-BRG1 (BRG1) into C33A cells. At 48 h after transfection, RNAPII occupancy at the indicated regions (see Fig 3A for position of the amplicons) was analysed by ChIP. Data (percentage of input) are the mean of at least n=6 quantitative PCR reactions from three independent experiments. Error bars represent ±s.d. values. (B) Constructs containing the indicated transcription units were co-transfected with empty vector (∅) or pTS-dnBRG1 (dnBRG1), together with pEGFP-N1 plasmid into 293T cells. Nuclei were untreated (−) or treated (+) with MNase. DNA was extracted and subjected to PCR with primers for 5S and CtSq5S regions. GFP was also amplified as a transfection control. Ct− denotes a PCR-negative control without DNA template. ChIP, chromatin immunoprecipitation; dnBRG1, dominant-negative Brahma-related gene 1; EF1α, elongation factor 1α; RNAPII, RNA polymerase II.
Discussion
In this study, we show that a positioned nucleosome in the body of a transcription unit impairs RNAPII progression in vivo and, as a consequence, provokes a reduction of transcription. Furthermore, we show that BRG1, the ATPase of the SWI–SNF complex, is recruited to the positioned nucleosome and helps RNAPII to overcome the nucleosomal barrier.
To rule out possible sequence effects, two different nucleosome-positioning sequences were used, and similar results were obtained with both of them. Interestingly, 601 sequence displays a much higher affinity for the octamer than 5S in in vitro nucleosome reconstitution assays (Lowary & Widom, 1998), but both showed a similar blocking effect in our in vivo assays. These data suggest that other chromatin-associated aspects, in addition to DNA-histone octamer affinity, affect the strength of the transcriptional blockage in vivo. For example, we cannot rule out the possibility that, in addition to DNA–histone octamer affinity, other factors are different between 601, 5S and other random nucleosomes and these might be affecting the rate of transcription. These include nucleosomal restoration behind advancing RNAPII, degree of histone modification or association to other chromatin structural factors.
In a series of elegant studies, Kingston and co-workers showed that the SWI–SNF complex is involved in the heat-shock factor protein 1-dependent activation of elongation of a promoter-proximal paused RNAPII at the mouse hsp70 gene (Brown et al, 1996; Sullivan et al, 2001). Now, we show that SWI–SNF facilitates the progression of RNAPII through nucleosomes during productive elongation in the middle of a transcription unit.
The mechanism by which the SWI–SNF complex enables RNAPII to overcome a nucleosomal barrier is unknown. Nucleosomes spontaneously undergo conformational fluctuations in which a stretch of their DNA transiently lifts off the histone surface (unwrapping; Li et al, 2005). In vitro studies carried out by Hodges et al, (2009) have recently reported that RNAPII does not seem to actively unwrap nucleosomes, but instead waits for these fluctuations spontaneous to occur. The SWI–SNF complex might help RNAPII by increasing the frequency or extent of these fluctuations. Thus, the transiently remodelled state provoked by the SWI–SNF complex would open a ‘window of opportunity' for RNAPII to cross the nucleosome. Does RNAPII require SWI–SNF to pass through every nucleosome? Even in vitro, nucleosomes are not an insurmountable barrier to elongation (Izban & Luse, 1991). Loosely positioned nucleosomes display a low core histone–DNA affinity and might adopt several positions, which represent a low barrier for RNAPII. We show that BRG1 was specifically required to increase transcription of templates with positioned nucleosomes. Furthermore, BRG1 occupancy was higher near to the strongly positioned nucleosome sequence. Therefore, we propose that traversal of RNAPII through strongly positioned nucleosomes is more dependent on the SWI–SNF complex than traversal of loosely positioned nucleosomes.
Methods
Cell lines and culturing conditions. The 293T and C33A cell lines were routinely grown in Dulbecco's modified Eagle's medium, supplemented with 7% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin.
Immunoblotting and antibodies. RNAPII (N-20, sc-899) and BRG1 (H-88, sc-10768) antibodies were obtained from Santa Cruz Biotechnology; γ-tubulin antibody from Sigma. Immunoblotting was carried out using standard procedures (see supplementary information online).
MNase digestion. Preparation of mononucleosomes by MNase digestion and analysis of nucleosomal positioning in vivo are described in detail in the supplementary information online. Briefly, 48 h after transfection, proteins were cross-linked to DNA by the addition of formaldehyde. Then, the cross-linking reaction was stopped by adding glycine, and nuclei were isolated and digested with MNase (supplementary Fig S1A online). To normalize, naked DNA was also digested with MNase (supplementary Fig S1B online). Then, DNA was extracted and equal amounts were run in a 1.2% agarose gel and visualized with ethidium bromide (supplementary Fig S1C,D online). The band corresponding to mononucleosome size (∼160 bp) was excised from the gel and DNA was purified. Equal amounts of DNA were used as a template for real-time PCR (qPCR) with the Applied Biosystems 7500 FAST Real-Time PCR System. The qPCR results were then normalized to the GFP gene qPCR as transfection control and to the MNase-digested naked DNA to avoid MNase digestion bias. Sequences of all oligonucleotides used for MNase mapping are listed in supplementary Table S1 online.
