Abstract
In this paper, we describe the development of an electrochemical DNA aptamer-based biosensor for detection of IFN-γ. A DNA hairpin containing IFN-γ-binding aptamer was thiolated, conjugated with Methylene Blue (MB) redox tag and immobilized on a gold electrode by self-assembly. Binding of IFN-γ caused the aptamer hairpin to unfold, pushing MB redox molecules away from the electrode and decreasing electron-transfer efficiency. The change in redox current was quantified using Square Wave Voltammetry (SWV) and was found to be highly sensitive to IFN-γ concentration. The limit of detection for optimized biosensor was 0.06 nM with linear response extending to 10 nM. This aptasensor was specific to IFN-γ in the presence of overabundant serum proteins. Importantly, the same aptasensor could be regenerated by disrupting aptamer-IFN-γ complex in urea buffer and re-used multiple times. Unlike standard sandwich immunoassays, the aptasensor described here allowed to detect IFN-γ binding directly without the need for multiple washing steps and reagents. An electrochemical biosensor for simple and sensitive detection of IFN-γ demonstrated in this paper will have future applications in immunology, cancer research and infectious disease monitoring.
INTRODUCTION
Interferon (IFN)-γ is an important cytokine that possesses antiviral, antiproliferative, differentiation inducing, and immunoregulatory properties.1 This cytokine is produced by a number of immune cell types including T-helper (CD4) cells and cytotoxic T-cells. The detection and quantification of IFN-γ is important for understanding what immune cells are taking part in the immune response and how vigorous this response is.2, 3 Conventional antibody (Ab)-based immunoassays involve multiple washing steps and employ reagents that are both temperature sensitive and expensive.4, 5 While a number of strategies for miniaturization and multiplexing of Ab-based cytokine immunoassays have been proposed,6–10 these approaches are still hindered by the high cost and low stability of Abs as well as by the relative complexity of transducing antibody-antigen binding events.
Aptamers are nucleic acid-based (DNA or RNA) affinity probes that provide a number of advantages over Abs including temperature stability, low cost and re-usability.11 Another significant advantage of aptamers stems from the relative simplicity with which nucleic acid sequences can be engineered to integrate affinity and signal transducing moieties into the same molecule. A number of aptasensors capable of both capturing the analyte and transducing the signal have been reported in the literature.12–14 Recently, our lab has demonstrated an aptamer beacon that emits fluorescence signal directly upon binding of IFN-γ molecules without the need for secondary labels or washing steps.15 However, fluorescence-based aptasensors suffer from problems related to stability and photobleaching of fluorophores.
Building on our previous work with an IFN-γ aptamer beacon,15 we wanted to develop a redox-labeled aptamers for electrochemical sensing of IFN-γ. Electrochemical aptasensors may offer greater signal stability, sensitivity and ease of calibration compared to fluorescence aptasensors.16, 17 Different electrochemically active labels (e.g. redox compounds, enzymes, or metal nanoparticles) have been employed to relay electrical signal resulting from aptamer-analyte binding.18–20 Of particular note is the work of Plaxco and colleagues who modified electrodes with redox tag-containing aptamers and demonstrated electrochemical detection of a range of analytes.21–23
The sensing strategy pursued in this study involved self-assembly of thiolated aptamer hairpin molecules on gold electrode surfaces (see Figure 1). The binding of IFN-γ molecules likely caused the hairpin to unfold, decreasing the efficiency of electron transfer from the redox label to the electrode. The change in redox current was quantified using square wave voltammetry (SWV) and was found to depend on the concentration of IFN-γ. Overall, this electrochemical aptasensor was found to be sensitive and specific to IFN-γ, pointing to future applications in immunology, cancer research and infectious disease monitoring.
Figure 1.
Schematic of aptamer-based electrochemical sensor for IFN-γ. (A) The aptamer hairpin was thiolated at the 5’ end allowing self-assembly on gold electrodes. Redox label was attached at the 3’ end of the hairpin and was in close proximity to the electrode surface. (B) Upon addition of IFN-γ aptamer hairpin changed confirmation and the redox label moved further away from the electrode, lowering the electron transfer efficiency. (C) The differences in faradaic current before and after addition of IFN-γ were quantified using square wave voltammetry (SWV).
