Abstract
Applications of next-generation nucleic acid sequencing technologies will lead to the development of precision diagnostics that will, in turn, be a major technology enabler of precision medicine. Terabyte-scale, multidimensional data sets derived using these technologies will be used to reverse engineer the specific disease networks that underlie individual patients’ conditions. Modeling and simulation of these networks in the presence of virtual drugs, and combinations of drugs, will identify the most efficacious therapy for precision medicine and customized care. In coming years the practice of medicine will routinely employ network biology analytics supported by high-performance supercomputing.
Introduction and context
In his book, The Innovator’s Prescription, Clayton Christenson describes a continuum of scientific progress from intuitive medicine, through empirical or evidence-based medicine to precision medicine, and describes the phenomenon of ‘disruptive’ innovation [1]. Precision medicine is characterized by three essential features: an understanding of what causes a disease; the ability to detect the causal factors; and the ability to treat the root causes effectively. In this essay, we equate understanding the cause of a disease with the ability to quantitatively model and simulate the operative disease ‘pathways’ or signaling networks. Detecting the causal factors will be defined as measuring the molecular biomarkers that allow us to analyze the flow of information through these networks. The ability to treat the root causes means that we have therapeutic agents that are able to selectively block or redirect the disease pathways.
Molecular biomarkers may consist of macromolecules and/or metabolites. Here we will only consider nucleic acid markers, from microRNAs to whole-genome associations. The ability to measure these markers, using so-called next-generation sequencing (NGS) technologies, is undergoing exponential improvements in data production rates accompanied by steadily declining costs [2,3]. These technologies could be ‘disruptive’ technology enablers that change the principles and practices embodied in many existing molecular diagnostic assays [4]. Furthermore, the ability to analyze and interpret large, multidimensional data sets [5] using biomedical informatics and systems biology techniques [6-8] will lead to unprecedented diagnostic precision and the ability to predict the efficacies of individual drugs and combination therapies in the clinical care setting. Diagnostic pathologists will have the central role as integrators and interpreters of the data.
Major recent advances
Second-generation DNA sequencing
A technical review of current (second-generation) sequencing and NGS technologies is beyond the scope of this report but several recent and excellent overviews are available [2,3,9]. For present purposes, the most important issues are data characteristics, diagnostic applications, and costs.
The key attributes of DNA or RNA sequence data are: ‘read lengths’; the accuracy of ‘calling’ individual bases in a sequence ‘read’; and the number of such reads per operational ‘run’ of a given sequencing technology. Read lengths are the average numbers of contiguous nucleotide bases in a polynucleotide sequence that are produced by a particular sequencing instrument. Using current technologies, read lengths vary from approximately 12-400 bases depending upon the particular technology [2,3,9]. Error rates and, in particular, error modalities are also technology-dependent. Errors consist of either inaccurate base calls, leading to erroneous substitutions in a sequence (an A for a G, for example), or erroneous insertions or deletions of one or more bases. Raw base-calling accuracy is on the order of 98.5-99.5%. To achieve 100% accuracy (which is critical for certain medical applications), varying degrees of oversampling of the data is a routine component of sequence data production. This oversampling is referred to as ‘coverage’, which indicates the number of times each base has been sampled in a sequence. Seven- to twelve-fold redundancies (or ‘7-12 × coverage’) are typical sampling frequencies in current practice. High coverage of diverse samples is possible because next-generation sequencers produce hundreds of thousands to tens of millions of reads, depending on the instrument, in a single operational cycle (which can last from hours to days).
Estimating the cost of sequencing is an accounting exercise that must take into consideration the costs of amortized capital equipment, consumable reagents, labor, and the operational cycle time - many assumptions are necessary to compare the costs of different technologies. Best estimates range from $1 to $60 per megabase (1 million bases) [9]. The human genome is about 3 billion bases, or 3,000 megabases, long. The current cost of determining one human genome at 10-fold coverage is continuously being revised but is probably on the order of $100,000. Reducing the per-genome cost to $10,000 and ultimately to $1,000 has been a stated goal of the US National Human Genome Research Institute since 2004 [10] and is also the goal of a $10 million competition sponsored by the X Prize Foundation [11]. Several attempts to win this competition are anticipated within a year [3].
