Mutated genes, pathways and processes in tumours
This analysis of genes mutated in cancers, drawn from small and large-scale genome studies, allows the authors to draw a map of pathways and processes altered by cancers, revealing both common and specific changes.
Keywords: cancer-mutated genes, large-scale resequencing, oncogenomics, pathways, systems biology
Abstract
Integration of the many available sources of cancer gene information—such as large-scale tumour-resequencing studies— identifies the ‘usual suspect' genes, mutated in many tumour types, as well as different sets of mutated genes according to the specific tumour type. Scaling-up the analysis reveals that this large collection of mutated genes cluster into a smaller number of signalling pathways and processes. From this, we draw a map of the altered processes, and their combinations, in more than 10 tumours types. Literature searches identify pathways and processes that are covered sparsely in the literature, and invite the proposal of new hypotheses to investigate cancer initiation and progression.
Introduction
Cancers are genetic diseases that originate from the accumulation of DNA alterations in cells (Stratton et al, 2009). These alterations can occur at the level of chromosomes—rearrangement and gain or loss of chromosomal regions—or genes—point mutations, copy number alterations and gain or loss of genomic regions. Such changes can affect coding or non-coding DNA, and might affect expression of the corresponding transcripts and proteins, or directly impair protein functions. Despite this diversity of genetic alterations, most important therapeutic advances in the past decade have come from the development of drugs that target proteins encoded by genes that are mutated in cancers (Vogelstein & Kinzler, 2004).
In this context, many types of cancer (in this study refered to by the tissue of origin, for instance breast or lung) are now being screened for somatic mutations, and an unprecedented number of cancer-associated mutations are being identified. It has been proposed that these mutated genes encode proteins that cluster in certain signalling pathways, and different sets of mutated genes in different tumours might affect the same functional pathways or processes (Copeland & Jenkins, 2009; Stratton et al, 2009). This pathway-mapping approach has been applied to recent large-scale tumour-resequencing results, which, along with a list of mutations and mutated genes, has generated a list of pathways enriched in mutations (for example, see Ding et al, 2008). Comparing the pathways enriched in mutations in breast and colorectal tumours, revealed few overlaps (Chittenden et al, 2008), but these studies did not integrate different sources of mutated gene data, or systematically assess different types of tumour. Further analysis of the cancer-mutated genes at the level of pathways and processes might provide an overview of the functional processes that are altered in the different tumours. This could also reveal new pathways or processes enriched in mutated genes. This should permit, by analogy with other areas in biology, the development of a classification to define common and specific mutated genes—and pathways and processes—in different types of tumour.
Results
A snapshot of cancer-mutated gene data
To obtain a snapshot of the genes mutated in cancer, we combined information from databases of cancer-mutated genes with data from recently published large-scale resequencing screens (Table 1; Methods section). Cancer-mutated genes were considered to be those that had been observed with at least one non-synonymous mutation (for example, a missense or nonsense point mutation, or a small insertion/deletion), either in the coding sequence or at a splice site.
Table 1. Sources of genes mutated in different cancer types.
| Name of study | Tumour type(s) screened |
|---|---|
| Databases | |
| Cancer Gene Census (Futreal et al, 2004) | Many |
| OMIM (Hamosh et al, 2005) | Many |
| COSMIC (Forbes et al, 2008) | Many |
| Gene-set resequencing studies | |
| Bardelli et al (2003) | Colorectal |
| Wang et al (2004) | Colorectal |
| Stephens et al (2005) | Breast |
| Davies et al (2005) | Lung |
| Greenman et al (2007) | Many |
| Thomas et al (2007) | Many |
| Loriaux et al (2008) | Acute myeloid leukaemia |
| Tomasson et al (2008) | Acute myeloid leukaemia |
| Cancer Genome Atlas (Research Network, 2008) | Glioblastoma |
| Ding et al (2008) | Lung |
| Whole-genome resequencing studies | |
| Wood et al (2007) | Breast and colorectal |
| Jones et al (2008) | Pancreas |
| Ley et al (2008) | Acute myeloid leukaemia |
| Parsons et al (2008) | Glioblastoma |
| Pleasance et al (2010b) | Lung |
| Pleasance et al (2010a) | Melanoma |
| Dalgliesh et al (2010) | Renal |
| COSMIC, Catalogue of Somatic Mutations in Cancer; OMIM, Online Mendelian Inheritance in Man. | |
Combining the above data identified 5,272 mutated genes, from more than 40 tumour types (supplementary Table S1 online). Some of these genes are mutated in many tumour types and correspond to the ‘usual suspect' oncogenes (BRAF, H-RAS or PI3KCA), tumour suppressors (APC, PTEN or TP53) or DNA repair proteins (BRCA2 or MSH2). These repeatedly mutated genes were not only catalogued by literature surveys, but also detected in large-scale resequencing studies, including those focused on whole genomes. Hence, they are likely to be involved in tumorigenesis generally, and mutated in many tumour types.