MATERIALS AND METHODS
Materials
The following reagents were used as received: 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), sodium bicarbonate (NaHCO3) (all reagent grade),6-mercapto-1-hexanol (MCH), (tris-(2-carboxyethyl) phosphine Hydrochloride (TCEP), Urea (98%) (all from Sigma-Aldrich, St. Louis, MO), IgG, Anti-Human IgG (from BD Pharmingen) and Methylene Blue (MB), Carboxylic Acid, Succinimidyl Ester (MB-NHS, Biosearch Technologies, INC, Novato, CA), Cell culture medium RPMI 1640: 1×, with L-Glutamine was purchased from VWR. Medium was supplemented with fetal bovine serum (FBS) and penicillin/streptomycin purchased from Invitrogen. Recombinant human IFN-γ was purchased from R&D systems (Minneapolis, MN). The 34-mer IFN-γ-binding aptamer sequence (IDT Technologies, San Diego, CA) was as follows: 5’-NH2-C6-GGGGTTGGTTGTGTTGGGTGTTGTGTCCAACCCC-C3-SH-3’. The aptamer was modified at the 5′-terminus with a C6-disulfide [HO(CH2)6-S-S-(CH2)6-] linker and at the 3′-end with an amine group for redox probe (MB) conjugation. The aptamer was dissolved in 10 mM HEPES buffer (pH 7.4 with 150 mM NaCl). This buffer was also employed in all IFN-γ sensor experiments.
Attachment of redox labels to aptamers
The MB-tagged aptamer was prepared using a procedure similar to that described by Plaxco and coworkers.23 Briefly, NHS-labeled MB was conjugated to the 3’-end of an amino-modified DNA aptamer through succinimide ester coupling. MB-NHS (1 µmol) was dissolved in 10 µl DMF/50 µl 0.5 M NaHCO3, then added to 10 µL of 200 µM aptamer solution, stirred, and allowed to react for 4 h at room temperature in the dark. After reaction, the sample was filtered using a centrifugal filter (Millipore, Amicon Ultra 3K 0.5mL) in order to purify and concentrate MB-modified aptamers. The stock aptamer solution (20µM) was stored at −20 °C for future use. Conjugation efficiency was estimated by MALDI-MS analysis to be ~30%. This efficiency is comparable to values reported previously.24
Assembly of aptamer molecules on gold electrodes
The IFN-γ sensor was fabricated using gold working electrode (BAS, Ø=1.6 mm). The electrodes were cleaned in “piranha” solution consisting of a 3:1 ratio of 30% w/v aqueous solutions of H2SO4 and H2O2 (caution: this mixture reacts violently with organic materials and must be handled with extreme care), then thoroughly washed with DI H2O and ethanol, and dried under nitrogen. Prior to modification of the electrodes aptamer stock solution (0.02 mM) was reduced in 10 mM TCEP for 1 h to cleave disulfide bonds. This solution was then diluted in HEPES buffer to achieve the desired aptamer concentration (from 0.5 µM to 8 µM). For aptamer immobilization, the gold electrodes were kept in solution of thiolated aptamer for 16 h in the dark at 4°C. Following incubation, the electrodes were rinsed with copious amounts of DI water and then immersed in an aqueous solution of 3 mM 6-mercapto-1-hexanol solution (MCH) for 1 h to displace nonspecifically adsorbed aptamer molecules and to passivate the electrode surface 25, 26. As a final step, the electrodes were rinsed with DI water, dried with nitrogen and stored at 4°C prior to use.
SPR analysis of aptamer assembly and IFN-γ binding
SPR experiments were performed using a four-channel BIAcore T3000 instrument (Uppsala, Sweden). All experiments were performed on bare gold chips obtained from BIAcore. Aptamer was heated to 37°C to form a hairpin, cooled down to room temperature and then dissolved in HEPES buffer (10mM HEPES, 150mM NaCl) to 0.1 µM concentration. This aptamer solution was injected into an SPR device at a flow rate of 5 µL/min and was allowed to interact with the chip for 20 min. Assembly of aptamer was followed by flowing 3 mM MCH in HEPES buffer at a flow rate of 20 µL/min for 30 sec. This step was needed to block gold surface after aptamer assembly in order to prevent non-specific adsorption. IFN-γ was dissolved in HEPES buffer and then introduced into an SPR instrument at flow rate of 20 µl/min, remaining in contact with SPR chip until saturation of the signal (typically 180 s). After injection of IFN-γ, the chip was washed with HEPES buffer for ~ 270 s. To dissociate IFN-γ and regenerate aptamer layer, the SPR chip was washed in 7M urea buffer. The process of surface regeneration did not compromise sensitivity of the aptamer layer and allowed us to challenge the same aptamer layer with multiple concentrations of IFN-γ.
SPR sensograms were analyzed using kinetic Langmuir 1:1 fitting model on BIAvaluation 4.0 software to determine association, dissociation and equilibrium (KD) constants of aptamer-IFN-γ binding. To eliminate the possibility of mass transport limitations obscuring kinetic measurements, SPR sensorgram for the same IFN-γ concentration was tested at different flow rates.
