MicroRNAs encoded by Kaposi's sarcoma-associated herpesvirus regulate viral life cycle
KSHV miR-K3 regulates viral latency by targeting nuclear factor I/B (NFIB). Lytic replication and gene expression were inhibited by overexpression of miR-K3 or depletion of NFIB.
Keywords: KSHV, miRNA, viral life cycle, RISC
Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) is linked with Kaposi's sarcoma and lymphomas. The pathogenesis of KSHV depends on the balance between two phases of the viral cycle: latency and lytic replication. In this study, we report that KSHV-encoded microRNAs (miRNAs) function as regulators by maintaining viral latency and inhibiting viral lytic replication. MiRNAs are short, noncoding, small RNAs that post-transcriptionally regulate the expression of messenger RNAs. Of the 12 viral miRNAs expressed in latent KSHV-infected cells, we observed that expression of miR-K3 can suppress both viral lytic replication and gene expression. Further experiments indicate that miR-K3 can regulate viral latency by targeting nuclear factor I/B. Nuclear factor I/B can activate the promoter of the viral immediate-early transactivator replication and transcription activator (RTA), and depletion of nuclear factor I/B by short hairpin RNAs had similar effects on the viral life cycle to those of miR-K3. Our results suggest a role for KSHV miRNAs in regulating the viral life cycle.
Introduction
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8, contains a linear double-stranded genome that encodes approximately 90 putative open reading frames (Pellett & Roizman, 2007). KSHV is associated not only with B-lymphoproliferative diseases, but also with Kaposi's sarcoma (Ganem, 2006; Laurent et al, 2008). Infection with KSHV normally results in a latent state in which the viral genome is maintained as an episome in the nuclei of infected cells. In KSHV-infected cells, lytic replication can be activated upon stimulation of cytokines or chemicals, and reactivation from latency to lytic replication is essential for viral transmission between hosts. Upon induction, KSHV gene expression follows a temporal and sequential order that can be divided into three phases: immediate-early, early and late.
In tumour, KSHVs are predominantly in latency stage with a small percentage of them undergoing lytic replication. Latent gene expression is believed to be essential for tumorigenesis; however, KSHV reactivation has been suggested to have a role in some malignancies, such as Kaposi's sarcoma (Ganem, 2006; Laurent et al, 2008). Viral pathogenesis depends on the balance between pathways that induce or suppress lytic replication. This balance is believed to be controlled by the lytic replication and transcription activator (RTA), and regulated by latent gene products, such as latency-associated nuclear antigen (LANA) and certain cell signalling pathways (Ganem, 2006).
The reactivation of KSHV was recently suggested to be regulated by herpesvirus-encoded microRNAs (miRNAs; Murphy et al, 2008; Umbach et al, 2008; Bellare & Ganem, 2009; Ziegelbauer et al, 2009). MiRNAs are noncoding RNAs comprising approximately 22 nucleotides that can regulate gene expression by inhibiting translation and/or destabilizing target messenger RNAs (mRNAs; Ambros, 2004; Bartel, 2004; Rana, 2007). KSHV encodes 12 pre-miRNAs that are processed into 17 mature miRNAs that are expressed during latency (Cai et al, 2005; Pfeffer et al, 2005; Grundhoff et al, 2006; Samols et al, 2007). The KSHV miRNAs miR-K5, miR-K9 and miR-K10 have been shown to induce viral reactivation by repressing Bcl-2-associated transcription factor 1, an apoptosis-inducing factor (Ziegelbauer et al, 2009). Computational analysis predicts that miR-K6-3p might target RTA to maintain viral latency by repressing viral lytic induction (Murphy et al, 2008), although further confirmation of this is needed. Thus, we hypothesized that some latent viral miRNAs might stabilize latency by functioning oppositely to miR-K5, miR-K9 and miR-K10. In this study, we show that KSHV miR-K3 can regulate viral latency by targeting nuclear factor I/B (NFIB). Both viral lytic replication and gene expression were inhibited by overexpressing miR-K3 or depleting NFIB. These findings suggest a role for KSHV miRNAs in regulating the viral life cycle.
