Abstract
Using PCR and alkaline gel electrophoresis, we show that, compared with DNA derived from virions used to establish infection, herpes simplex virus DNA derived from quiescently infected rat pheochromocytoma (PC12) cells in culture accumulates alkaline-labile lesions. That is, compared with equivalent amounts of virion DNA, viral DNA from nerve growth factor-differentiated long-term infected cells in culture is consistently 3 to 10 times more refractory to amplification by PCR. Despite using equal mole amounts of DNA isolated from quiescently infected cells (determined by quantitative Southern blots), DNA from quiescently infected cells could not be detected by PCR under conditions in which the virion-derived DNA was easily detected. Refractoriness to PCR was confirmed by analysis with a ligation-mediated PCR technique. The refractoriness was not the result of genomic circularization. The refractoriness was, however, related to the time that the quiescently infected cells had been maintained in culture. The refractoriness to PCR was taken as an indication that the viral DNA was damaged. This hypothesis was confirmed by showing that viral DNA from quiescently infected PC12 cells accumulated alkaline-labile DNA lesions, as determined by alkaline gel electrophoresis. The phenomenon was not limited to tissue culture, because viral DNA derived from the ganglia of latently infected mice is also 3 to 10 times more refractory to amplification than are equivalent amounts of virion-derived genomes. Taken together, these results represent the first evidence that herpes simplex virus DNA is physically damaged as a function of long-term infection. Implications for viral reactivation and pathogenesis are discussed.
Introduction
Herpes simplex virus (HSV) establishes long-term infections in neuronal cells in vivo and in vitro from which infectious viruses can be reactivated (Roizman et al, 2007; Roizman and Sears, 1987; Wagner and Bloom, 1997; Whitley and Gnann, 1993). Because reactivation can result in a productive infection, at least some viral genomes must be maintained intact. The mechanism by which genome integrity is maintained and the extent to which it is actually maintained are not well understood. The work described here attempts to address the latter part of that question.
The mammalian cell genome is constantly exposed to endogenous metabolic byproducts and environmental factors that can alter its chemical structure. The oxidation and alkylation of bases, the formation of bulky chemical adducts, and the cross-linking of adjacent nucleotides occur thousands of times per day in the DNA of human cells (Cline and Hanawalt, 2003; Nouspikel and Hanawalt, 2002). It is often necessary for these lesions to be removed to ensure complete and accurate DNA replication and even gene expression.
The pathways involved in recognizing and managing (repairing or eliminating) damaged DNA are complex and highly evolved. More than 130 genes are now known to be involved in DNA repair in the mammalian genome (Wood et al, 2001). The fact that host DNA repair pathways play crucial roles in HSV DNA replication during the productive infection has been documented extensively (Iwahori et al, 2007; Lilley et al, 2005; Lilley et al, 2007; Muylaert and Elias, 2007; Shirata et al, 2005; Wilkinson and Weller, 2004; Wilkinson and Weller, 2005; Wilkinson and Weller, 2006). Little is known, however, about how the host repair system maintains the integrity of the latent viral genome or its role in reactivation. It has been suggested that the virus might differentially manipulate the DNA repair machinery to favor lytic cycle or latency (Lilley et al, 2007).
Repair of damaged DNA in neuronal cells is apparently much more limited than is repair in mitotically active cells. Indeed, global repair of non-expressed genes is significantly reduced in terminally differentiated neurons (Brooks, 2002; Francisconi et al, 2005; Jensen and Linn, 1988; Nouspikel and Hanawalt, 2000; Nouspikel and Hanawalt, 2002). It has been reasoned that terminally differentiated cells need not repair “non-expressed” genes to maintain function, because, other than in rare or extraordinary circumstances (olfactory neurons), DNA replication does not occur. In fact, brain tissue, when compared with other nonneuronal tissues, was shown to accumulate twice as many DNA lesions (Abner and McKinnon, 2004; McMurray, 2005). Because latency occurs in neuronal cells, as do important pathological phases of infection, understanding how the viral genome is managed in these cells is critical to a full understanding of HSV pathobiological characteristics.