Electrochemical characterization of aptasensor response to IFN-γ
Electrochemical measurements were made using a CHI 842B Electrochemical Workstation (CHInstruments, Austin, TX) with a three electrode system consisting of Ag/AgCl (3 M KCl) reference electrode, Pt wire counter electrode and a gold working electrode. Electrochemical experiments were performed in 10 mM HEPES buffer using square wave voltammetry (SWV) with a 40 mV amplitude signal at a frequency of 60 Hz, over the range from −0.10 to −0.50 V versus Ag/AgCl references.
Electrodes were modified with aptamer molecules as described above and were placed into a custom-made Plexiglass electrochemical cell. The sensor was allowed to equilibrate, as determined by stable faradaic current, and was then challenged with human recombinant IFN-γ dissolved in HEPES buffer. Before conducting voltammetric measurements, the sensor was allowed to react with analyte for 15 min. After cytokine detection experiments, aptasensors could be regenerated by immersion in 7M urea buffer for 1 min followed by copious rinsing with DI water. Effectiveness of regeneration protocol was verified by SWV.
For measurements in RPMI and RPMI/serum solutions, electrochemical experiments were performed using square wave voltammetry (SWV) under the same condition as described above.
Effects of aptamer surface density on IFN-γ detection
Studies were carried out to determine how packing density of aptamer molecules affected sensitivity and limit of detection of the biosensor. In these SWV experiments, potential was scanned from −0.1V to −0.5V with a step potential of 4 mV, frequency of 60 Hz, and amplitude of 40 mV. Gold electrodes were immersed in 0.5, 1.0 and 2.0 µM aptamer solution in 10 mM HEPES buffer (pH 7.4). Aptamer-coated electrodes were then incubated in 10 mM HEPES buffer (pH 7.4) for 30 min to equilibrate. The surface density of MB-labeled aptamer molecules was calculated by integrating the area under the SVW curve and converting charge to the number of surface-bound DNA molecules. Gold electrodes modified with aptamer layers of different densities were challenged with a series of concentrations of IFN-γ (from 0.06 nM to 50 nM) and analyzed by SWV. The results were presented as the difference in faradaic current before and after target binding divided by the initial faradaic current. Because binding of IFN-γ causes a decrease in current, the sensitivity of the biosensor was reported in terms of signal loss or suppression, with the greatest signal loss per given IFN-γ concentration being the most sensitive sensor.
RESULTS AND DISCUSSION
SPR analysis of aptamer assembly and IFN-γ binding
Aptamer molecules were thiolated to ensure self-assembly on gold electrodes. The assembly of aptamer molecules and subsequent binding of IFN-γ were investigated using SPR. Both unmodified and MB-tagged aptamers were tested to verify that attachment of a redox tag did not alter cytokine binding to the aptamer. SPR sensogram (Figure 2A) shows rapid binding of aptamer (0.1 µM solution in HEPES buffer) on gold surface. Subsequent injection of MCH (3 mM solution in HEPES buffer) resulted in additional SPR signal increase, suggesting deposition of alkanethiol molecules in between DNA hairpin molecules. Passivation of aptamer-containing surfaces with mercapto hexanol (MCH) has been reported to decrease non-specific binding to aptasensors,25, 26 presumably by filling in the gold regions left exposed after aptamer assembly.
Figure 2.
SPR analysis of aptamer/IFN-γ interactions. (A) SPR sensorgram showing assembly of 0.1 µM aptamer solution, followed by blocking with 3 mM MCH. Both solutions were prepared in HEPES buffer. (B) SPR responses of an aptamer layer challenged with different concentrations of IFN-γ to determine equilibrium constant KD.
Upon assembly, the sensing layer was challenged with varying concentrations (1 to 16 nM) of human recombinant IFN-γ in order to determine the equilibrium binding constant KD. Figure 2B shows representative SPR signals of different IFN-γ concentrations binding to an aptamer layer. Incorporating results of several SPR experiments (n=3) into a kinetic (Langmuir 1:1) fitting model (BIAcore software 4.0) allowed to determine aptamer affinity for IFN-γ (KD). The affinity constants of unmodified aptamer and MB-tagged aptamer for IFN-γ were found to be KD= 3.11±0.84×10−10 and KD=3.40±0.47×10−10 respectively. These values demonstrate that MB attachment did not interfere with cytokine binding to the aptamer. The affinity constants described here are significantly lower than KD of 3 nM reported by us for linear IFN-γ aptamer.15
Electrochemical detection of IFN-γ using aptamer-modified electrodes
Given its excellent affinity for IFN-γ, we sought to convert aptamer-carrying hairpin into an electrochemical biosensor. This was done by conjugating a redox tag (MB) at the 5’ end of DNA sequence. The choice of a redox tag was based on previous reports in the literature describing MB-conjugated nucleic acid molecules as offering better shelf-life and performance in serum compared to other redox molecules (e.g. ferrocene).24, 27 As shown in Figure 1A, in the absence of the analyte, hairpin molecules were closed and MB redox tags were positioned next to the electrode surface. Binding of IFN-γ molecule likely caused hairpins to open, moving MB redox tags away from the electrode surface and decreasing electron transfer/current at the electrode (see Figure 1B). SWV was used to monitor changes in the intensity of the reduction current of MB as shown in Figure 1C. The peak current intensity dropped as a function of increasing IFN-γ concentration.