By contrast, 73% of the 5,272 mutated genes have only been detected in one type of tumour (supplementary Table S1 online). The majority (more than 70%) of these ‘tumour-specific' mutated genes have been identified in genome-wide resequencing studies. Many of them might not have a direct role in the specific tumour or might be passengers, that is, cancer-neutral variants that are retained during the evolution of cancer (Futreal et al, 2004). Nevertheless, initial studies have addressed the problem of tumour specificity by showing that some of the genes mutated in large-scale breast and colorectal cancer studies are not mutated in glioblastoma, melanoma and pancreatic carcinoma (Balakrishnan et al, 2007). In our integrated data set, a subset of more than 200 tumour-type-specific genes was detected from more than one of the information sources. For example, two isoforms of casein kinase 1, CKIα and CKIδ, were observed to be mutated in breast cancer cells according to two and three resequencing studies, respectively (supplementary Table S1 online). In addition, the CKIɛ isoform was also mutated in breast cancer (Fuja et al, 2004). Interestingly, CKIδ was recently shown to modulate the transcriptional activity of the oestrogen receptor (Giamas et al, 2009), well known for its involvement in breast cancer.
To fully assess the existence of tumour-specific genes and their role in promoting tumorigenesis, more detailed validation studies will be necessary.
Processes enriched in mutated genes
In this study, we focus on 14 tumour types for which more than 50 mutated genes have been identified from our integration of the different mutation sources (Methods section, supplementary Table S1 online, Online Visualisation Tool (http://contexts.bioinfo.cnio.es/cancer-processes)). For each mutated gene, we first examined the functional roles of the corresponding proteins in about 600 cellular pathways gathered from Kegg, Biocarta and Reactome. This analysis was then extended to more than 5,000 Gene Ontology biological processes, and more than 19,000 entries from the Interpro domains database (Methods section). All of these pathways, processes and protein domains were challenged in each tumour type to identify those that contained greater than chance numbers of mutated genes (Methods section, supplementary Table S2 online). The complete results of associations between pathways, processes and protein domains and the different tumour types can be browsed and searched through the Online Visualisation Tool, which will be updated with newly published results of large-scale resequencing mutation screens.
We used a probability threshold of 0.01 (including pathways with a less than 1 in 100 chance of containing that number of mutant genes), and focused on cellular pathways from Kegg, Biocarta and Reactome (Methods section). Eleven tumour types were associated with significantly mutated cellular pathways (Fig 1). For example, in brain tumours—for which 1,692 mutated genes are catalogued—we identified 18 cellular pathways and processes that contained more mutated genes than would be expected by chance (Fig 1). One of these pathways is the Kegg ErbB signalling pathway. Eighty-seven genes that have been screened for mutations in brain tumours were annotated for this pathway, of which 26 genes were observed to be mutated (Q-value <0.001) in at least one of the mutated gene data sources, which suggests the involvement of this pathway in brain tumours. ErbB receptor tyrosine kinases are known to be involved in many cancers (Mitsudomi & Yatabe, 2010). The ATP-binding cassette transporter family also contains 15 genes that have been observed to be mutated in the brain tumour large-scale resequencing study, among the 39 that were screened (Q-value <0.001). This association is well described in the brain cancer literature, as ATP-binding cassette transporters are involved in the blood–brain barrier and their mutational status might reflect their role in drug resistance (Begley, 2004).