Results And Discussion
To test our hypothesis that latent viral miRNAs might stabilize viral latency by repressing viral lytic replication, we investigated whether blocking the miRNA biogenesis pathway could induce replication. As the key components of the miRNA biogenesis machinery are Dicer and Argonaute 2 (AGO2; Chu & Rana, 2007), we knocked down human Dicer and AGO2 using short hairpin RNAs (shRNAs) in latently infected BC-3 cells. These are derived from human primary effusion (body-cavity based) lymphoma, and only a few per cent spontaneously reactivate (Sun et al, 1999).
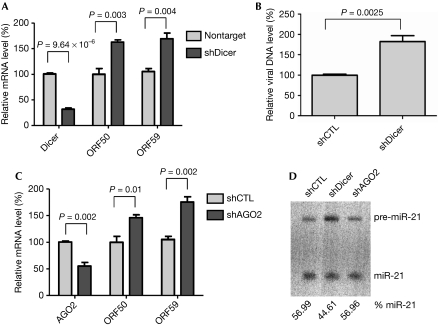
Dicer was efficiently knocked down in BC-3 cells transduced with shRNAs (Fig 1A). Northern blot analysis showed that knocking down Dicer decreased the production of a cellular miRNA marker—miR-21—confirming that the miRNA biogenesis pathway was affected in BC-3 cells with Dicer shRNAs (Fig 1D). Quantitative analysis of the KSHV lytic immediate-early gene ORF50, and the early gene ORF59, showed that knocking down Dicer in BC-3 cells increased viral lytic gene expression (Fig 1A). This was also associated with an increased viral DNA level (Fig 1B). Similarly, depleting AGO2 in BC-3 cells increased the expression of viral lytic genes, ORF50 and ORF59 (Fig 1C). Together, these results suggest that miRNAs are involved in inhibiting the KSHV lytic cycle.
Figure 1.
Blocking microRNA biogenesis induces Kaposi's sarcoma-associated herpesvirus lytic gene expression. BC-3 cells were transduced with lentiviral shRNAs targeting Dicer or AGO2 and selected with puromycin. (A) Knocking down Dicer induces viral lytic genes expression. The mRNA levels of Dicer, the KSHV lytic genes, ORF50 and ORF59, were quantified by RT–qPCR. (B) Knocking down Dicer induces viral lytic DNA replication. BC-3 cells were transduced with lentiviral Dicer shRNA and selected with puromycin. KSHV DNA was quantified by real time–PCR to detect ORF26 genomic sequences. Viral DNA levels are normalized to samples transduced with control shRNA. (C) Knocking down AGO2 induces viral lytic gene expression. The mRNA levels of AGO2, the KSHV lytic genes, ORF50 and ORF59, were quantified by RT–qPCR. Data are normalized to the mRNA level of cells transduced with control shRNA. All data are normalized to GAPDH. Results represent the standard deviation of three independent experiments. (D) Depleting Dicer decreases production of mature miRNA. Northern blot analysis of miR-21 RNA from BC-3 cells transduced with lentiviral shRNAs targeting control shRNA, Dicer and AGO2. The relative levels of mature miR-21 RNA are shown below the blot. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; KSHV, Kaposi's sarcoma-associated herpesvirus; mRNA, messenger RNA; miRNA, microRNA; RT–qPCR, reverse transcriptase–quantitative PCR; shCTL, control short hairpin RNA; shRNA, short hairpin RNA.