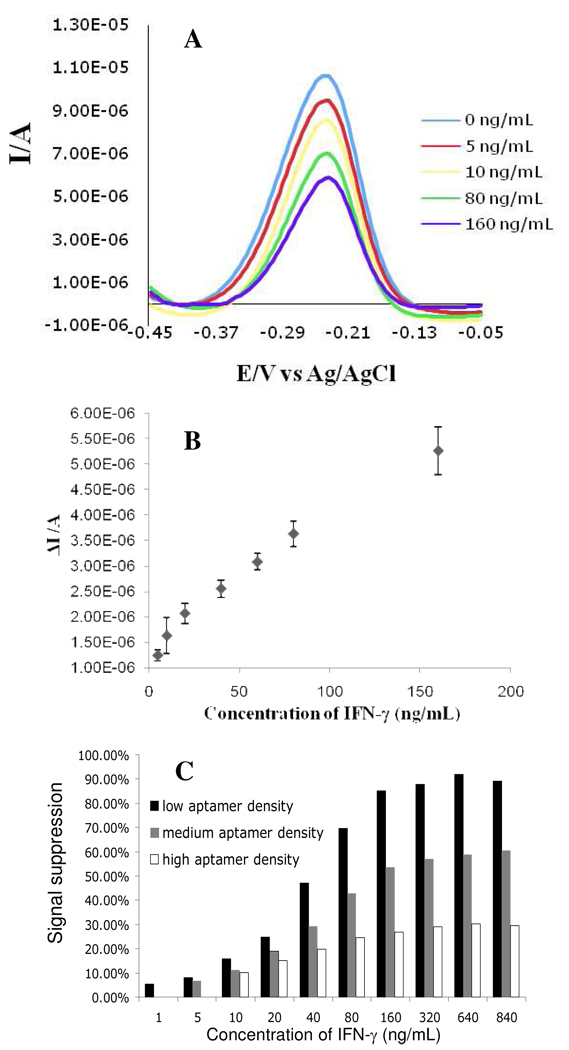
The choice of frequency for SWV analysis has been noted as important parameter in defining sensitivity of the aptasensor.28 We characterized behavior of aptamer-containing electrodes at frequencies ranging from 3 to 1000 Hz in the presence of 10 nM IFN-γ (saturated concentration) and determined that the signal change to be greatest for frequencies ranging from 60 to 100 Hz (see Figure 1 in Supporting Information). Therefore, all subsequent SWV experiments were performed at 60 Hz sampling frequency. Figure 3A shows a typical SWV voltammogram with MB reduction peak appearing at −0.25 V (vs. Ag/AgCl). Upon increasing the concentration of IFN-γ in the vicinity of the biosensor, the peak current decreased – suggesting unfolding of the hairpin molecules. As shown in Figure 3A, the loss in signal due to binding of IFN-γ molecules was proportional to the cytokine concentration. This loss in signal could be replotted as the change in reduction current (ΔI) before and after addition of IFN-γ. Figure 3B shows a calibration curve of signal change vs. cytokine concentration and demonstrates that our aptasensor had a limit of detection of 0.06 nM with linear range extending to 10 nM. The detection limit of our aptasensor is in line with expected IFN-γ concentration in serum29 or in the proximity of activated immune cells.30
Figure 3.
Characterization of sensor response to IFN-γ. (A) Voltammograms obtained with SWV showed that decrease in faradaic current was proportional to solution concentration of IFN-γ. These results were obtained with gold electrodes modified with 2 µM concentration of aptamer (medium aptamer density). (B) Calibration curve of current vs. IFN-γ concentration for aptasensor prepared using 2 µM aptamer concentration. The calibration curve was fitted to a line with the equation of y = 2×10−8 X + 1E-06 and R2 of 0.9801. The data points and error bars represent average and standard deviations of measurements from three different aptamer-modified electrodes (n=3). (C) Sensitivity of electrodes with different aptamer packing densities was expressed as the loss or suppression of signal (current) upon binding of 50 nM IFN-γ. The larger the signal suppression (or current loss) the higher sensitivity of the biosensor. Electrodes with low, medium and high packing density were prepared using 0.5, 2 and 8 µM aptamer concentrations. Aptasensors with low packing density were found to be most sensitivity with detection limit of 0.06 nM or 1 ng/ml IFN-γ.
