The seventeenth EMBO Conference on the Molecular and Developmental Biology of Drosophila covered a broad range of topics. Much progress was made by combining two or more fields of study including quantitative approaches to cell and developmental biology, dissecting interrelations of physiology and development and integrated genomic analysis.
Abstract
The seventeenth EMBO Conference on the Molecular and Developmental Biology of Drosophila took place in Kolymbari, Crete, between 20 and 26 June 2010. The conference covered a broad range of topics and much progress was made by combining two or more fields of study. Such combinations included quantitative approaches to cell and developmental biology, dissecting interrelations of physiology and development and integrated genomic analysis.
Introduction
This year, the biennial EMBO conference on ‘The Molecular and Developmental Biology of Drosophila' convened in its traditional venue, the Orthodox Academy of Crete, Greece. This special meeting—which has been taking place for over 30 years—is part of what makes the Drosophila community communicative and vibrant. More than 110 participants—including many young scientists who attended the meeting for the first or second time—assembled for an intense week of presentations and informal discussions, carefully arranged by A. Ephrussi, W. Gelbart, C. Delidakis, D. Andrew, A. Bejsovec, S. Bray, C. Desplan, A. Giangrande, J. Knoblich, M. Leptin, H. Lipshitz, S. Parkhurst, P. Rørth, R. Saint and M. van Doren.
From genes to genomes
An important topic at this year's meeting was the regulation of gene expression. Presentations ranged from comprehensive dissections of a single enhancer to genome scale analyses. Scott Barolo (U. Michigan) reported on the structure, function and evolution of sparkling, a Notch- and EGFR/MAPK-regulated, cone-cell-specific enhancer of the Drosophila Pax2 gene. His results suggest that the enhancer's organization determines the correct expression pattern by enabling certain short-range regulatory interactions at the expense of others. Debbie Andrew (Johns Hopkins U.) showed that the CrebA family of bZip transcription factors function as direct regulators of at least 200 genes, encoding either components of the secretory pathway or secreted cargo; a function that is conserved in human cells.
Key questions in genomics are how to build comprehensive models of gene regulatory networks based on sequence and transcription factor binding data, and how to predict temporal and spatial gene expression on a global scale, based on cis-regulatory motifs. Francois Schweisguth (Institut Pasteur) has developed computational methods to identify the cis-regulatory modules (CRMs) that control a set of co-regulated genes. Starting with a training set of known Drosophila CRMs and orthologous sequences from 11 other Drosophila genomes, he reported computational prediction of transcription factor binding sites and novel genomic CRMs underlying co-regulation of gene expression programmes, in sensory organ precursor cells. Eileen Furlong (EMBL) reported a high-resolution map of the occupancy patterns of key transcription factors during consecutive stages of mesoderm development. By using a machine learning approach, her group demonstrated that transcription factor binding alone is sufficient to predict CRM spatio-temporal activity, without prior knowledge of the expression or sequence preferences of the transcription factors themselves. Her results demonstrate that similar spatio-temporal expression patterns can be caused by a range of CRMs, suggesting a lack of consistent motif grammar within global gene regulatory networks. Manolis Kellis (MIT) found that combining regulatory motif sequences and predicted regulator activity with chromatin marks is predictive of downstream gene expression. He also described the integrative analysis efforts for the NIH modENCODE project.
Chromatin function in germ cells
The role of chromatin modifiers in promoting germ cell identity and function was also discussed at the meeting. The main function of a germ cell is to pass its intact genetic material to the next generation. This might be daunting considering transposons make up approximately 20% of the Drosophila genome and mobilization of these elements leads to genetic instability. Piwi-interacting RNAs (piRNAs) encoded by heterochromatic clusters silence transposons in germ cells. William Theurkauf (U. Massachusetts) reported that the HP1 homologue Rhino binds to these clusters and promotes piRNA precursor production. The rhi gene is rapidly evolving and under strong positive selection, suggesting that it might be co-evolving with the transposon targets of piRNA silencing.