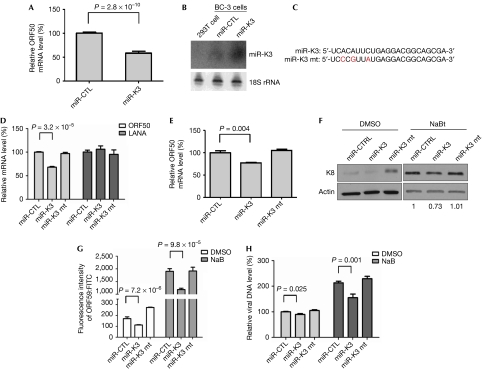
KSHV encodes 12 miRNAs in latently infected cells (Cai et al, 2005; Pfeffer et al, 2005; Grundhoff et al, 2006; Samols et al, 2007). We hypothesized that these might function to maintain the viral latent state and repress lytic gene expression. To identify which miRNAs are involved in this process, individual viral miRNAs were overexpressed by lentiviral vectors in BC-3 cells. Cells were further selected with zeocin and assessed for levels of both viral miRNAs and ORF50 mRNA, which is a marker for lytic gene expression (Fig 2A,B; supplementary Figs S1,S2 online). Among the 12 miRNAs examined, expression of miR-K9, miR-K10 and miR-K12 increased ORF50 expression, as found in other recent research (Ziegelbauer et al, 2009). Four miRNAs (K1, K3, K7 and K11) were able to decrease ORF50 expression; the effects of miR-K3 were most significant and consistent (Fig 2A). To confirm this result, we examined ORF50 levels in cells stably expressing an miR-K3 mutant (miR-K3 mt; Fig 2C) and observed that only the expression of miR-K3, but not of miR-K3 mt, can decrease ORF50 expression (Fig 2D). In addition, the expression of the latent gene LANA is not affected by miR-K3 overexpression (Fig 2D), suggesting that miR-K3 only inhibits lytic gene expression and functions to maintain viral latency. We also analysed the expression of additional lytic genes, including K8 (early gene) and ORF59 (delayed early gene), and viral DNA copy number. We observed that levels of all of these were decreased by miR-K3 overexpression, but not by the overexpression of miR-K3 mt (Fig 2E,F,G,H). Taken together, the above results indicate that miR-K3 is the stabilizer of KSHV latency.
Figure 2.
miR-K3 stabilizes Kaposi's sarcoma-associated herpesvirus latency. (A) BC-3 cells were transduced with lentiviruses expressing KSHV miR-K3, selected with zeocin, and analysed by using RT–qPCR for mRNA levels of the viral lytic gene ORF50. Data are normalized to the mRNA level in control miR-transduced cells. All data are normalized to GAPDH. The results represent the mean±s.e.m. of seven independent experiments. (B) BC-3 cells stably express miR-K3. RNAs from cells stably expressing miR-K3 and miR-CTL were used for northern blot analysis of miR-K3. 18S rRNA is shown as the loading control. (C) Sequences of miR-K3 and miR-K3 mt. (D) miR-K3 specifically downregulates viral lytic gene expression but not viral latent gene. mRNA levels for ORF50 and LANA were analysed by using RT–qPCR. Data represent mRNA levels normalized to miR-CTL. (E) miR-K3 decreases chemical-induced viral lytic gene expression. BC-3 cells stably expressing miR-K3 and miR-K3 mt were induced to lytic replication by sodium butyrate and ORF50 mRNA levels were analysed by using RT–qPCR. (F) miR-K3 decreases viral lytic early gene K8 expression. BC-3 cells were treated with DMSO or sodium butyrate, and K8 protein levels were examined by using western blotting. (G) miR-K3 decreases ORF59 expression. BC-3 cells were treated with DMSO or sodium butyrate, and ORF59 protein levels were determined by FACS analysis. Data represent the mean±s.e.m. of three independent experiments. (H) miR-K3 supresses viral lytic DNA replication. KSHV DNA was quantified by real-time–PCR to detect ORF26 genomic sequences. Data are normalized to the viral DNA level of cells transduced with miR-CTL and represent the mean±s.e.m. of three independent experiments. CTL, control; DMSO, dimethylsulphoxide; FACS, fluorescence-activated cell sorting; KSHV, Kaposi's sarcoma-associated herpesvirus; LANA, latency-associated nuclear antigen; miRNA, microRNA; mRNA, messenger RNA; mt, mutant; rRNA, ribosomal RNA; RT–qPCR, reverse transcriptase–quantitative PCR.