ChIP assays. The ChIP assays were performed as described previously (Strutt & Paro, 1999). Briefly, chromatin was sonicated to an average fragment size of 400–500 bp using Diagenode Bioruptor. Rabbit- or mouse-purified IgG (Sigma) was used as a control for nonspecific interaction of DNA. The human ACTB gene amplicon was used as a control for nonspecific binding of genomic DNA to beads or IgGs, and GFP gene PCR was used for transfection control. Quantification of immunoprecipitated DNA was performed by qPCR. At least three independent experiments were carried out. Each sample was quantified using three real-time PCR determinations. Sequences of all oligonucleotides used for ChIP are listed in supplementary Table S2 online.
Luciferase reporter assays. Luciferase activity was determined with a luciferase assay kit (Promega), according to the manufacturer's instructions.
RNA extraction and reverse transcriptase–qPCR. Total RNA was prepared by using the RNAsy Kit (Qiagen), as described in the manufacturer's instructions. Quantification of gene products was performed by qPCR. Oligonucleotides used in reverse transcription–qPCR experiments are listed in supplementary Table S2 online.
Additional experimental procedures are described in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank K. Zhao, C. L. Peterson, J. Widom, P. Charnay, C. Muchardt, M. Yaniv and G. Hager for generously providing plasmids. We also thank A. Aguilera, F. Prado, S. Chávez, R. Luna and M. García-Domínguez for critical reading of the paper and discussion. This study was supported by Ministerio de Educación y Ciencia (BFU2008-00238, CSD2006-00049) and by Junta de Andalucía (P06-CVI-01400).
Footnotes
The authors declare that they have no conflict of interest.
References
- Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM (2006) Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell 24: 469–479 [DOI] [PubMed] [Google Scholar]
- Brown SA, Imbalzano AN, Kingston RE (1996) Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev 10: 1479–1490 [DOI] [PubMed] [Google Scholar]
- Bushnell DA, Cramer P, Kornberg RD (2002) Structural basis of transcription: alpha-amanitin-RNA polymerase II cocrystal at 2.8 A resolution. Proc Natl Acad Sci USA 99: 1218–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M, Li B, Workman JL (2006) RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell 24: 481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh LA, Fire A, Samuels M, Sharp PA (1989) 5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J Biol Chem 264: 2250–2257 [PubMed] [Google Scholar]
- Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- de La Serna IL, Carlson KA, Hill DA, Guidi CJ, Stephenson RO, Sif S, Kingston RE, Imbalzano AN (2000) Mammalian SWI–SNF complexes contribute to activation of the hsp70 gene. Mol Cell Biol 20: 2839–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, Begemann M, Crabtree GR, Goff SP (1994) The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79: 119–130 [DOI] [PubMed] [Google Scholar]
- FitzGerald PC, Simpson RT (1985) Effects of sequence alterations in a DNA segment containing the 5S RNA gene from Lytechinus variegatus on positioning of a nucleosome core particle in vitro. J Biol Chem 260: 15318–15324 [PubMed] [Google Scholar]
- Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C (2009) Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science 325: 626–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban MG, Luse DS (1991) Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev 5: 683–696 [DOI] [PubMed] [Google Scholar]
- Kulaeva OI, Gaykalova DA, Studitsky VM (2007) Transcription through chromatin by RNA polymerase II: histone displacement and exchange. Mutat Res 618: 116–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langle-Rouault F, Patzel V, Benavente A, Taillez M, Silvestre N, Bompard A, Sczakiel G, Jacobs E, Rittner K (1998) Up to 100-fold increase of apparent gene expression in the presence of Epstein–Barr virus oriP sequences and EBNA1: implications of the nuclear import of plasmids. J Virol 72: 6181–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Levitus M, Bustamante C, Widom J (2005) Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol 12: 46–53 [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J (1998) New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol 276: 19–42 [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M (1993) A human homologue of Saccharomyces cerevisiae SNF2–SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J 12: 4279–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H, Paro R (1999) Mapping DNA target sites of chromatin proteins in vivo by formaldehyde crosslinking. Methods Mol Biol 119: 455–467 [DOI] [PubMed] [Google Scholar]
- Sullivan EK, Weirich CS, Guyon JR, Sif S, Kingston RE (2001) Transcriptional activation domains of human heat shock factor 1 recruit human SWI/SNF. Mol Cell Biol 21: 5826–5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiriet C, Hayes JJ (2005) Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev 19: 677–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetsuki T, Naito A, Nagata S, Kaziro Y (1989) Isolation and characterization of the human chromosomal gene for polypeptide chain elongation factor-1 alpha. J Biol Chem 264: 5791–5798 [PubMed] [Google Scholar]
- van der Vlag J, den Blaauwen JL, Sewalt RG, van Driel R, Otte AP (2000) Transcriptional repression mediated by polycomb group proteins and other chromatin-associated repressors is selectively blocked by insulators. J Biol Chem 275: 697–704 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.