In addition to genome applications, NGS will also dramatically enhance our ability to analyze transcriptomes. Since the mid-1990s, when they were first invented, gene expression microarrays have been the dominant technology for transcriptome analysis. Microarray technologies rely on sequence-specific probe hybridization and fluorescence detection. This is an analog technology subject to noise from background fluorescence and cross-hybridization and yields only the relative abundances of mRNAs. Furthermore, array design is restricted to known sequence entities and, by definition, not capable of detecting novel transcripts. Recent studies have observed that digital gene expression measurements (that is, actual direct enumeration of mRNAs) using currently available sequencing technologies offer significant advantages in resolution and reproducibility [12]. It should be feasible to use digital gene expression profiling longitudinally to study the natural history of disease as well as monitor response to therapy (see below).
Network biology
There are a number of different approaches to interpreting large sequencing datasets and, increasingly, these fall within the realm of systems or network biology [6-8]. Two contrasting approaches for unraveling the behavior of underlying genetic circuits and biochemical pathways are: (a) the curation of well-known biological pathways from the literature and conversion to descriptive visual displays depicting them as maps to visualize molecular profile changes in their context [for example, Kyoto Encyclopedia of Genes and Genomes (KEGG) [13], Ingenuity Systems [14], and GeneGo Metacore [15] pathway analysis]; and (b) the construction of systems of differential equations that model the time evolution of gene products and their connection to phenotypic changes [16-18]. While many of these efforts are impressive and in some rare cases have the ability to make non-intuitive predictions supported by experiment, these approaches address only a tiny fraction of the total possible circuitry of human biology and disease. Classical engineering solutions depend on a complete and accurate blueprint of the system [19]. Black box models also work, provided the inputs and outputs are robustly validated. We are still far from having comparable blueprints for the pathophysiologies of disease: connections are still missing, and systematic measurements to parameterize and otherwise validate models are still hard to make. But some exciting progress has been made.
Sieberts and Schadt [20] pioneered the creation and application of methods that linked genetic alterations to transcriptional changes and physiological outcomes in a genetic cross of two inbred mouse strains, one with a propensity for obesity and diabetes, and the other with a genetic resistance to obesity and diabetes. Here the genetic diversity acted as the ‘perturbation’ or ‘driver’ that allowed correlation between various components to be interpreted as ‘causal’ network interactions. Bayesian network inference algorithms applied to these well-designed data sets revealed a complex interconnected circuitry of several hundred genes that drive the low density lipoprotein, high density lipoprotein, and triglyceride metabolism, and body mass index of these mice that was subsequently validated in a large human cohort [21,22]. This work led to the discovery of several novel drug targets and drug candidates (E Schadt, personal communication). Groups such as ours have developed technology that formalizes these approaches and applies them to the discovery of biomarkers and drug targets. The key aspects of these methods and their growing success and future challenges include: the use of genetic variation and/or drug perturbation combined with transcriptional or other molecular profiling changes and clinical outcomes to go beyond correlation to extract causal interactions; the use of large-scale supercomputing involving thousands or tens of thousands of processors to score billions of possible network fragments and evaluate billions of system-wide network hypotheses that can best explain the data; having a sufficient number of patients or biological samples of appropriate genetic diversity and/or the use of appropriate animal models as surrogates for human diseases.
