Abstract
Autophagy is a universally conserved metabolic program of catabolism that plays important roles in energy homeostasis and impacts both normal physiology and multiple disease processes, including cancer. Autophagy has been documented as a pro-survival mechanism used to maintain viability under starvation conditions; however, conflicting findings have also implicated autophagy in the control of cell death. Adding to the controversy, central mediators of autophagy have been implicated in both pro-survival and pro-death processes. This report highlights recent insights into our understanding of how autophagy is regulated and newly discovered physiological roles for autophagy in normal biology and disease.
Introduction and context
Cellular systems maintain homeostatic equilibrium through a constant balance between biosynthetic (anabolic) processes and catabolism. Macroautophagy, herein referred to as autophagy, is an evolutionarily conserved, catabolic metabolic program that is a key pathway for cellular adaptation to metabolic stresses such as nutrient withdrawal (amino acids and glucose) or hypoxia. During autophagy, internal cellular components, including bulk cytoplasm and organelles, are sequestered into double-membrane structures known as autophagic vesicles (AVs). Following fusion of AVs to lysosomes, the internal contents are degraded, and the degradation products are used to fuel catabolic metabolic processes for energy generation [1]. Starvation-induced autophagy is an important process by which cells ‘recycle’ existing contents for fuel to promote cell viability, while basal levels of autophagy play a critical role in protein and organelle quality control [2].
Autophagy is induced through a stepwise process culminating in the assembly of the autophagosome by core autophagy machinery. A distinct family of autophagy-related genes that mediate the assembly and processing of the autophagosome have been identified [3]. At the molecular level, the induction of autophagy is linked to signal transduction pathways involved in nutrient sensing (Figure 1). Signalling by the phosphatidylinositol 3′-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) pathway downstream of growth receptors engages cellular programs of growth and proliferation and inhibits catabolic metabolic pathways, including autophagy [4]. Inhibition of mTOR, which integrates growth factor signals and amino acid availability to regulate cap-dependent protein translation, is associated with the induction of autophagy [5]. Low cellular energy levels can stimulate autophagy by inhibiting mTOR, a process regulated in part by an LKB1/AMPK (AMP-activated protein kinase)-mediated energy checkpoint [6-8].
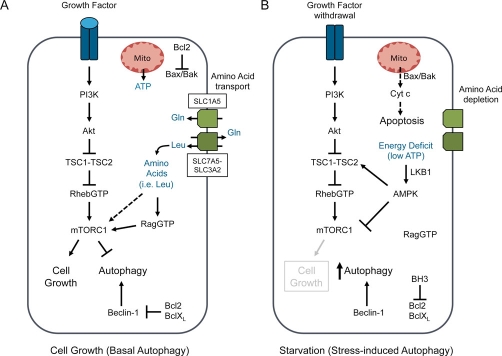
Figure 1. Pathways of autophagy control in mammalian cells.
(a) Under normal growth conditions, signal transduction downstream of growth factor receptors activates mTORC1 via the PI3K/Akt pathway. Activation of mTORC1 promotes cell growth through the regulation of cap-dependent protein translation and the simultaneous inhibition of autophagy. Glutamine (Gln) is transported into cells via the SLC1A5 glutamine transporter and is subsequently used to import leucine (Leu) via the SLC7A5-SLC3A2 complex. Intracellular leucine activates mTORC1 Rag GTPases or a second undefined pathway. Basal autophagy is maintained under these conditions by the activity of Beclin-1, which is inhibited by anti-apoptotic Bcl-2 and Bcl-XL. Bcl-2 and Bcl-XL also function to maintain viability by antagonizing Bax/Bak-dependent mitochondrial apoptosis. (b) Under conditions of metabolic stress, including nutrient depletion (glucose and amino acid), growth factor withdrawal, or energy deficit, the mTOR pathway is inhibited, resulting in autophagy induction. Under conditions of glutamine depletion, the resulting decline in leucine import reduces amino acid-dependent activation of mTORC1. Activation of the LKB1-AMPK pathway by energetic imbalance results in mTORC1 inhibition through activation of the TSC1-TSC2 complex and inhibition of the mTOR-binding partner Raptor. Extended periods of nutrient withdrawal can induce mitochondrial-dependent apoptosis through activation of caspases downstream of Bax/Bak-dependent cytochrome c (Cyt c) release. Antagonism of Bcl-2 family members by BH3-only proteins may trigger either autophagy or apoptosis, depending on the context. AMPK, AMP-activated protein kinase; mito, mitochondrion; mTOR, mammalian target of rapamycin; mTORC1, mTOR complex 1; PI3K, phosphatidylinositol 3′-kinase; TSC, tuberous sclerosis protein.