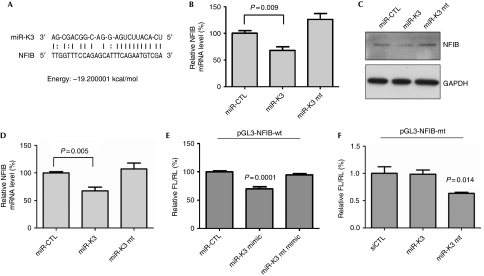
It has been previously reported that viral RTA could be targeted by KSHV miRNAs (Bellare & Ganem, 2009). We, however, hypothesized that cellular genes that reactivate KSHV could be targeted by viral miRNAs, such as miR-K3, to stabilize viral latency. To identify host targets of miR-K3, we used sequence analysis (the miRanda algorithm; Enright et al, 2003) to look for potential miR-K3 targets. A cellular transcription factor, NFIB, was predicted to have one potential miR-K3 target site in its 3′ untranslated region (3′UTR), suggesting that NFIB could be a direct target of miR-K3 (Fig 3A). NFIB belongs to the NFI gene family, also known as CAAT box transcription factors (CTFs), which have a broad role in DNA replication, cell proliferation and development (de Jong & van der Vliet, 1999; Gronostajski, 2000). Recently, murine NFIB was observed in a genome-wide complementary DNA library screen to reactivate KSHV (Yu et al, 2007). To examine whether NFIB is regulated by miR-K3, mRNA and levels of human NFIB protein were analysed by using reverse transcriptase–quantitative PCR and western blotting. Compared with miR-CTL-infected cells, both NFIB mRNA (Fig 3B) and NFIB protein (Fig 3C) were decreased in BC-3 cells expressing miR-K3, but not in those expressing miR-K3 mt. Similar results were found in 293T cells transiently transfected with miR-K3 mimics (Fig 3D). To determine whether NFIB is the direct target of miR-K3, its 3′UTR was constructed in the pGL-3 vector and a mutant 3′UTR was cloned with a miR-K3 mt-binding site. HeLa cells co-transfected with miR-K3 and the reporter plasmid containing the wild-type NFIB 3′UTR showed a 25% decrease in luciferase activity (Fig 3E), whereas cells co-transfected with the miR-K3 mt did not have altered luciferase activity of the reporter plasmid. Similar results were found in experiments with NFIB 3′UTR mt (Fig 3F). Together, these data indicate that the NFIB is directly targeted by miR-K3.
Figure 3.
MicroRNA-K3 suppresses nuclear factor I/B expression. (A) miR-K3 sequence and seed-matching site in the NFIB 3′UTR. (B) miR-K3 downregulates NFIB mRNA level. mRNA levels for NFIB were analysed by RT–qPCR. Data are normalized to mRNA levels for the miR-control (miR-CTL) and represent the means±s.e.m. of three independent experiments. (C) miR-K3 suppresses NFIB protein levels. Total protein from BC-3 cells stably expressing miR-K3 was analysed by using western blot for NFIB expression. GAPDH is shown as a loading control. (D) miR-K3 expression suppresses NFIB in 293T cells. 293T cells were transfected with 50 nM miR-CTL, miR-K3 or miR-K3 mt. NFIB mRNA was quantified by RT–qPCR at 48 h. Data are normalized to mRNA level of cells transduced with miR-CTL and represent the mean±s.e.m. of three independent experiments. (E) miR-K3 downregulates luciferase activity of reporter plasmid containing 3′UTR of NFIB. HeLa cells were co-transfected with the firefly luciferase vector containing the NFIB 3′UTR, internal control Renilla reporter and miR-K3 mimic or miR-K3 mt mimic. Firefly/Renilla ratios are normalized to those of miR-CTL. (F) Mutant miR-K3 downregulates luciferase activity of reporter plasmid containing corresponding mutant 3′UTR of NFIB. Experiments and data analysis were performed as described above in (E). CTL, control; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; KSHV, Kaposi's sarcoma-associated herpesvirus; miRNA, microRNA; mRNA, messenger RNA; mt, mutant; NFIB, nuclear factor I/B; RT–qPCR, reverse transcriptase–quantitative PCR; UTR, untranslated region; wt, wild-type.