Effects of aptamer surface density on biosensor performance
An important factor affecting sensitivity and performance of an aptasensor is the packing density of aptamer molecules on the surface.31, 32 Given that the binding of IFN-γ likely induces conformational changes in the hairpin structure of the aptamer, the surface density of probe molecules was hypothesized to be related to sensitivity of the aptasensor. The surface density of MB-aptamer surface density was calculated based on the intensity of the reduction peaks collected during SWV experiments. Such an approach is frequently used to determine grafting density of DNA strands.33, 34 In our calculations, we assumed that one MB molecule was conjugated to one aptamer - a reasonable assumption for MB-NHS to NH2-aptamer conjugation chemistry. Another point to note is that MB labeling efficiency was ~30%; so that the surface density values reported below refer only to MB-modified aptamer molecules.
For determining aptamer surface density, the gold electrodes were incubated with MB-aptamer solution ranging in concentration from 0.5, 1.0, and 2.0 µM and were subsequently passivated with MCH. The amount of total charge passed during the redox reaction was calculated by integrating the current with respect to time. The relationship between the total charge passed (Q, in coulombs) and the surface density of the MB moiety (Γ) is described as Q = nFAΓ, where n is the number of electrons transferred per MB moiety (= 2), A is the electrode surface area, and F is the Faraday constant. The MB-aptamer surface densities for were estimated to be (6.57±0.55) ×1013 molecules/cm2, (4.17±0.59) ×1012 molecules/cm2 and (8.53±0.56) ×1012 molecules/cm2 for 0.5, 1.0 and 2.0 µM aptamer solution concentrations. Our surface density is in general agreement with other reports employing MB-tagged hairpin aptamers31, 32. Increasing aptamer solution concentration above 2.0 µM did not result in higher surface density of the probe – suggesting steric hindrance or electrostatic repulsion of negatively charged DNA molecules at higher concentrations. Because binding of analyte results in unfolding of aptamer probes and decrease in redox current, the sensor response was reported in terms of signal loss or suppression. Signal suppression was calculated by determining the difference in faradaic current before and after addition of IFN-γ. For a given cytokine concentration, biosensors with larger signal suppression were deemed to be more sensitive. When challenged with 20 nM of IFN-γ, electrodes with lowest aptamer density (4.17±0.59×1012 molecules/cm2) produced signal suppression of ~85% whereas electrodes with highest aptamer probe density (6.57±0.55×1013 molecules/cm2) had signal suppression of ~25%. Therefore, electrodes with lower aptamer packing density were ~3.4 times more sensitive than electrodes with higher density aptamer layer. For the biosensors with highest aptamer density, the detection limit was 0.6 nM compared to detection limits of 0.3 nM and 0.06 nM for medium and lower aptamer surface density respectively (see Figure 3C). We hypothesize that sparse packing provides more space for unfolding of aptamer hairpin structures. Conversely, at higher packing, some of the aptamers may not be able to unfold due to steric hindrance of neighboring molecules. Fewer probes changing conformation upon cytokine binding would mean lower redox currents and a less sensitive biosensor.
Specificity and re-usability of IFN-γ aptasensor
To demonstrate specificity, aptasensors were challenged with non-specific proteins such as IgG, anti-IgG or BSA as well as with the analyte of interest - IFN-γ. Figure 4A compares SWV responses of our biosensor to IgG (100 nM), anti-IgG (100 nM), BSA (100 nM) and IFN-γ (3.6 nM) and shows that the aptasensor did not respond to a high concentration of non-specific proteins but did respond to a low concentration of IFN-γ. Given the importance of IFN-γ as a clinical marker of infectious diseases such as malaria, we also wanted to test response of our aptasensor a complex serum-containing environment. As shown in Figure 4B, addition of 10% serum into RPMI media resulted in signal loss of ~70% compared to media alone. However, despite this loss in signal, spiking IFN-γ into serum-containing media resulted in measurable changes in redox current as demonstrated in Figure 4B. The ability to selectively detect IFN-γ molecules in the presence of overabundant non-specific proteins bodes well for future use of this aptasensor in clinical diagnostics.
Figure 4.