Major recent advances
Regulation by amino acids
Several recent breakthroughs have advanced our understanding of the molecular mechanisms governing the regulation of autophagy. mTOR activity has long been known to be responsive to nutrient levels; amino acid depletion is a potent stimulator of autophagy. However, the mechanisms linking amino acid levels to mTOR activation and autophagy inhibition have remained one of the key outstanding questions in the field. Three independent studies by the groups of Guan, Sabatini, and Murphy have provided new mechanistic insight into how nutrients direct autophagy. The first insight was the identification of the Rag family of small GTPases as key stimulators of mTOR complex 1 (mTORC1) activity in response to amino acids [9,10]. Amino acids stimulate the association of Rag GTPases with the mTOR-binding partner Raptor, resulting in mTOR activation (Figure 1). Constitutively active Rag mutants mimic a ‘nutrient replete’ state, conferring resistance to starvation-induced autophagy triggered by amino acid withdrawal. Second, Nicklin et al. [11] provided evidence for a coupled glutamine-leucine amino acid shuttle system involved in mTOR regulation and autophagy induction. They demonstrated that glutamine import by the glutamine transporter SLC1A5 is coupled to the import of leucine via the SLC7A5-SLC3A2 antiporter; intracellular leucine is then sensed by intracellular mediators (possibly Rag GTPases) to stimulate mTOR activity (Figure 1). Knockdown of expression of either SLC1A5 or SLC7A5-SLC3A2 results in the induction of autophagy and a reduction in cell size [11]. Thus, amino acid transporters upstream of mTOR play a key role in autophagy regulation by dictating amino acid availability.
Autophagy and cell death
Autophagy has been ascribed both cytoprotective and pro-apoptotic functions, and as such the role of autophagy in cell death has remained controversial. ‘Autophagic’ cell death is loosely defined by the presence of autophagosomes in dying cells [12]. The use of this classification is poor as autophagy, like apoptosis, is a cellular morphology, and many, if not most, cells increase their rate of autophagy under conditions of stress that promote cell death. Thus, defining cell death as ‘autophagic’ based on the presence of AVs may not be accurate [13]. Confounding this classification, autophagy can induce cell death directly through both conventional apoptotic machinery [14] and caspase-independent processes [15], depending on the context. Moreover, when metabolic stress is induced in cells lacking the function of conventional apoptotic pathways, autophagy ultimately results in energetic crisis, leading to necrosis [1]. Thus, under pathophysiological conditions of nutrient or oxygen limitation (that is, a growing tumour lacking vasculature), autophagy may promote necrosis instead of apoptosis. It remains unclear whether autophagic cell death functions as a central mediator of programmed cell death or is simply a mechanism of ‘last resort’ when conventional apoptosis pathways are impaired.
Recent findings in lower organisms have suggested a physiological role for autophagy in cell death control during normal development. Autophagy is specifically induced in Drosophila melanogaster at two developmental stages - germarium and mid-oogenesis - and induction of autophagy at these stages promotes starvation-induced cell death [16,17]. Interestingly, these developmental stages are highly influenced by nutritional status, which may suggest that ‘autophagy as executioner’ is primarily linked to cellular bioenergetics rather than other apoptotic pathways such as those triggered by DNA damage. To date, the demonstration of a required role for autophagy in cell death control during development in vivo has been limited to experimental systems in the fruit fly. Whether autophagy plays a similar role in mammals remains to be determined.
While its role in cell death control remains unclear, autophagy has been implicated in coordinating the clearance of dying cells and cellular debris. The ‘recycling’ function of autophagy serves an important role in the clearance of apoptotic cells [18]. Deregulation of autophagy has been implicated in various pathological conditions, including neurodegeneration [19] and tumourigenesis (discussed in the following section). The contribution of autophagic cell death to these processes remains an open question.