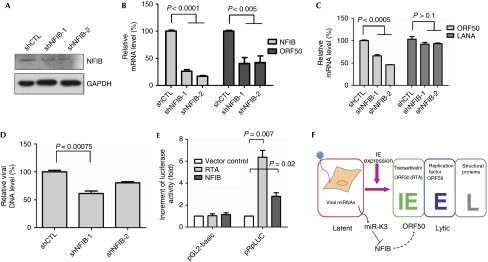
To test whether NFIB is involved in regulating viral life cycle, BC-3 cells were transduced with lentiviral shRNAs targeting NFIB. The expression of NFIB in BC-3 cells was efficiently silenced by shRNAs (Fig 4A,B). Quantitative analysis of viral latent and lytic gene expression showed that depleting NFIB decreased expression of the lytic gene ORF50, but not of the latent gene LANA (Fig 4C). Furthermore, viral DNA copy number was also reduced in NFIB-depleted cells induced with sodium butyrate (Fig 4D). As members of the NFI family are site-specific DNA-binding transcription factors that bind to the dyad symmetrical consensus sequence, 5′-TGGC(N5)GCCA-3′, or with lower affinity to TTGGC and TGGCA (Gronostajski, 2000), we analysed the sequence of ORF50 promoter and observed that it had one mismatched NF1-binding site—5′-TGGC(N5)GCCC-3′—and several other lower-affinity sites (supplementary Fig S3 online), indicating that the ORF50 promoter region contains the putative NFIB-binding site. We further examined the effect of NFIB on ORF50 promoter activity by co-transfecting 293T cells with NFIB and a promoter construct containing a 3-kb upstream ORF50 promoter sequence in the pGL-2 basic luciferase reporter pRpLuc (Deng et al, 2000). RTA was used as a positive control and could stimulate its own promoter approximately sixfold and NFIB activated the ORF50 promoter approximately 2.5-fold (Fig 4E).
Figure 4.
Nuclear factor I/B modulates viral lytic replication and ORF50 promoter activity. BC-3 cells were transduced with lentiviruses expressing shRNA targeting NFIB and selected with puromycin. shNFIB-1 and shNFIB-2 target different sites of NFIB. (A) shNFIB knocks down NFIB protein levels. Western blot was used to analyse decreased NFIB expression by shRNAs. GAPDH is shown as a loading control. (B,C) Knocking down NFIB suppresses viral lytic gene expression. mRNA levels for (B) NFIB, the KSHV lytic gene ORF50, and (C) the latent gene LANA, were quantified by RT–qPCR. mRNA data are normalized to mRNA levels of cells transduced with control shRNA. All data are normalized to GAPDH. The results represent the means±s.e.m. of three independent experiments. (D) Knocking down NFIB suppresses viral lytic DNA replication. KSHV DNA was quantified by real-time PCR to detect ORF26 genomic sequences. Viral DNA data are normalized to viral DNA levels of cells transduced with control shRNA (shCTL). (E) NFIB turns on ORF50 promoter activity. 293T cells were co-transfected with pRpLuc and RTA (ORF50) or NFIB and collected for luciferase assay at 48 h. Relative luciferase activity of individual transfections was normalized to total protein concentration. (F) Role of miR-K3 in suppressing viral lytic replication. Expression of miR-K3 downregulates NFIB, which activates the promoter activity of the viral immediate-early (IE) gene, Rta, which in turn suppresses viral lytic replication. The model of viral life cycle is modified from that of Tsurumi et al (2005). GAPDH, glyceraldehyde 3-phosphate dehydrogenase; KSHV, Kaposi's sarcoma-associated herpesvirus; miRNA, microRNA; mRNA, messenger RNA; NFIB, nuclear factor I/B; RT–qPCR, reverse transcriptase–quantitative PCR; shCTL, control short hairpin RNA.
In this study, we hypothesized that KSHV-encoded miRNAs regulate viral latency and screened all 12 miRNAs for potential regulators of viral lytic replication. We have demonstrated that miR-K3 was able to inhibit viral lytic replication by directly targeting cellular transcription factor NFIB. NFIB can activate ORF50 promoter activity, and decreased expression of NFIB inhibits lytic replication. As ORF50 is a key immediate-early viral transactivator that can induce KSHV reactivation, miR-K3 inhibition of ORF50 expression by downregulating the NFIB transcription factor might establish and maintain viral latency (Fig 4F).
The NFI/CTF activity is observed in a wide range of species and binds to the sequence 5′-TGGCNNNNNGCCA-3′. The NFI/CTF has been observed to interact with DNA elements in the promoter region of herpesvirus immediate-early antigens, including immediate-early 1 (IE1) of human cytomegalovirus (Hennighausen & Fleckenstein, 1986) and RTA of Epstein–Barr virus (Glaser et al, 1998). We also found that NFIB—a member of the NFI/CTF family—activated the promoter activity of RTA—the KSHV IE gene. This activation could be through direct binding to the Rta promoter region. These findings suggest that interactions between the promoter of IE and NFI/CTF are important in regulating herpes viral lytic replication.