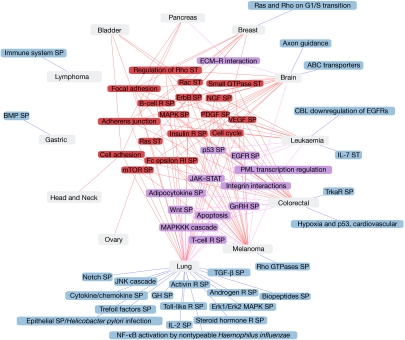
Figure 1.
Altered cellular pathways and processes in tumours. Graphic representation of cellular pathways/processes containing a significant number of mutated genes (Q-values <0.01) among tumour types (Methods section). Cellular pathways containing a significant number of mutated genes in four or more tumour types are depicted in red, in two or three tumour types in violet, and in only one tumour type in blue. ECM, extracellular matrix; R, receptor; SP, signalling pathway; ST, signal transduction.
The number of significant cellular pathways and processes associated with each tumour type ranged from 1 (for example, gastric or ovary) to 41 (lung) and allowed us to compare the pathways affected by mutated genes in different tumour types. In total, 17 cellular pathways contained a significant number of genes that were mutated in four or more tumour types, including the MAPK, mTOR, PDGF, VEGF and ErbB signalling pathways (Fig 1, in red). Other common cellular pathways included those related to cellular adhesion, involving genes coding for proteins active in focal adhesions and adherens junctions. Expanding the search to Gene Ontology processes revealed the same trends: small GTPases, Rho, Ras and Rac cellular pathways and processes related to signalling, cellular adhesion and proliferation contained a significant number of mutated genes in many tumour types (Fig 1). Protein domain annotations confirmed these results; among the mutated genes were many kinases, as well as proteins with immunoglobulin- and EGF-like domains, known to be involved in signal transduction and cell communication. These cellular pathways, processes and protein domains are often altered and known to be involved in tumorigenesis. As such, they would be expected to contain a significant number of mutated genes (Methods section; Hanahan & Weinberg, 2000). However, this overall picture could not have been obtained from separate studies of the mutated genes in each tumour type; different sets of genes identified the same significant pathways in different tumour types. For instance, the mammalian target of rapamycin—mTor—signalling pathway contains a significant number of genes that are mutated in brain, lung, melanoma, colorectal and ovarian cancers, but only 48% of the mutated genes are shared among these tumour types.
Processes sparsely described in the literature
Our survey of cellular pathways, processes and protein domains, identified about 20 results that are only sparsely described in the literature associated with each tumour type (Methods section, Table 2).
Table 2. Examples of cellular pathways, processes and protein domains.
| Tumour type | Mutated genes | |
|---|---|---|
| Cellular pathways | ||
| Adipocytokine signalling pathway (hsa04920) | Colorectal | ACSL4 ACSL5 AKT1 CAMKK2 CHUK MTOR IRS2 IRS4 JAK1 MAPK9 PRKCQ PTPN11 STK11 TYK2 |
| Lung | AKT1 CAMKK2 CHUK MTOR IKBKB IRS1 IRS4 JAK1 JAK2 JAK3 PPARGC1A PRKCQ PTPN11 RELA STAT3 STK11 TYK2 | |
| CBL-mediated ligand-induced downregulation of EGF receptors (h_cblPathway) | Leukaemia | CBL CSF1R EGFR MET PDGFRA |
| Epithelial cell signalling in Helicobacter pylori infection (hsa05120) | Lung | CHUK CSK CXCR2 EGFR IKBKB MAP2K4 MAP3K14 MAPK11 MAPK14 MAPK8 MET PLCG1 PLCG2 PTPN11 PTPRZ1 RELA SRC |
| NF-κB activation by non-typeable Hemophilus influenzae (h_nthiPathway) | Lung | CHUK CREBBP EP300 IKBKB MAP2K6 MAP3K14 MAPK11 MAPK14 RELA SMAD4 TGFBR1 TGFBR2 |