Autophagy and tumour suppression
The involvement of autophagy in cancer development and progression has been an important recent advance in the field of cancer biology. The upregulation of autophagy has been correlated with differing stages of cancer progression. In particular, autophagy is believed to be upregulated in cancerous lesions marked by environments of decreased oxygen or nutrient stress or both. Multiple lines of evidence suggest that oncogene and tumour suppressor networks exert opposing effects on autophagy. When activated, several oncogenes, including PI3K/Akt, mTOR, and Bcl-2, function largely as inhibitors of autophagy, while tumour suppressors [that is, PTEN (phosphatase and tensin homologue), Beclin-1, tuberous sclerosis protein 2 (TSC2), LKB1, and p53] stimulate autophagy [20]. This dichotomy has remained controversial though, as autophagy can promote cell survival in response to cellular stress and thus autophagy could potentially contribute to oncogenesis. However, the involvement of autophagy in tumour suppression may actually stem from its role in the degradation of damaged proteins and organelles, including mitochondria, rather than its role in stress responses [18].
Beclin-1 remains the primary autophagy regulator associated with tumourigenesis. Haploinsufficiency of Beclin-1 promotes tumourigenesis in mouse models and is associated with breast and ovarian tumours in humans [21,22]. This may be due in part to a still poorly defined role for Beclin-1 in the maintenance of chromosome integrity [23]. Modifiers of Beclin-1 activity can alter tumourigenic potential; positive regulators of Beclin-1, including UVRAG (UV radiation-associated gene) and Bif, display tumour suppressor properties [24,25], while Beclin-1 function is inhibited by Bcl-2 [26], a known oncogene. Together, these data suggest that regulation of basal levels of autophagy through Beclin-1 is an important gateway to tumourigenesis. Another regulator of autophagy is the tumour suppressor p53, although its role in autophagy induction remains controversial. The ability of p53 to induce autophagy appears to depend on its cellular localization; nuclear localized p53 triggers stress-induced autophagy through transcriptional control of autophagy mediators, including DRAM (damage-regulated autophagy modulator) [27], while cytoplasmic p53 appears to function as a negative regulator [28]. Growing tumours require p53-dependent autophagy to survive metabolic stress in vivo [7] and may represent an important avenue for therapeutic intervention, particularly in p53-deficient tumours.
Future directions
Much insight has been gained into the mechanisms and biology of autophagy, but many questions remain. Although we have focussed solely on macroautophagy in this report, autophagy exists in several distinct forms (that is, microautophagy and chaperone-mediated autophagy) and can target specific cellular organelles (that is, pexophagy and mitophagy). Understanding the differential regulation of these processes remains a major challenge for the field. In addition, despite recent advances, our knowledge of the signalling networks and layers of regulation that govern autophagy is limited. For example, what are the mechanisms by which amino acids signal to Rag GTPases (or other mediators) to limit autophagy? How do other non-metabolic stressors such as DNA-damaging agents signal to the autophagy machinery? Finally, recent studies have implicated autophagy as an integral biological process involved in a number of pathophysiological conditions, including cancer, neurodegeneration, aging, and infectious disease. The challenge will be to identify the exact role - positive or negative - that autophagy plays in these conditions, to determine the underlying mechanisms that regulate autophagy in each case, and to translate this knowledge into autophagy-based therapeutics to treat disease.
Acknowledgments
The author thanks Julian Lum for helpful discussions during the completion of this manuscript. The author acknowledges the support of the McGill University Faculty of Medicine and the Canadian Institutes of Health Research (MOP-93799).
Abbreviations
- AMPK
adenosine monophosphate (AMP) -activated protein kinase
- AV
autophagic vesicle
- DRAM
damage-regulated autophagy modulator
- mTOR
mammalian target of rapamycin
- mTORC1
mTOR complex 1
- PI3K
phosphatidylinositol 3′-kinase
- PTEN
phosphatase and tensin homologue
- SLC1A5
solute carrier family 1 (neutral amino acid transporter), member 5
- TSC2
tuberous sclerosis protein 2
- UVRAG
UV radiation-associated gene
Competing interests
The author declares that he has no competing interests.