The NFI/CTF transcription factors have also been shown to enhance initiation of adenovirus DNA replication, by recruiting adenovirus DNA polymerase and protein primer pTP to the origin of DNA replication (de Jong & van der Vliet, 1999). As in other viral and cellular replication origins—such as Epstein–Barr virus—transcription factors were proposed to help the initiation of DNA replication, not only by directly interacting and recruiting initiator protein to the origin, but also by altering local chromatin structure (Kohzaki & Murakami, 2005). In KSHV-infected cells, cellular transcription factors were also observed to bind to sequences within the KSHV origin of lytic DNA replication (ori-Lyt); these factors include NFI/CTF, activator protein 1, and CCAAT/enhancer binding protein (C/EBP; Wang et al, 2006; Wu et al, 2003). We suggest that NFIB might have a similar role, to enhance KSHV lytic DNA replication. Furthermore, C/EBPα induces viral reactivation not only by binding to ori-Lyt, but also by contributing to the transactivation of promoters for RTA and p21, a cyclin-dependent kinase inhibitor (Wu et al, 2002, 2003). Increased expression of p21 might result in host G1 cell cycle arrest, which seems to be an important early step in the lytic cycle for most herpesviruses (Flemington, 2001). Expression of the C/EBPa gene can be activated both by C/EBPα autoactivation and by C/EBPβ transactivation (Tang & Lane, 1999), which has recently been identified as the target of the KSHV-encoded miRNAs, miR-K3 and miR-K7 (Qin et al, 2009). We observed that miR-K3 inhibits viral lytic replication, possibly by downregulating C/EBPβ. Interestingly, a recent study reported that a KSHV miRNA, miR-K12-4-5p, enhanced DNA methyl transferase 3a and 3b mRNA levels by reducing retinoblastoma-like protein 2 expression, which is a repressor of DNA methyl transferase mRNA transcription (Lu et al, 2010). Taken together, these studies suggest that multiple pathways, such as NFIB activation and epigenetic regulations, are modulated by KSHV miRNAs to maintain viral latency.
In summary, we observed that viral-encoded miRNAs function as regulators, both by suppressing (miR-K1, miR-K3, miR-K7 and miR-K11) and by inducing (miR-K9, miR-K10 and miR-K12) viral lytic replication. KSHV pathogenesis depends on regulation of the balance between two phases: latency and lytic replication. Our results support the role of viral-encoded miRNAs in controlling this balance.
Methods
Reporter assay. For the promoter reporter assay, 293T cells were co-transfected with 50 ng pRpLuc and 100 ng RTA or NFIB in 12-well plates. Cells were collected at 48 h for luciferase assay (Promega). Relative luciferase activity of individual transfections was normalized to total protein concentration. For the 3′UTR reporter assay, HeLa cells were transfected with 50 nM miRNAs for twenty four hours, and co-transfected with 50 nM miRNAs, 200 ng of reporter plasmid containing 3′UTR of NFIB and 50 ng of renilla luciferase reporter driver by the thymidine kinase promoter. Twenty-four hours post-transfection, cells were collected for dual-luciferase assay (Promega). Relative firefly luciferase activity was normalized to Renilla luciferase activity. The P-values were calculated using Student's t-test with multiple independent samples.