Specificity of aptasensor response to IFN-γ. (A) SWV measurements of aptasensor response to 100 nM concentration of non-specific proteins (IgG, anti-IgG, BSA) as well as to 3.6 nM (60 ng/ml) of IFN-γ. These results show that biosensor did not respond to non-specific protein but did respond to IFN-γ. This electrode had medium packing density of aptamer molecules. (B) Aptasensor response in HEPES buffer, RPMI media and RPMI supplemented with 10% serum. Varying concentrations of IFN-γ were spiked into serum-containing media as highlighted by the dashed box. (C) Close-up view of the aptasensor response to IFN-γ spiked into serum-containing media. These data show that while changes in current were dampened in the presence of serum, sensor response to varying IFN-γ concentration was still discernable. This electrode had the lowers packing density of aptamer molecules.
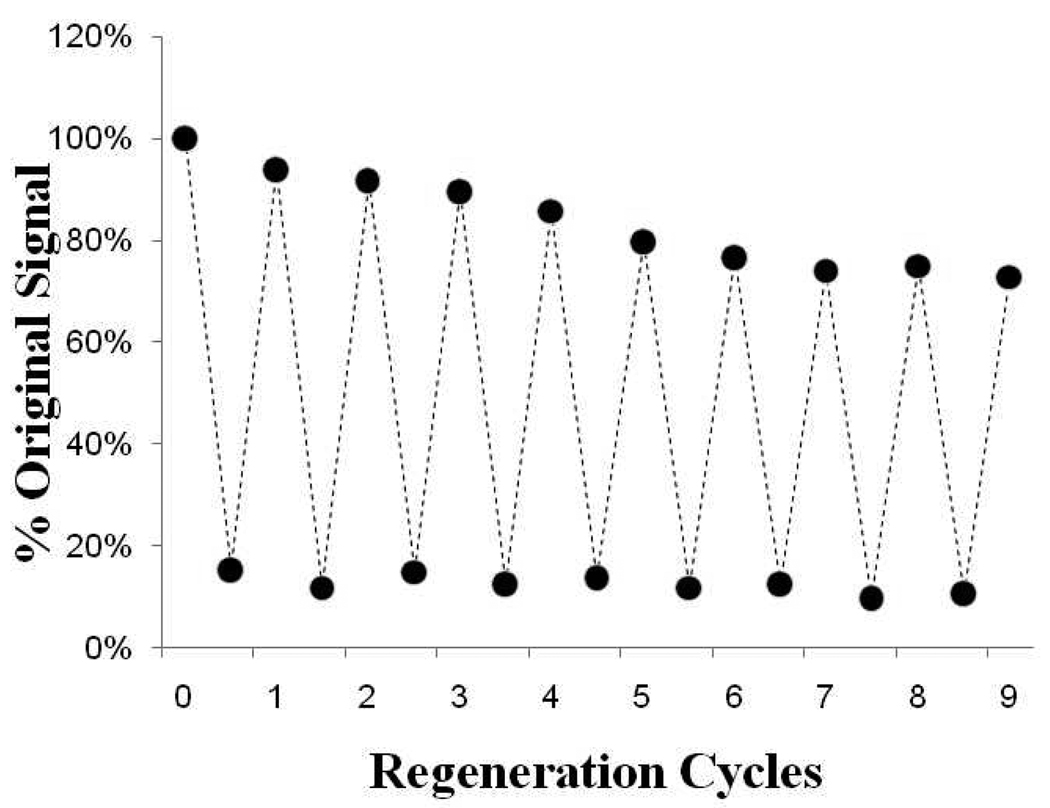
Compared to antibody-based affinity biosensors, aptasensors provide an advantage of chemical stability. The chemical stability of nucleic acids ensures that aptasensors may be regenerated under conditions that disrupt aptamer-protein complex. Figure 5 highlights re-usability of IFN-γ aptasensor. In this experiment, a biosensor was challenged with 10 nM IFN-γ and then regenerated by treatment with urea buffer. Our data show that the biosensor retained significant levels of activity after 10 cycles of cytokine exposure and sensor regeneration.
Figure 5.
Re-usability of the biosensor challenged with 10 nM IFN-γ. The cycle of challenge with cytokine, regenerate, challenge with cytokine was repeated 10 times without significant loss in biosensor signal.