Chromatin regulation also features in germ-line stem cell biology. Ruth Lehmann (New York U.) reported that transcriptional silencing of a select number of loci by chromatin-modifying enzymes is needed not only for germ-line stem cell maintenance, as has been previously demonstrated, but also for their differentiation. The Lehmann lab also analysed primordial germ cell (PGC) formation. Using 4D multi-photon imaging analysis, they showed that PGCs form by a highly specialized mode of cytokinesis, in which coordinated restriction of two contractile furrows leads to the formation of two PGCs.
Mark van Doren (Johns Hopkins U.) reported on the role of chromatin regulators in promoting germ cell sexual identity—a poorly understood phenomenon. The group studied Phf7, a protein that contains a PHD-finger motif common in chromatin regulators. Phf7 is expressed preferentially in male germ cells. Loss of phf7 function appears to feminize XY germ cells, whereas its overexpression masculinizes XX germ cells. Thus, male sexual identity of germ cells depends on chromatin regulation.
Robustness and stochasticity
In order to support normal development, gene expression must be carefully controlled. One phenomenon that has puzzled the field is the existence of apparently redundant modules of control. Work presented by several groups begins to unravel this mystery at the transcriptional and post-transcriptional level.
Small RNAs—such as microRNAs and endo-siRNAs—regulate gene expression, yet development is often unaffected by their absence. Richard Carthew (Northwestern U.) presented two instances of small RNAs helping embryos compensate for variations in growth temperature by normalizing expression of segmentation or neurogenic genes. David Stern (Princeton U.) deleted two of the apparently redundant enhancers from the shavenbaby (svb) gene, which is required for proper specification of trichomes. Flies carrying this deletion produced fewer trichomes than wild-type flies, but only when reared at extreme temperatures or when combined with a wingless mutation. Taken together, it seems that what is referred to as ‘redundancy' can be dependent on environmental conditions; when animals are reared under optimal conditions, certain enhancers, small RNAs, or other regulators might seem superfluous. Exposed to environmental perturbations or genetic variation, they can confer robustness.
Claude Desplan (New York U.) described a mechanism whereby robustness and stochasticity coexist. Colour vision in Drosophila depends on two subtypes of photoreceptors that are arranged randomly throughout the eye and express different rhodopsin genes. Despite this stochasticity, the overall number of photoreceptors expressing a given rhodopsin is constant. This is controlled by the PAS-bHLH transcription factor Spineless. Competition between two promoters for an enhancer element controls the stochastic expression of each spineless allele and thereby the arrangement of photoreceptor subtypes, similarly to the human colour vision system.
Cell biology and development
Cell biology and developmental biology can relate to each other in several ways, each field promoting the other. Sometimes, investigating a developmental phenotype can lead to novel discoveries in cell biology. For example, Kenji Matsuno (U. of Tokyo) described a novel cell behaviour in the hindgut. Epithelial cells in this tubular organ adopt a planar cell shape that is chiral. This leads to an asymmetrical left–right directional rotation of the gut epithelial tube. The group found that MyosinID and DE-Cadherin are required for this novel cell behaviour, which they term ‘planar cell-shape chirality'.
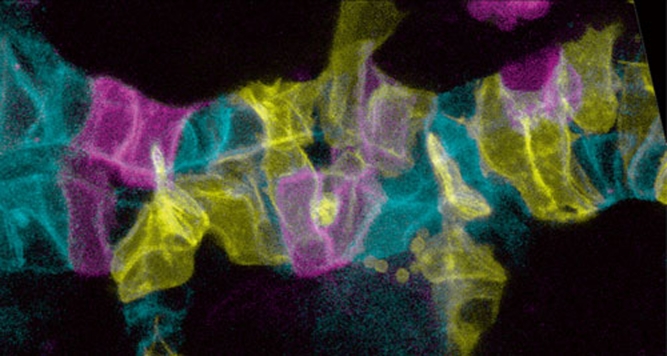
Several presentations demonstrated how investigating questions of cell biology can promote understanding of developmental processes. This strategy is particularly useful when applied with novel computational or imaging approaches. Stefan Luschnig (U. Zurich) presented analyses of tracheal tube expansion, during which the tracheae dilate their narrow lumen. The group showed that a cell-autonomous secretion-dependent programme—rather than extrinsic (luminal) cues—drives tube expansion, suggesting a critical role for membrane growth in this process. To track and analyse the behaviour of individual epithelial cells during morphogenesis, the Luschnig group adapted the ‘brainbow' system—originally developed to study neuronal connectivity in the mouse—for applications in Drosophila (Fig 1).