miRNA and shRNA expression. Lentiviral shRNA vectors were from purchased Open biosystems. The miRNA lentiviral expression vectors were cloned into pMIF-cGFP-Zeo-miR with 250 bp of flanking sequence surrounding individual miRNA vectors (System Biosciences). Lentiviruses were produced from 293FT cells by co-transfecting with packaging plasmids. The BC-3 cells were transduced and selected with either puromycin or zeocin.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank S. Chanda, J. Reed and members of the Rana laboratory for helpful discussions, encouragement and support. This study was supported by National Institutes of Health grants and institutional funds to T.M.R.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ambros V (2004) The functions of animal microRNAs. Nature 431: 350–355 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bellare P, Ganem D (2009) Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 6: 570–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR (2005) Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci USA 102: 5570–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Rana TM (2007) Small RNAs: regulators and guardians of the genome. J Cell Physiol 213: 412–419 [DOI] [PubMed] [Google Scholar]
- de Jong RN, van der Vliet PC (1999) Mechanism of DNA replication in eukaryotic cells: cellular host factors stimulating adenovirus DNA replication. Gene 236: 1–12 [DOI] [PubMed] [Google Scholar]
- Deng H, Young A, Sun R (2000) Auto-activation of the Rta gene of human herpesvirus-8/Kaposi′s sarcoma-associated herpesvirus. J Gen Virol 81: 3043–3048 [DOI] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS (2003) MicroRNA targets in Drosophila. Genome Biol 5: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemington EK (2001) Herpesvirus lytic replication and the cell cycle: arresting new developments. J Virol 75: 4475–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D (2006) KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu Rev Pathol 1: 273–296 [DOI] [PubMed] [Google Scholar]
- Glaser G, Vogel M, Wolf H, Niller HH (1998) Regulation of the Epstein–Barr viral immediate early BRLF1 promoter through a distal NF1 site. Arch Virol 143: 1967–1983 [DOI] [PubMed] [Google Scholar]
- Gronostajski RM (2000) Roles of the NFI/CTF gene family in transcription and development. Gene 249: 31–45 [DOI] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS, Ganem D (2006) A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12: 733–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L, Fleckenstein B (1986) Nuclear factor 1 interacts with five DNA elements in the promoter region of the human cytomegalovirus major immediate early gene. EMBO J 5: 1367–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohzaki H, Murakami Y (2005) Transcription factors and DNA replication origin selection. Bioessays 27: 1107–1116 [DOI] [PubMed] [Google Scholar]
- Laurent C, Meggetto F, Brousset P (2008) Human herpesvirus 8 infections in patients with immunodeficiencies. Hum Pathol 39: 983–993 [DOI] [PubMed] [Google Scholar]
- Lu F, Stedman W, Yousef M, Renne R, Lieberman PM (2010) Epigenetic regulation of Kaposi′s sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2–DNMT pathway. J Virol 84: 2697–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ (2008) Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc Natl Acad Sci USA 105: 5453–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett PE, Roizman B (2007) The family herpesviridae: A brief introduction. In Fields Virology, Knipe DM et al. (eds) pp 2381–2398. Philadelphia, PA, USA: Lippincott Williams & Wilkins [Google Scholar]
- Pfeffer S et al. (2005) Identification of microRNAs of the herpesvirus family. Nat Methods 2: 269–276 [DOI] [PubMed] [Google Scholar]
- Qin Z, Kearney P, Plaisance K, Parsons CH (2009) Pivotal advance: Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded microRNA specifically induce IL-6 and IL-10 secretion by macrophages and monocytes. J Leukoc Biol 87: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana TM (2007) Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol 8: 23–36 [DOI] [PubMed] [Google Scholar]
- Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R (2007) Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog 3: e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Lin SF, Staskus K, Gradoville L, Grogan E, Haase A, Miller G (1999) Kinetics of Kaposi′s sarcoma-associated herpesvirus gene expression. J Virol 73: 2232–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang QQ, Lane MD (1999) Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev 13: 2231–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi T, Fujita M, Kudoh A (2005) Latent and lytic Epstein–Barr virus replication strategies. Rev Med Virol 15: 3–15 [DOI] [PubMed] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR (2008) MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454: 780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang Q, Maul GG, Yuan Y (2006) Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: dual role of replication and transcription activator. J Virol 80: 12171–12186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FY, Tang QQ, Chen H, ApRhys C, Farrell C, Chen J, Fujimuro M, Lane MD, Hayward GS (2002) Lytic replication-associated protein (RAP) encoded by Kaposi sarcoma-associated herpesvirus causes p21CIP-1-mediated G1 cell cycle arrest through CCAAT/enhancer-binding protein-alpha. Proc Natl Acad Sci USA 99: 10683–10688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FY, Wang SE, Tang QQ, Fujimuro M, Chiou CJ, Zheng Q, Chen H, Hayward SD, Lane MD, Hayward GS (2003) Cell cycle arrest by Kaposi′s sarcoma-associated herpesvirus replication-associated protein is mediated at both the transcriptional and posttranslational levels by binding to CCAAT/enhancer-binding protein alpha and p21(CIP-1). J Virol 77: 8893–8914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F et al. (2007) Systematic identification of cellular signals reactivating Kaposi sarcoma-associated herpesvirus. PLoS Pathog 3: e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelbauer JM, Sullivan CS, Ganem D (2009) Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet 41: 130–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.