The genome of HSV can persist in neuronal cells in a quiescent state for long periods of time, perhaps even for the lifetime of the host. Reactivation of the infectious virus from latently infected cells in the ganglia is a characteristic of the natural history of infection as well as of experimental systems (Batterson et al, 1983; Fraser et al, 1981; Mitchell et al, 2003; Renlund et al, 1979; Roizman and Sears, 1987; Wagner and Bloom, 1997; Whitley and Gnann, 1993). For example, following the explant of trigeminal ganglia (TG) from latently infected animals such as rabbits and mice, co-cultivation with permissive tissue culture cells usually results in appearance of the productive virus (Maggioncalda et al, 1996; Wagner and Bloom, 1997). Although the number of viral genomes within the latently infected ganglia that can result in progeny is unknown, it appears to be a small subpopulation of the total, because the number of primary cytopathic foci resulting from explant cocultivation is much smaller than the number of genomes contained within the ganglia (Kennedy et al, 1983). The possible reasons for the relative inefficiency of HSV reactivation are many. The possibility that damage accumulated within the viral genome plays a role has not been explored.
We have developed a tissue culture model of quiescent HSV infection. PC12 cells respond to nerve growth factor (NGF) by exhibiting many properties of the peripheral nervous system. For example, in response to nanogram amounts of NGF, PC12 cells cease division, extend long neuritic-like processes that can support action potentials, and secrete catecholamine (Greene and Tischler, 1976). We have shown that infection of NGF-differentiated PC12 (NGF-PC12) cells with HSV-1 results in a well-tolerated, long-term quiescent infection in which the viral genome is “endless” or circular, transcription of key viral genes becomes undetectable or very low by reverse-transcriptase PCR and northern hybridization, and no evidence of productive infection is observed (Moxley et al, 2002; Su et al, 1999; Su et al, 2002). In contrast, transcription of cellular genes such as GAPDH, persists in long-term quiescently-infected PC12 cells (Su et al, 1999).
In this study, we observed a striking phenomenon: Viral DNA derived from long-term (2 – 5 weeks post infection) infected NGF-PC12 cells or latently infected mouse TG was far more refractory to the polymerase chain reaction (PCR) than was a similar amount of viral DNA isolated from virions. Moreover, the viral DNA derived from the quiescently infected cells was much more sensitive to alkali than was viral DNA from virions or recently infected cells, suggesting inherent damage. This is the first report indicating that HSV-1 DNA derived from quiescently infected neuronal-like NGF-PC12 cells is physically damaged.
Materials and Methods
Cells, viruses, and media
PC12 cells were originally obtained from the American Type Culture Collection. The PC12 cells were maintained in an RPMI medium supplemented with 10% horse serum, 5% fetal bovine serum, and 1% antibiotic–antimycotic agent. HSV-1, strains 17 and F, were prepared in Vero cells.
Differentiation of PC12 cells and establishment of quiescent infection
NGF-differentiated PC12 cultures were established as described previously (Su et al, 1999). PC12 cells were dispersed from clusters by syringe, and 6 × 105 cells were seeded onto T-75 flasks coated with poly-L-ornithine hydrobromide (Sigma, St. Louis, MO) in the same media for PC12 cell culture with an additional supplement of 100 ng/ml of 2.5S NGF (BD Bioscience, Franklin Lakes NJ). On the 7th day after addition of NGF, the cells were treated with 20 μM of 5'-fluoro-2-deoxyuridine (Sigma, St. Louis, MO) for 3 days to remove the undifferentiated cells. Fresh NGF-containing media was supplied thereafter.
Differentiated PC12 cultures were infected with 1.2 × 107 PFU of HSV-1 strain 17. Following 1 hour (h) incubation at 37°C, the cultures were treated with 3 ml of sodium citrate buffer (pH=3) for 30 seconds to 1 minute to inactivate residual virus modified as described (Su et al, 2000). After low pH treatment, cultures were incubated at 37°C with fresh medium containing NGF.