Figure 1.
A group of tracheal epithelial cells. Using the ‘brainbow' method, each cell can be labelled differently. Image courtesy of Dominique Förster and Stefan Luschnig.
Anne Ephrussi (EMBL) discussed how oskar messenger ribonucleoproteins (mRNPs) use microtubules and their associated motor proteins—kinesin heavy chain (KHC) and cytoplasmic dynein—to reach the posterior pole of the developing oocyte. They developed an ex vivo assay that allows accurate spatio-temporal tracking of individual oskar mRNPs. Preliminary results suggest a cryptic tug-of-war between the two opposite polarity motors, and that the dynein intermediate chain—an obligatory dynein subunit—is involved in maintaining KHC processivity, illustrating the function of dynein in oskar mRNP transport.
Christian Dahmann (MPI Cell Biology and Genetics) and colleagues have used physical approaches and quantitative imaging to show that actomyosin-dependent mechanical tension on cellular junctions is increased along the anteroposterior compartment boundary in the developing Drosophila wing. Simulations show that this increase in tension maintains a stable interface between two proliferating cell populations, suggesting that the increase in local mechanical tension directs cell sorting at compartment boundaries.
Mechanical tension also drives polarized cell movements during germ band extension in the Drosophila embryo. Proteins involved in actomyosin contractility and cell adhesion are asymmetrically localized in intercalating cells, which form ‘rosettes'. Recent work from Jennifer Zallen (Memorial Sloan–Kettering Cancer Center) shows that rosette formation involves a mechanical feedback loop in which tension recruits myosin to the cortex, triggering a wave of myosin localization that coordinates cell behaviour in multicellular populations.
Benny Shilo (Weizmann Institute of Science) described an extension of studies on muscle cell fusion in the embryo, to adult muscle formation in the pupa. Myoblasts from the wing imaginal disc are attracted to the flight muscles and remain semi-differentiated until they encounter the myotube, with which they will fuse. Many of the genes implicated in embryonic muscle fusion are also required in the adult, indicating the use of a common machinery.
Signalling in development
Several talks demonstrated that the signalling field can still hold surprises. Francois Payre (U. Toulouse/CNRS) presented work done in collaboration with Yuji Kageyama (Okazaki Institute). They showed that small peptides (11–32amino acids) encoded by the polycistronic polished rice/tarsal-less sORF RNA control the differentiation of embryonic epidermal cells, by triggering the amino-terminal truncation of the shavenbaby protein. Truncated shavenbaby switches its transcriptional activity from a repressor to an activator. Additional developmental processes are likely to be regulated by sORF-encoded small peptides.
Utpal Banerjee (U. California Los Angeles) discussed the homeostatic maintenance of blood stem/progenitor cells by signals originating from two different sources. Stem cells are maintained by a niche-derived Hedgehog signal. In addition, the differentiating daughter cells of blood stem cells regulate their maintenance through a backward signal that activates adenosine-derived growth factor A. Adenosine-derived growth factor A functions as an adenosine deaminase and inhibits proliferation and differentiation of the progenitors by maintaining low levels of adenosine.
Physiology and development
With many regulators conserved between mammals and flies, and with the growing sense that physiology is linked to and affects many clinical conditions, Drosophila physiology received much attention this year. In particular, the roles of the insulin and the steroid hormone Ecdysone pathways, and the connections between them, were a focus of interest.
Stephen Cohen (Institute of Molecular and Cell Biology, Singapore) described a role for the micro RNA miR14 in the control of insulin production in neurons. The Cohen lab is in the process of knocking out all miRNAs in Drosophila. So far, about 80% of the miRNA genes have been deleted, effecting many phenotypes that are associated with the central nervous system.