Future directions
Sequence-based diagnostic applications
Based upon current research applications of second-generation sequencing [5,9] and a knowledge of current molecular diagnostic methods and their uses [4], many future diagnostic applications of NGS can be anticipated. For example, sequencing is being used for polymorphism and mutation detection and cataloging, measurement of copy number variation and other structural variations such as insertions, deletions, and gene rearrangements, epigenetic analysis of DNA methylation and chromatin remodeling, and digital transcriptional profiling and metagenomic analysis of heterogeneous microbial populations [5,9]. It is likely that various applications of NGS technology will eventually supplant a wide range of classical techniques used in clinical laboratories. Such techniques currently support diagnosis of cancer and many non-neoplastic diseases, human leukocyte antigen (HLA) typing and infectious disease and include cytogenetics, PCR-based assays, Southern blot analysis, analog microarray technologies, and microbiology [4,23]. Unlike the current spectrum of labor-intensive, heterogeneous methods, NGS diagnostics will be completely automated. Furthermore, the ability of a single technology to produce multilayer, integratable data will enable precision diagnostics and the medical applications of network biology.
Diagnostic and therapeutic applications of network biology
We are still in the discovery and validation phases of network biology but feel confident that over the next 10 years we will have elucidated and modeled a number of disease pathways to the point where realistic in silico simulations of therapeutic interventions will not only be a routine process for drug and biomarker discovery but also useful for personalized medicine. We envision the following scenario (Figure 1) in which pathologists will extend their traditional role as integrators of data to diagnose and classify disease to become modelers of disease processes [24] who will provide clinicians and their patients with customized care information and therapeutic recommendations. Customized care information should include the specific aberrantly regulated genes, RNAs, and proteins that are responsible for driving the disease process, the number of signaling circuits that are involved, and the drug targets that they contain. Therapeutic recommendations should include the drug or combination that is predicted to be most effective in a particular patient, the best biomarkers to monitor response to therapy - that is, the additional diagnostic tests that are the most valuable surrogate markers for predicting treatment outcomes - and, if the patient becomes resistant to initial therapy, the compensatory or parallel pathways that need to be drugged.
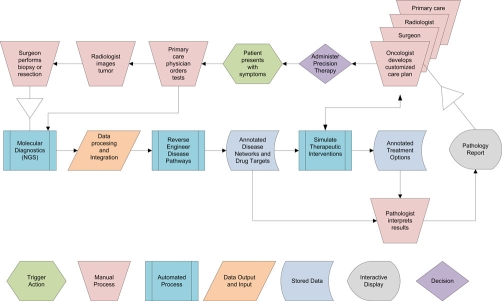
Figure 1. Workflow for precision diagnostics in personalized medicine.
Processes in the top row depict a patient interacting with clinical specialists. The middle and bottom rows depict the largely automated processes that will occur in the pathology department. The natures of the individual processes are as indicated in the key below the figure. The large arrowheads depict control transfers of biospecimens and data between clinical departments and pathology. Pathology reports of the future will be interactive, containing links to underlying databases and parameter sets that clinical teams can use to perform their own simulations for developing a customized treatment plan for the individual patient.
The pathology report of the future will provide precision diagnoses that are at the core of personalized medicine and will be an interactive software tool for clinical teams to design a customized care regimen and monitor its efficacy during treatment.
Acknowledgments
We thank Jeffrey E Saffitz for his vision, enthusiasm, encouragement, and support.
Abbreviations
- HLA
human leukocyte antigen
- NGS
next-generation (nucleic acid) sequencing
Competing interests
MSB is a consultant to Gene Network Sciences (Cambridge, MA, USA).