| GnRH signalling pathway (hsa04912) | Colorectal | ADCY8 ADCY9 CAMK2A CAMK2B EGFR GNAS HRAS KRAS MAP2K4 MAP2K7 MAP3K2 MAP3K3 MAPK9 MMP2 NRAS PLA2G2A PRKCA PRKCB SRC |
| Lung | CAMK2G EGFR GNAS HRAS ITPR2 KRAS MAP2K4 MAP2K6 MAP3K2 MAP3K3 MAP3K4 MAPK11 MAPK14 MAPK7 MAPK8 MMP2 NRAS PLCB1 PRKACA PRKACB PRKCB PRKCD PRKX RAF1 SRC | |
| Regulation of transcriptional activity by PML (h_pmlPathway) | Leukaemia | CREBBP HRAS KRAS NRAS PML RARA RB1 TP53 |
| Melanoma | CREBBP DAXX HRAS KRAS NRAS RB1 TNF TNFRSF1A TP53 | |
| Trefoil factors initiate mucosal healing (h_tffPathway) | Lung | AKT1 CSH2 CTNNB1 CYCS EGFR ERBB2 GHR HRAS KRAS MUC2 NRAS PTK2 SHC1 SOS1 |
| Bioactive peptide induced signalling pathway (h_biopeptidesPathway) | Lung | CAMK2G CDK5 FYN HRAS JAK2 KRAS MAPK14 MAPK8 NRAS PLCG1 PRKCB RAF1 SHC1 SOS1 STAT5A |
| Processes | ||
| Huntington disease (hsa05040) | Leukaemia | CLTC CLTCL1 CREBBP EP300 HIP1 TP53 |
| Base-excision repair (GO:0006284) | Lung | CKN2 MBD4 MSH6 NTHL1 PCNA POLG RAD51L3 TP53 |
| Double-stranded break repair through non-homologous end joining (GO:0006303, GO:0000726) | Melanoma | POLA1 PRKDC XRCC4 XRCC6 |
| Cytokinesis (GO:0000910) | Lung | ARHGEF11 AURKB AURKC BRCA2 DIAPH2 ESPL1 INCENP MYH9 ROCK1 |
| Protein import into nucleus, translocation (GO:0000060) | Leukaemia | ARNT BCL3 BCL6 JAK2 |
| Protein import into nucleus, translocation (GO:0000060) | Leukaemia | ARNT BCL3 BCL6 JAK2 |
| Regulation of protein export from nucleus (GO:0046825) | Lung | BARD1 GSK3B PTPN11 |
| Protein domains | ||
| BRK (IPR006576) | Lung | CHD5 CHD6 CHD9 SMARCA4 |
| Bromodomain (IPR001487) | Brain | BRD2 EP300 KAT2B MLL PHIP SMARCA2 SMARCA4 SP100 TAF1 TAF1L TRIM24 TRIM33 ZMYND8 |
| Lung | BAZ1A CREBBP EP300 MLL SMARCA4 TAF1 TAF1L TRIM24 TRIM33 | |
| Helicases (IPR014001, IPR001650, IPR014021) | Breast | ATRX BRIP1 CHD1 CHD5 CHD7 CHD8 CHD9 DDX10 DDX18 DDX3X DDX59 EIF4A2 ERCC3 ERCC6 FANCM HELQ RAD54L SKIV2L SMARCAD1 SMARCAL1 |
| Melanoma | ATRX CHD6 CHD8 DDX24 DDX28 DDX3X DDX4 DDX54 DICER1 POLQ RAD54L SMARCA1 SMARCA4 SMARCA5 SMARCAD1 SMARCAL1 SNRNP200 | |
| Diacylglycerol kinase (IPR000756, IPR001206) | Melanoma | CERKL DGKB DGKD DGKG DGKZ |
| Laminin G (IPR001791, IPR012680) | Pancreas | CELSR1 CNTNAP4 COL11A1 COL5A1 FAT1 FAT3 FAT4 LAMA1 LAMA4 LAMA5 NRXN3 |
| CBL, Casitas B-lineage lymphoma; EGF, epidermal growth factor; GnRH, gonadotropin-releasing hormone; GO, gene ontology; PML, promyelocyclic leukaemia. | ||
Some relevant examples include the Biocarta pathway called ‘regulation of transcriptional activity by PML', that contains eight different genes that are mutated in leukaemia (Q-value <0.000001, Table 2). This result reflects the well-known t(15;17) translocation between the PML and RARA genes that occurs in the majority of acute promyeloid leukaemias (Salomoni et al, 2008). Other studies have shown that these genes can also be mutated in leukaemia, as can other genes in the pathway, such as those for the transcriptional co-activator cyclic AMP REBP-binding protein and the tumour suppressors p53 and retinoblastoma (Table 2). Interestingly, the ‘regulation of transcriptional activity by PML', pathway also contains a significant number of mutated genes in melanoma, with nine genes annotated to be involved (Table 2). Five mutated genes are common to both tumour types, but there are also interesting differences: the PML and RARA genes have not yet been observed to be mutated in melanoma, and three other genes are mutated in melanoma but currently not associated with leukaemia (TNF and its receptor, TNFRSF1A1, and the DAXX transcription repressor).