Latent infection of mice
After corneal scarification, 4- to 6-week-old female Balb/c mice (Charles River Breeding Laboratories, Kingston, N.Y.) were inoculated in each eye with approximately 3 × 104 or 3 × 105 PFU of HSV-1 strain 17 or 4 × 107 PFU of HSV-1 strain F. Animals were sacrificed at day 45 after infection; latently infected TGs were removed, snap-frozen, and stored at −80°C. These tissues were homogenized for DNA isolation using SDS/proteinase K digestion, phenol/chloroform extraction, and ethanol precipitation.
Quantification and analysis of viral DNA by Southern blot hybridization
Nuclear DNA was isolated as previously described (Su et al, 2002). To quantify the amount of viral DNA, DNA samples isolated from NGF-treated PC12 cultures at D0, D1, D10, D20, and D30 post infection (or CV-1 cells when applicable) and virion DNA (described below) were digested with BamHI. The digested DNA was resolved on a 1% TAE agarose gel and transferred to a nylon membrane. The membrane was hybridized with the selected 32P-radiolabeled HSV-1 probes of interest. The autoradiographic image was generated and quantified by a Bio-Rad phosphoimager using the virion DNA to generate a standard curve.
To prepare virion DNA used as southern blot standards and PCR controls, Vero cells were infected with HSV-1 strain 17 (MOI = 0.5) for approximately 24 hours, followed by PBS wash and centrifugation of infected cells at 2000 RPM for 10 min at 4°C. Infected cells were subjected to 3 freeze/thaws and sonicated for 30 seconds. The pellet was resuspended in 1 ml virion lysis bufer (0.25% Triton X-100, 10 mM EDTA, 10 mM Tris, pH 8.0) and transferred to a 1.5 ml microcentrifuge tube. To this, 88 ul of 5 M NaCl was mixed in and the mixture sat on ice for 10 min followed by centrifugation at 13,000 rpm for 4 min at 4°C. The supernatant was transferred to a Sorvall tube and virions were pelleted by centrifugation at 16,300 rpm for 1 hr at 4°C (SS34 rotor). Virions were digested with proteinase K overnight and and DNA was purified by phenol/chloroform extraction followed by ethanol precipitation. DNA pellets were dissolved in TE and checked for conentration and purity by spectrophotometry and agarose gel electrophoresis. A260/A280 readings were checked to be within 1.8 to 2.0 range. An example of cell loading control is shown in Supplemental Figure 1.
To detect alkaline-labile lesions, DNA was digested with BamHI. One-third of the digested DNA was resolved on a 1% TAE agarose gel and the other two-thirds of the digest was resolved on a 1% alkaline agarose gel (50 mM NaOH and 1 mM EDTA) and was transfered to a nylon membrane. The membrane was hybridized with the selected 32P-radiolabeled HSV-1 probes of interest. Quantification of the hybridized image was performed as described above.
PCR of viral DNA
PCR was performed with the puReTaq Ready-To-Go PCR beads according to the manufacturer's instructions (GE Healthcare UK, Little Chalfont Buckinghamshire). Based on Southern blot quantification, 1 ng of viral DNA from each sample was used for PCR with primers specific for HSV-1 TK (forward: 5' GGTATCGCGCGGCCGGGTA 3'; reverse: 5'ATGGCTTCGTACCCCTGCCAT 3'; 531 bp product), ICP27 (forward: 5' TTTCTCCAGTGCTACCTGAAGG 3'; reverse: 5'TCAACTCGCAGACACGACTCG 3'; 282 bp product), gC (forward: 5' GAAACTGCCTCCACCGGGC 3'; reverse: 5'GGCGTCACCTCGCCGATAATC 3'; 628 bp product), POL (forward: 5' TACGGGTTCACGGGAGTGCAGC 3'; reverse: 5' CTGAGGTGCGGTTGATAAACGC 3'; 461 bp product) and LAT (forward: 5' GACAGCAAAAATCCCCTGAG 3'; reverse: 5'ACGAGGGAAAACAATAAGGG 3'; 194 bp product) at annealing temperatures of 55°C, 55°C, 60°C, and 55°C, respectively.