The electronic version of this article is the complete one and can be found at: http://www.F1000.com/Reports/Biology/content/1/73
References
- 1.Christensen CM, Grossman JH, Hwang J. The Innovator's Prescription: a Disruptive Solution for Health Care. New York: McGraw-Hill; 2009. [Google Scholar]
- 2.Hert DG, Fredlake CP, Barron AE. Advantages and limitations of next-generation sequencing technologies: a comparison of electrophoresis and non-electrophoresis methods. Electrophoresis. 2008;29:4618–26. doi: 10.1002/elps.200800456. [DOI] [PubMed] [Google Scholar]
- 3.Pettersson E, Lundeberg J, Ahmadian A. Generations of sequencing technologies. Genomics. 2009;93:105–11. doi: 10.1016/j.ygeno.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Mais DD, Nordberg M. Quick Compendium of Molecular Pathology. Chicago: American Society for Clinical Pathology; 2008. [Google Scholar]
- 5.Kahvejian A, Quackenbush J, Thompson JF. What would you do if you could sequence everything? Nat Biotechnol. 2008;26:1125–33. doi: 10.1038/nbt1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kriete A, Eils R. Computational Systems Biology. Amsterdam, Boston: Elsevier; 2006. [Google Scholar]
- 7.Palsson B. Systems Biology: Properties of Reconstructed Networks. Cambridge, New York: Cambridge University Press; 2006. [Google Scholar]
- 8.Szallasi Z, Stelling J, Periwal V. System Modeling in Cell Biology: from Concepts to Nuts and Bolts. Cambridge, MA: MIT Press; 2006. [Google Scholar]
- 9.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–45. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 10.Mardis ER. Anticipating the 1,000 dollar genome. Genome Biol. 2006;7:112. doi: 10.1186/gb-2006-7-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.X PRIZE Foundation Archon X PRIZE FOR Genomics. [http://genomics.xprize.org/ ]
- 12.'t Hoen PA, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RH, de Menezes RX, Boer JM, van Ommen GJ, den Dunnen JT. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.0 Must ReadEvaluated by Bernd Weisshaar 10 Nov 2008
- 13.KEGG Kyoto Encyclopedia of Genes and Genomes. [http://www.genome.jp/kegg. ]
- 14. Ingenuity Pathways Analysis [http://www.ingenuity.com/trial/start.html. ]
- 15.GeneGo Bioinformatics software for systems biology ' drug discovery. [http://www.genego.com/metacore.php. ]
- 16.Chen WW, Schoeberl B, Jasper PJ, Niepel M, Nielsen UB, Lauffenburger DA, Sorger PK. Input-output behavior of ErbB signaling pathways as revealed by a mass action model trained against dynamic data. Mol Syst Biol. 2009;5:239. doi: 10.1038/msb.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haberichter T, Mädge B, Christopher RA, Yoshioka N, Dhiman A, Miller R, Gendelman R, Aksenov SV, Khalil IG, Dowdy SF. A systems biology dynamical model of mammalian G1 cell cycle progression. Mol Syst Biol. 2007;3:84. doi: 10.1038/msb4100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaffer CA, Zwolak JW, Randhawa R, Tyson JJ. Modeling molecular regulatory networks with JigCell and PET. Methods Mol Biol. 2009;500:81–111. doi: 10.1007/978-1-59745-525-1_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazebnik Y. Can a biologist fix a radio? Or, what I learned while studying apoptosis. Cancer Cell. 2002;2:179–82. doi: 10.1016/S1535-6108(02)00133-2. [DOI] [PubMed] [Google Scholar]; F1000 Factor 8.4 ExceptionalEvaluated by Bino John 01 Nov 2006, Yi-Kuo Yu 01May 2007, Winston Hide 11 Feb
- 20.Sieberts SK, Schadt EE. Moving toward a system genetics view of disease. Mamm Genome. 2007;18:389–401. doi: 10.1007/s00335-007-9040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–35. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Emmanouil Dermitzakis 29 May 2008
- 22.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, Mouy M, Steinthorsdottir V, Eiriksdottir GH, Bjornsdottir G, Reynisdottir I, Gudbjartsson D, Helgadottir A, Jonasdottir A, Jonasdottir A, Styrkarsdottir U, Gretarsdottir S, Magnusson KP, Stefansson H, Fossdal R, Kristjansson K, Gislason HG, Stefansson T, Leifsson BG, Thorsteinsdottir U, Lamb JR, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–8. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]; F1000 Factor 4.9 Must ReadEvaluated by Vivian Cheung 28 Mar 2008, Eileen Dolan 02 Apr, Emmanouil Dermitzakis 29 May 2008
- 23.Shendure J. The beginning of the end for microarrays? Nat Methods. 2008;5:585–7. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- 24.Walk EE. The role of pathologists in the era of personalized medicine. Arch Pathol Lab Med. 2009;133:605–10. doi: 10.5858/133.4.605. [DOI] [PubMed] [Google Scholar]