The gonadotropin-releasing hormone (GnRH) signalling pathway is significantly associated with lung and colorectal cancers, as there are 26 and 19 genes in this pathway that are mutated in these tumour types, respectively (the mutated gene data are extracted from seven and six, respectively, different and partly overlapping mutated genes sources; Q-value <0.001; Fig 1; Table 2). The GnRH receptor is expressed in tumours derived from reproductive tissues, such as breast and prostate tumours, and agonists/antagonists of this receptor can be used to treat hormone-dependent tumours. GnRH can also control metastasis in melanoma (Cheung & Wong, 2008). However, apart from the report that cytotoxic analogues of the GnRH receptor inhibit growth in colorectal cell lines (Szepeshazi et al, 2007), to our knowledge no previous research can account for the association between GnRH signalling and lung or colorectal tumours. The adipocytokine signalling pathway also contains a significant number of genes that are mutated in lung and colorectal cancers (19 and 14 mutated genes, respectively; Q-value <0.01; Fig 1; Table 2). Obesity is a risk factor for cancer, and recent studies have shown that the underlying mechanisms for disease development could involve adipocytokines (Tilg & Moschen, 2006). This is consistent with the link between the adipocytokine signalling pathway and lung and colorectal cancers proposed in this study.
Other examples have come from the study of Interpro protein domains (Table 2). For instance, 13 proteins that are mutated in brain tumours contain a bromodomain (Q-value <0.01), a potentially relevant association given the role of this domain in histone binding and transcriptional regulation (Mujtaba et al, 2007). Another example is the laminin G protein domain, involved in cell attachment, that was contained in 12 different proteins, the genes of which are mutated in pancreatic cancer (Q-value <0.01). Indeed, to the best of our knowledge, the relationship between pancreatic cancer and the laminin G protein domain has not been described previously or identified as relevant (F.X. Real, personal communication). This link should be investigated further by direct experiments.
Discussion
The analysis of 5,272 mutated genes at the level of cellular function showed that cancer-mutated genes cluster into specific cellular processes. This work should be extended in the future to account for other types of genomic alteration (including changes in copy number or alterations of noncoding regions), and refined to address spatiotemporal issues (such as tumour subtypes, stages or grades).
Combined analysis of the mutated genes revealed set of genes that are mutated in many tumour types (201 genes are mutated in four or more different tumour types), and contribute to core cellular pathways and processes that are common to many tumour types. For example, the cell-cycle process contains a significant number of mutated genes in five tumour types, and cell-adhesion-related processes are associated with more than six tumour types. Overall, the analysis identified well-known associations between about 15 different pathways and processes and nine tumour types.
Many mutated genes are associated with only few or one tumour type. Three thousand eight hundred and sixty genes have been observed mutated in only one tumour type, and 952 in two tumour types. These specific genes might be non-relevant passenger variants, and might decrease in number with the progressive incorporation of new large-scale tumour resequencing data. Nevertheless, the associations between specific tumour types, genes, pathways and processes are a very interesting source of information in the search for biological sources of tumour specificities. We have detected 25 relevant cellular pathways and processes (Fig 1) including the association between the promyelocyclic leukaemia (PML) pathways of transcription regulation and melanoma—that are not well covered in the associated literature.