Quantitative real-time PCR
To determine the amount of HSV-1 DNA, two HSV-1 primer sets were used: Tk5' (forward: 5'GCAGCGACCCGCTTAACA 3', reverse: 5' GAAGAGGTGCGGGAGTTTCAC 3') and ICP0 (forward: 5' CCCACTATCAGGTACACCAGCTT 3', reverse: 5' CTGCGCTGCGACACCTT 3'). These real-time PCR quantification reactions were performed using the LightCycler (Roche Biochemical, Germany), at 95°C for 10 min, then 45 cycles at 95°C for 5 s, 55°C for 10 s, and 72°C for 10 s. SYBR green Mastermix reagents (Roche Biochemical, NJ) were used to test for double-stranded DNA products resulting from the PCRs, and melting curve analysis was used to test the specificities of these PCR products. Standard PCR conditions for cycling and dissociation curve analysis as well as data analysis using the relative standard curve method were used as recommended by the manufacturer. Standard curves were made for each of the virus-specific primer sets using known dilutions of viral DNA.
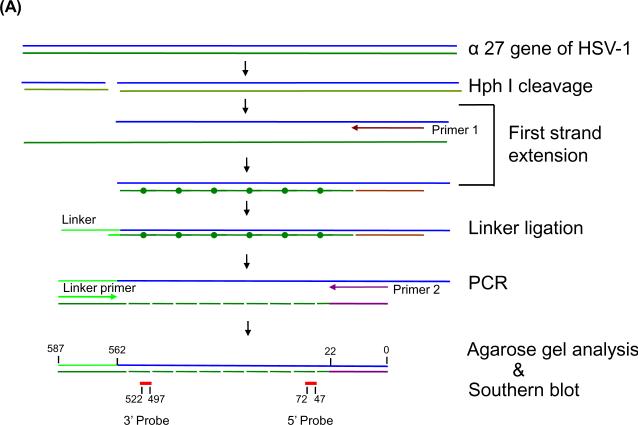
Ligation-mediated PCR (LM-PCR) of HSV-1 ICP27 gene and Southern blot to determine the specificity of the PCR product
HSV-1 DNA was first digested with restriction enzyme Hph I overnight at 37°C and purified by phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation. The digested sample was subjected to the first strand extension with the first ICP27 primer (5'-GCTCTTCGTGGTGTGGTGTCCTTTCTCCAGTGCTACCTGAAGG-3') with an annealing temperature of 63°C for extension and then ligated with a unidirectional linker (5'-GCGGTGATTTAAAAGATCTGAATTC-3'and 5'-GAATTCAGATC-3') to the 3'-blunt end by T4 DNA ligase at 15°C overnight. After ligation, a PCR amplification was performed with a second ICP27 primer (forward primer 5'-TTTCTCCAGTGCTACCTGAAGG-3' and reverse primer 5'-GCGGTGATTTAAAAGATCTGAATTC-3') (linker primer), with an annealing temperature at 63°C. After amplification for 30 cycles, the PCR products were resolved on a 1.5% agarose gel. The resolved PCR products were transferred by capillary to a nylon membrane. The membrane was hybridized by 32P-labeled internal 5' probe (5'-AACTGTGTTCGCGGCGGCGTCTGGC-3') or internal 3' probe (5'-GGTGTCATAGTGCCCTTAGGAGCTT-3'). The autoradiographic image was generated and quantified by a Bio-Rad phosphoimager.
Results
HSV DNA isolated from quiescently infected PC12 cells is refractory to PCR
To study viral DNA structure in quiescently infected neuronal-like tissue culture cells, PC12 cells were differentiated with NGF, infected with strain 17 at an MOI of 5, and maintained in culture for 24 days beyond the time at which infectious virus could no longer be recovered, as described elsewhere (Su et al, 1999). DNA was isolated from the infected cells, digested with restriction endonuclease BamHI, resolved by agarose gel electrophoresis, analyzed by Southern blot, and hybridized to radioactive probes specific for the terminal sequences. Care was taken to load the samples from both virions and quiescently infected cells with equal amounts of viral DNA. This was achieved by preparing serial dilutions of known amounts of virion-derived DNA, followed by Southern blot analysis of the dilutions plus cell-derived DNA. Only the diluted samples with equal amounts of viral DNA are shown in Figure 1. With DNA equivalence established by this method, the DNA samples were tested and compared for differences in PCR efficiency.