Nutrition and development programmes are interconnected by complicated feedback loops that might change in a temporal and cell-specific manner. Andrea Brand (U. Cambridge) presented data showing that the Insulin/IGF receptor pathway is necessary for neuroblasts to exit quiescence and embark on a post-embryonic phase of proliferation to generate the adult nervous system. She showed that glial cells neighbouring the neuroblasts produce Insulin/IGF-like peptides in response to nutrition, and that this expression is sufficient to reactivate neuroblasts. Aurelio Teleman (German Cancer Research Center) presented the identification and characterization of the Drosophila homologue of the Diabetes and Obesity Regulated gene (dDOR), which acts as a co-activator for the ecdysone receptor. dDOR regulates pupation as well as lipid metabolism, and might connect the Insulin and ecdysone pathways in fat body cells. Pierre Leopold (U. Nice) also discussed the connections between Insulin and ecdysone signalling. His lab showed that rising levels of ecdysone at the end of juvenile development act specifically on fat body cells to inhibit dMyc. This local inhibition leads by an unknown relay mechanism to the systemic downregulation of Insulin signalling and to growth arrest, determining animal size at the end of the juvenile period.
Research into physiology can be improved by novel biomarkers. A case in point was Irene Miguel-Aliaga's (U. Cambridge) presentation. She has analysed fly faeces as an experimental measure of food intake, intestinal transit and water/acid–base balance. She reported that these processes are modulated by the reproductive state of the fly and mediated by a novel subset of hindgut-innervating neurons. Thus, enteric neurons function to couple nutritional and reproductive states with intestinal homeostasis.
Community resources and novel tools
A particular strength of Drosophila as a model organism has always been the wealth of resources that are available to the whole community. Much of the ground-breaking work in the field would not have been possible without continued focus on the generation of novel tools, and the vision of the people who design and share them. William Gelbart (Harvard U.) described the current status of FlyBase and his future plans (Tweedie et al, 2009). One of the challenges faced by researchers in the field is to keep pace with the large amounts of genomic sequence data emerging from both modENCODE and the wider community. Making FlyBase more accessible to the broader research community—especially those working on the human genome and arthropod disease—is another priority. The importance of acknowledging community resources in publications in order to emphasize their value to funding agencies, was also discussed. Hugo Bellen (Baylor College of Medicine) reported on a modified Minos element (MIMIC) with sequences for recombination-mediated cassette exchange (RMCE) that will facilitate the integration of any DNA fragment for in vivo protein tagging, gene disruption and more. Norbert Perrimon (Harvard Medical School) reported that his group designed an optimized vector that delivers hairpin RNAs to the germline using the endogeneous miRNA pathway. This will facilitate the study of oogenesis and early embryogenesis, which have so far not been amenable to silencing by RNAi. Spyros Artavanis-Tsakonas (Harvard Medical School) presented the progress on the DPiM project, which is a collaboration of the laboratories of Steven Gygi, Susan Celniker and Vijay Raghavan. They are generating a Drosophila protein interaction map, based on the affinity purification of tagged proteins and mass spectrometric analysis of protein complexes. Within the next year, the project will complete the examination of 8,000 proteins. Pavel Tomancak (MPI Cell Biology and Genetics) described the FlyFos system that generates in vivo reporters of gene expression by combining a library of genomic fosmid clones and an efficient liquid culture recombineering pipeline. His lab intends to generate a genome-wide collection of tagged fosmids to study the dynamics of tissue-specific gene expression and sub-cellular protein localization.
Summary
This year marked the one-hundredth anniversary of the first Drosophila research paper by Thomas Hunt Morgan, which discussed the sex-linkage of the white mutation—the first mutation that was isolated in the fly (Morgan, 1910). This discovery was instrumental in establishing Drosophila as an important model organism for developmental genetics. The EMBO conference assured us that with its unparalleled genetic tools and novel high-throughput methodologies, fly research is bound to produce many more exiting discoveries for a long time to come.
References
- Morgan TH (1910) Science 32: 120–122 [DOI] [PubMed] [Google Scholar]
- Tweedie S et al. (2009) Nucleic Acids Res 37: D555–D55918948289 [Google Scholar]