The discovery of cancer-mutated genes is rapid, and this pathway/process approach could extend beyond the traditional analyses of frequently mutated genes, and enable the investigation of underlying causative relationships. We provide a resource that will facilitate this type of analysis with the progressive incorporation of data from high-throughput resequencing studies, and re-evaluation of the associations between cancer types, mutated genes and pathways/processes. The tumours associated with many cellular pathways and processes (leukaemia, lung, brain and colorectal tumours) have all been screened on a large or genome-wide scale, demonstrating that such experiments can capture new features. In the future, as more data become available for different tumour types or subtypes, the map of pathways, processes and protein domains containing significant numbers of mutated genes will become increasingly precise.
Methods
Mutated gene data. Data on mutations and mutated genes were retrieved from three databases, 17 whole-genome resequencing studies and resequencing screens focusing on gene subsets (Table 1). All genes with at least one non-synonymous mutation in their coding sequence or at a splice-site were taken into account, and matched to Ensembl IDs. The mutated genes were mapped to the tumour type in which their mutations had been described (when possible, following the International Classification of Diseases-10 topographic neoplasm classification, www.who.int/classifications/icd).
Functional annotations. Functional annotations were retrieved (directly or through the DAVID knowledgebase; Huang et al, 2009) for pathway-dedicated databases: Kegg (Kanehisa et al, 2008), Biocarta (http://www.biocarta.com/) and Reactome (Matthews et al, 2009); in Gene Ontology biological process (Ashburner et al, 2000); and in Interpro Protein Domains database (Hunter et al, 2009).
Statistical test. A Fisher's exact test was performed with the R multtest package to find pathways, processes or protein domains significantly enriched for mutated genes, in each tumour type. The test measures the significance of the association between the number of proteins annotated for a given cellular process, the number of mutated proteins in this process and the number of mutated proteins in a given tumour type. This statistical test implies the definition of a background as the total set of genes that have been screened for mutations, to avoid bias in the results (Chittenden et al, 2008). For each tumour type, the union of the gene sets that had been screened for mutations on a large-scale was taken into account as background (supplementary information online). All the tests were adjusted for multiple testing according to the Benjamini and Hochberg false discovery rate-controlling procedure (Benjamini & Hochberg, 1995).
Results presented in this study use the significance threshold Q-value <0.01 and, for the sake of clarity, Fig 1 only displays cellular pathways and processes. Other processes, such as Kegg cancer pathways or Gene Ontology general biological processes have been manually removed, but can be viewed as part of the complete results of tumour-associated pathways, processes and protein domains, together with Q-value <0.1 in supplementary Table S2 online and in the Online Visualisation Tool (for updated data).
Literature search for enriched pathways, processes and protein domains. PubMed abstracts associated with each tumour type were searched with the National Cancer Institute cancer topics keywords to collate the literature set. Each of these bibliomes was then manually mined for keywords representing pathways, processes and protein domains significantly enriched in mutated genes in the corresponding tumour types.
Online Visualisation Tool. The Online Visualisation Tool provides a graphical representation of the complete results of pathways, processes and protein domains significantly associated with tumour types. The tool is available at http://contexts.bioinfo.cnio.es/cancer-processes. Users can select, display and filter the results for specific tumour types and annotation databases with three different Q-value thresholds. Each tumour type, pathway, process or gene can be searched by keywords. Data on cancer-mutated genes will be updated with the release of new large-scale resequencing studies, and the pathway/process/domain associations will be re-evaluated.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank F.X. Real and D. Rico for valuable comments and suggestions. This study is supported by a grant from the Instituto de Salud Carlos III (ISCIII), COMBIOMED (rD07/0067/0014) and Bio2007-66855 from the Spanish Ministry of Ciencia e Innovación. A.B. is supported by a ‘Juan de la Cierva fellowship' from the Spanish Ministry of Ciencia e Innovación.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ashburner M et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan A, Bleeker FE, Lamba S, Rodolfo M, Daniotti M, Scarpa A, van Tilborg AA, Leenstra S, Zanon C, Bardelli A (2007) Novel somatic and germline mutations in cancer candidate genes in glioblastoma, melanoma, and pancreatic carcinoma. Cancer Res 67: 3545–3550 [DOI] [PubMed] [Google Scholar]