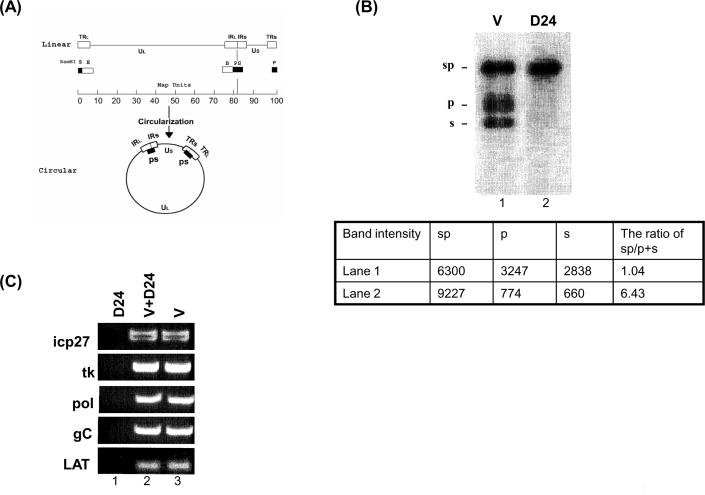
Figure 1.
PCR amplification of HSV-1 DNA from virions and quiescently infected PC12 cells. (A) Maps of the HSV-1 genome as linear and circular forms and of the probes used in this study. The linear map of the HSV-1 genome is divided into unique long (UL) and unique short (US) segments represented as lines. Open boxes represent repeated sequences (TRL, TRS, terminal repeat long and short, respectively; IRL, IRS, inverted repeat long and short, respectively). Solid boxes drawn under the map show the genomic locations of BamHI fragments P, S, and the internal joint PS fragment, which hybridizes to radioactive probes that are generated from BamHI PS fragments. Probes generated from BamHI B fragments hybridize to two BamHI fragments (B and E) because they both contain repeat sequences with different UL terminal sequences. Note that BamHI P and S fragments exist only as linked PS fragments (2 copies) in the circular form. (B) Southern blot quantification of HSV-1 DNA derived from virions (V) produced in productively infected cells (linear form) and quiescently infected NGF-PC12 cultures (circular form). HSV-1 DNA was isolated from quiescently infected NGF-PC12 culture 24 days post infection (D24). HSV DNA was quantified by southern blot using the BamH1 B probe and virion DNA as a reference for a standard curve. 2 ng of each was digested with BamHI, resolved by agarose gel electrophoresis, blotted, and hybridized with 32P-labeled Bam PS fragments. Blots were scanned and quantified using the Bio-Rad phosphoimager as shown in panel B. Lane 1 is 2 ng of virion DNA and lane 2 is D24 HSV DNA. (C) PCR amplification of virion DNA (V; lane 3), quiescent HSV DNA (D24; lane 1) or a combination of both virion and D24 DNA (V + D24; lane 2) with primers specific to icp27, tk, pol, gC, and LAT. Approximately 0.2 ng of HSV DNA (D24 or V) was used in each PCR reaction for 35 cycles. Amplified DNA fragments were analyzed by gel electrophoresis, stained with ethidium bromide, and photographed under an ultraviolet transilluminator.
As shown in Figure 1B, the viral DNA is readily detected by southern blot (lane 2), and is primarily in an “endless” state. That is, compared with the DNA isolated from virions, the terminal fragments “s” and “p,” as indicated in Figure 1A and shown in Figure 1B, are barely detectable in the DNA derived from quiescently infected cells when the BamHI-digested HSV-1 DNA was probed by the joined fragment, Bam sp fragment. The reduction in terminal “s” and “p” bands was quantified using a phosphoimager, with the numerical results being consistent with the assumption of an endless configuration (lack of “s” and “p” terminal fragments in viral DNA derived from quiescently infected cells). This result is expected, because viral DNA in latently infected ganglia from animals and people assumes an endless configuration. Hence, the terminal fragments of the viral genome become ligated. We observed this phenomenon in the NGF-differentiated PC12 model (Su et al, 2002). Thus, the amounts of HSV-1 DNA in the samples from virion and quiescently infected cells (Figure 1), are similar.