CONCLUSIONS
In this paper we describe the development of an electrochemical aptamer-based biosensor for detection of IFN-γ. Biorecognition element of the biosensor consisted of thiolated DNA hairpin carrying a redox tag – MB. Gold electrodes modified with aptamer probes showed Faradaic current signature consistent with reduction potential of MB. Binding of IFN-γ caused current to decrease, thus providing basis for detection. The biosensor was specific to IFN-γ, and showed limit of detection of 0.06 nM with linear range extending to 10 nM. While traditional IFN-γ ELISA is 5 to 10 times more sensitive,35 aptamer-based electrochemical biosensor described here offers several advantages: 1) unlike antibody-based immunoassays, our aptasensor responds directly to the presence of cytokine without the needed for multiple labeling/washing steps, 2) aptamer-based biosensor is suitable for dynamic monitoring of IFN-γ whereas antibody-based immunoassay provides only one time point (end-point) measurement, 3) aptamer layer is chemically stable so that the biosensor may be reused multiple times. In addition, further modifications in the geometry or length of the aptamer molecules may lead to future development of more sensitive IFN-γ aptasensors.
Compared to other electrochemical biosensors for IFN-γ reported in the literature, our aptsensor has similar sensitivity to the biosensor described by Min et al. who used impedance spectroscopy to detect pM levels of IFN-γ binding to aptamer-modified electrodes.36 In another study, van Bennekom and colleagues reported detecting attamolar levels of IFN-γ binding to Ab-modified electrodes using impedance spectroscopy37 which is considerably lower than 60 pM concentration detected in our study. While extremely promising, impedance-based measurements have a significant drawback in that they are difficult to interpret, particularly in complex physiological samples where a large number of factors can contribute to resistance/capacitance components of impedance signal. In contrast, IFN-γ aptasensor described here utilizes a redox tag (MB) and therefore relies on measurement of changes in faradaic current that occur upon analyte binding. This makes for a much more reliable electrical measurement. We found this biosensor to be sensitive and specific to IFN-γ even in the presence of a high concentration of serum proteins.
Our laboratory is interested in monitoring immune cell function10,30,38 and in the future, we envision utilizing electrochemical aptasensor for detecting IFN-γ release from small groups of cells or single cells. Beyond cell monitoring, electrochemical IFN-γ aptasensor may be used in clinical diagnostics, immunology or cancer research.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Yohei Yokobayshi in the Department of Biomedical Engineering at UC Davis for helpful discussions and comments. We also thank Dr. Eugene Kamarchik, Dr. Dianlu Jiang and Dr. Weitao Jia for their help and advice. This work was supported financially by an NSF EFRI award and by NIH grant (REB006519Z) awarded to AR.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional information is noted in the text and is available online at http://pubs.acs.org
REFERENCES
- 1.Boehm U, Klamp T, Groot M, Howard JC. Annu. Rev. Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 2.Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. J. Exp. Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reece WHH, Pinder M, Gothard PK, Milligan P, Bojang K, Doherty T, Plebanski M, Akinwunmi P, Everaere S, Watkins KR, Voss G, Tornieporth N, Alloueche A, Greenwood BM, Kester KE, McAdam KPWJ, Cohen J, Hill AVS. Nat. Med. 2004;10:406–410. doi: 10.1038/nm1009. [DOI] [PubMed] [Google Scholar]
- 4.Karlsson AC, Martin JN, Younger SR, Bredt BM, Epling L, Ronquillo R, Varma A, Deeks SG, McCune JM, Nixon DF, Sinclair E. J. Immunol. Methods. 2003;283:141–153. doi: 10.1016/j.jim.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Cox JH, Ferrari G, Janetzki S. Methods. 2006;38:274–282. doi: 10.1016/j.ymeth.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Fan R, Vermesh O, Srivastava A, Yen BKH, Qin LD, Ahmad H, Kwong GA, Liu CC, Gould J, Hood L, Heath JR. Nat. Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.FitzGerald SP, McConnell RI, Huxley A. J. Proteome Res. 2008;7:450–455. doi: 10.1021/pr070409o. [DOI] [PubMed] [Google Scholar]