- Bardelli A et al. (2003) Mutational analysis of the tyrosine kinome in colorectal cancers. Science 300: 949. [DOI] [PubMed] [Google Scholar]
- Begley DJ (2004) ABC transporters and the blood–brain barrier. Curr Pharm Des 10: 1295–1312 [DOI] [PubMed] [Google Scholar]
- Benjamini J, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Method 57: 289–300 [Google Scholar]
- Cancer Genome Atlas Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung LW, Wong AS (2008) Gonadotropin-releasing hormone: GnRH receptor signaling in extrapituitary tissues. FEBS J 275: 5479–5495 [DOI] [PubMed] [Google Scholar]
- Chittenden TW, Howe EA, Culhane AC, Sultana R, Taylor JM, Holmes C, Quackenbush J (2008) Functional classification analysis of somatically mutated genes in human breast and colorectal cancers. Genomics 91: 508–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA (2009) Deciphering the genetic landscape of cancer—from genes to pathways. Trends Genet 25: 455–462 [DOI] [PubMed] [Google Scholar]
- Dalgliesh GL et al. (2010) Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463: 360–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H et al. (2005) Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res 65: 7591–7595 [DOI] [PubMed] [Google Scholar]
- Ding L et al. (2008) Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR (2008) The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr Protoc Hum Genet Chapter 10: Unit 10.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuja TJ, Lin F, Osann KE, Bryant PJ (2004) Somatic mutations and altered expression of the candidate tumor suppressors CSNK1 epsilon, DLG1, and EDD/hHYD in mammary ductal carcinoma. Cancer Res 64: 942–951 [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR (2004) A census of human cancer genes. Nat Rev Cancer 4: 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamas G, Castellano L, Feng Q, Knippschild U, Jacob J, Thomas RS, Coombes RC, Smith CL, Jiao LR, Stebbing J (2009) CK1delta modulates the transcriptional activity of ERalpha via AIB1 in an estrogen-dependent manner and regulates ERalpha-AIB1 interactions. Nucleic Acids Res 37: 3110–3123 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Greenman C et al. (2007) Patterns of somatic mutation in human cancer genomes. Nature 446: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA (2005) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 33: D514–D517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hunter S et al. (2009) InterPro: the integrative protein signature database. Nucleic Acids Res 37: D211–D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S et al. (2008) Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321: 1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M et al. (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ et al. (2008) DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456: 66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loriaux MM et al. (2008) High-throughput sequence analysis of the tyrosine kinome in acute myeloid leukemia. Blood 111: 4788–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L et al. (2009) Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res 37: D619–D6122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsudomi T, Yatabe Y (2010) Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J 277: 301–308 [DOI] [PubMed] [Google Scholar]
- Mujtaba S, Zeng L, Zhou M (2007) Structure and acetyl-lysine recognition of the bromodomain. Oncogene 26: 5521–5527 [DOI] [PubMed] [Google Scholar]
- Parsons DW et al. (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED et al. (2010a) A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED et al. (2010b) A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomoni P, Ferguson BJ, Wyllie AH, Rich T (2008) New insights into the role of PML in tumour suppression. Cell Res 18: 622–640 [DOI] [PubMed] [Google Scholar]
- Stephens P et al. (2005) A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet 37: 590–592 [DOI] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA (2009) The cancer genome. Nature 458: 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szepeshazi K, Schally AV, Halmos G (2007) LH-RH receptors in human colorectal cancers: unexpected molecular targets for experimental therapy. Int J Oncol 30: 1485–1492 [PubMed] [Google Scholar]
- Thomas RK et al. (2007) High-throughput oncogene mutation profiling in human cancer. Nat Genet 39: 347–351 [DOI] [PubMed] [Google Scholar]
- Tilg H, Moschen AR (2006) Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nat Rev Immunol 6: 772–783 [DOI] [PubMed] [Google Scholar]
- Tomasson MH et al. (2008) Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood 111: 4797–4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW (2004) Cancer genes and the pathways they control. Nat Med 10: 789–799 [DOI] [PubMed] [Google Scholar]
- Wang Z et al. (2004) Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science 304: 1164–1166 [DOI] [PubMed] [Google Scholar]
- Wood LD et al. (2007) The genomic landscapes of human breast and colorectal cancers. Science 318: 1108–1113 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.