The electronic version of this article is the complete one and can be found at: http://www.F1000.com/Reports/Biology/content/1/68
References
- 1.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–48. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Charles Brenner 09 Feb 2005
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 4.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelárová H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–6. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 5.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–6. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 6.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–90. doi: 10.1016/S0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]; F1000 Factor 4.8 Must ReadEvaluated by Iswar Hariharan 03 Dec 2003, Angus Nairn 09 Dec 2003
- 7.Buzzai M, Jones RG, Amaravadi RK, Lum JJ, DeBerardinis RJ, Zhao F, Viollet B, Thompson CB. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 2007;67:6745–52. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]
- 8.Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, Slingerland JM, Mills GB. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–24. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6.0 Must ReadEvaluated by Robert Abraham 19 Feb 2007
- 9.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.0 Must ReadEvaluated by John Kyriakis 19 Jun 2008
- 10.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–45. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, Myer VE, MacKeigan JP, Porter JA, Wang YK, Cantley LC, Finan PM, Murphy LO. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–34. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.5 Must ReadEvaluated by Robert Abraham 18 Feb 2009, Christian Meyer 20 Feb 2009, Peter Taylor 09 Apr 2009
- 12.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, Hengartner M, Knight RA, Kumar S, Lipton SA, Malorni W, Nuñez G, Peter ME, Tschopp J, Yuan J, Piacentini M, Zhivotovsky B, Melino G, Nomenclature Committee on Cell Death 2009 Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.4 Must ReadEvaluated by Ray Rodgers 07 Jan 2009, Judith S Eisen 07 Apr 2009
- 14.Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.2 RecommendedEvaluated by Daniel Klionsky 23 Jan 2007, Eric Baehrecke 07 Feb 2007
- 15.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–48. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Robert Abraham 08 Jan 2008
- 16.Nezis IP, Lamark T, Velentzas AD, Rusten TE, Bjørkøy G, Johansen T, Papassideri IS, Stravopodis DJ, Margaritis LH, Stenmark H, Brech A. Cell death during Drosophila melanogaster early oogenesis is mediated through autophagy. Autophagy. 2009;5:298–302. doi: 10.4161/auto.5.3.7454. [DOI] [PubMed] [Google Scholar]
- 17.Hou YC, Chittaranjan S, Barbosa SG, McCall K, Gorski SM. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol. 2008;182:1127–39. doi: 10.1083/jcb.200712091. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Harald Stenmark 08 Oct 2008
- 18.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–46. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6.4 Must ReadEvaluated by Eric Baehrecke 21 Mar 2007, Christoph Borner 26 Apr 2007
- 19.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]; F1000 Factor 8.4 ExceptionalEvaluated by Eric Baehrecke 04 May 2006, Daniel Klionsky 09 May 2006, Sharad Kumar 09 May 2006, Nektarios Tavernarakis 02 Aug 2006
- 20.Maiuri MC, Tasdemir E, Criollo A, Morselli E, Vicencio JM, Carnuccio R, Kroemer G. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ. 2009;16:87–93. doi: 10.1038/cdd.2008.131. [DOI] [PubMed] [Google Scholar]
- 21.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI200320039. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Daniel Klionsky 20 Feb 2004
- 22.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 23.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.4 Must ReadEvaluated by Daniel Klionsky 25 Jun 2007, Eric Baehrecke 09 Jul 2007
- 24.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mulé JJ, Pledger WJ, Wang HG. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–51. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.4 Must ReadEvaluated by Harald Stenmark 22 Oct 2007, Daniel Klionsky 07 Nov 2007
- 25.Liang C, Feng P, Ku B, Oh BH, Jung JU. UVRAG: a new player in autophagy and tumor cell growth. Autophagy. 2007;3:69–71. doi: 10.4161/auto.3437. [DOI] [PubMed] [Google Scholar]
- 26.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–39. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]; F1000 Factor 9.8 ExceptionalEvaluated by Robert Abraham 04 Oct 2005, Vojo Deretic 04 Oct 2005, Albert La Spada 31 Jan 2006
- 27.Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–34. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6.5 Must ReadEvaluated by Atan Gross 21 Jul 2006, Scott Lowe 18 Aug 2006, Daniel Klionsky 14 Sep 2006
- 28.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, Criollo A, Morselli E, Zhu C, Harper F, Nannmark U, Samara C, Pinton P, Vicencio JM, Carnuccio R, Moll UM, Madeo F, Paterlini-Brechot P, Rizzuto R, Szabadkai G, Pierron G, Blomgren K, Tavernarakis N, Codogno P, Cecconi F, Kroemer G. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–87. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8.0 ExceptionalEvaluated by W Brent Derry 08 May 2008, David Sassoon 20 May 2008