- 8.Hermann M, Veres T, Tabrizian M. Anal. Chem. 2008;80:5160–5167. doi: 10.1021/ac800427z. [DOI] [PubMed] [Google Scholar]
- 9.Wang CC, Huang RP, Sommer M, Lisoukov H, Huang RC, Lin Y, Miller T, Burke J. J. Proteome Res. 2002;1:337–343. doi: 10.1021/pr0255203. [DOI] [PubMed] [Google Scholar]
- 10.Zhu H, Stybayeva G, Silangcruz J, Yan J, Ramanculov E, Dandekar S, George MD, Revzin A. Anal. Chem. 2009;81:8150–8156. doi: 10.1021/ac901390j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 12.Liu JW, Cao ZH, Lu Y. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song SP, Wang LH, Li J, Zhao JL, Fan CH. Trac-Trends Anal. Chem. 2008;27:108–117. [Google Scholar]
- 14.Drummond TG, Hill MG, Barton JK. Nat. Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 15.Tuleuova N, Jones CN, Yan J, Ramanculov E, Yokobayashi Y, Revzin A. Anal. Chem. 2010;82:1851–1857. doi: 10.1021/ac9025237. [DOI] [PubMed] [Google Scholar]
- 16.Hianik T, Wang J. Electroanalysis. 2009;21:1223–1235. [Google Scholar]
- 17.Sassolas A, Blum LJ, Leca-Bouvier BD. Electroanalysis. 2009;21:1237–1250. [Google Scholar]
- 18.Li D, Song S, Fan C. Acc. Chem. Res. 2010 doi: 10.1021/ar900245u. [DOI] [PubMed] [Google Scholar]
- 19.Lu Y, Li XC, Zhang LM, Yu P, Su L, Mao LQ. Anal. Chem. 2008;80:1883–1890. doi: 10.1021/ac7018014. [DOI] [PubMed] [Google Scholar]
- 20.Radi AE, Acero Sanchez JL, Baldrich E, O'Sullivan CK. J. Am. Chem. Soc. 2006;128:117–124. doi: 10.1021/ja053121d. [DOI] [PubMed] [Google Scholar]
- 21.Ricci F, Bonham AJ, Mason AC, Reich NO, Plaxco KW. Anal. Chem. 2009;81:1608–1614. doi: 10.1021/ac802365x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lubin AA, Plaxco KW. Acc. Chem. Res. 2010;43:496–505. doi: 10.1021/ar900165x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao Y, Lai RY, Plaxco KW. Nat. Protoc. 2007;2:2875–2880. doi: 10.1038/nprot.2007.413. [DOI] [PubMed] [Google Scholar]
- 24.Kang D, Zuo X, Yang R, Xia F, Plaxco KW, White RJ. Anal. Chem. 2009;81:9109–9113. doi: 10.1021/ac901811n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steel AB, Herne TM, Tarlov MJ. Anal. Chem. 1998;70:4670–4677. doi: 10.1021/ac980037q. [DOI] [PubMed] [Google Scholar]
- 26.Herne TM, Tarlov MJ. J. Am. Chem. Soc. 1997;119:8916–8920. [Google Scholar]
- 27.Ferapontova EE, Gothelf KV. Langmuir. 2009;25:4279–4283. doi: 10.1021/la804309j. [DOI] [PubMed] [Google Scholar]
- 28.White RJ, Plaxco KW. Analytical Chemistry. 2010;82:73–76. doi: 10.1021/ac902595f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Metz J, Sprangers F, Endert E, Ackermans MT, ten Berge IJM, Sauerwein HP, Romijn JA. J. Appl. Physiol. 1999;86:517–522. doi: 10.1152/jappl.1999.86.2.517. [DOI] [PubMed] [Google Scholar]
- 30.Zhu H, Stybayeva G, Macal M, Ramanculov E, George MD, Dandekar S, Revzin A. Lab Chip. 2008;8:2197–2205. doi: 10.1039/b810244a. [DOI] [PubMed] [Google Scholar]
- 31.Ricci F, Lai RY, Heeger AJ, Plaxco KW, Sumner JJ. Langmuir. 2007;23:6827–6834. doi: 10.1021/la700328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White RJ, Phares N, Lubin AA, Xiao Y, Plaxco KW. Langmuir. 2008;24:10513–10518. doi: 10.1021/la800801v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reeves JH, Song S, Bowden EF. Anal Chem. 1993;65:683–688. [Google Scholar]
- 34.Yang WR, Ozsoz M, Hibbert DB, Gooding JJ. Electroanal. 2002;14:1299–1302. [Google Scholar]
- 35.Siawaya JFD, Roberts T, Babb C, Black G, Golakai HJ, Stanley K, Bapela NB, Hoal E, Parida S, van Helden P, Walzl G. Plos One. 2008;3 doi: 10.1371/journal.pone.0002535. Article No.: e2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Min K, Cho M, Han S-Y, Shim Y-B, Ku J, Ban C. Biosens. Bioelectron. 2008;23:1819–1824. doi: 10.1016/j.bios.2008.02.021. [DOI] [PubMed] [Google Scholar]
- 37.Dijksma M, Kamp B, Hoogvliet JC, Van Bennekom WP. Anal. Chem. 2001;73:901–907. doi: 10.1021/ac001051h. [DOI] [PubMed] [Google Scholar]
- 38.Yan J, Sun YH, Zhu H, Marcu L, Revzin A. Biosens. Bioelectron. 2009;24:2604–2610. doi: 10.1016/j.bios.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.