A different situation is seen in the results of PCR amplification of the two DNA samples. As shown in Figure 1C, although viral DNA corresponding to various regions of the genome is readily amplified and detected when derived from the virions, the PCR product from a similar amount of DNA (as determined by equal band intensity in the Southern blots, see Figure 1B) derived from the quiescently infected PC12 cells was virtually undetectable. Indeed, using the same amounts of virion DNA and quiescently infected viral DNA, as determined by Southern blot analysis, it took an additional 2 to 3 cycles of amplification of the quiescently infected viral DNA to achieve similar amounts of amplified product (not shown). The refractoriness to amplification was not restricted to one region of the genome. Primers specific for ICP27, tk, pol, gC, and LAT genes were all far less effective in mediating amplification of DNA from quiescently infected cells compared to virion-derived DNA.
Circular and linear viral DNAs are amplified by PCR similar efficiency
Because DNA from the quiescently infected cells used in these experiments is predominantly in the endless, presumably circular, structure, it was possible that the circular nature of the viral DNA from quiescently infected cells was responsible for the refractoriness to PCR. This possibility was tested by comparing the efficiency of amplification of viral DNA derived from virions and CV-1 cells, 2 hours after infection, with strain 17 in the presence of phosphonoacetic acid (PAA, 400 μg/ml), an inhibitor of viral DNA replication.
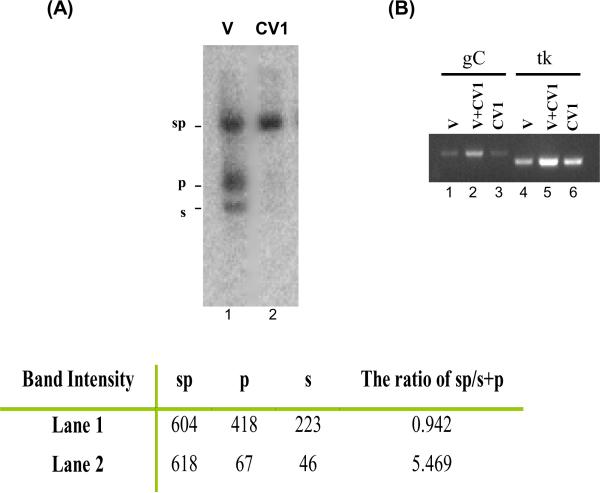
As previously observed (Su et al, 2002) and shown in Figure 2A, HSV DNA accumulates in an endless, presumably, circular configuration, shortly after infection of CV-1 cells and in the presence of PAA (compare Figure 2A, lanes 1 and 2). Care was taken to load similar amounts of infected cell and virion derived DNA based on quantitative Southern blotting from multiple dilutions and using only the samples that yielded similar amounts of viral DNA, as described in Methods. As the figure also shows, similar amounts of viral DNA were present in the virion and in the CV-1 cell-derived samples used for the analysis. Although the viral DNA present in the CV-1 cells was predominantly in the endless, or circular, state, it was as efficiently amplified by PCR as was the linear, virion-derived viral DNA. Thus, a circular or endless state of viral DNA is not in itself refractory to amplification by PCR. From a technical perspective, this data also suggests that isolation of infected cell DNA per se, is not the cause of the reduced PCR of DNA derived from quiescently-infected PC12 shown in Fig. 1C.
Figure 2.
PCR amplification of linear virion DNA (V) and circular HSV DNA derived from 2 hr infected CV-1 cells. (A) CV-1 cells were infected with HSV-1 strain 17 at an MOI of 5. Two hours after infection, nuclei were harvested and treated with 0.1% deoxycholate. DNA was then isolated from deoxycholate-treated nuclei, digested with BamHI, resolved by 1% agarose gel electrophoresis, blotted, and hybridized with 32P-labeled Bam PS fragments. The blot was scanned and quantified using the Bio-Rad phosphoimager. Lane 1 is 2 ng of virion DNA (V), and lane 2 is infected CV-1 DNA (CV1). (B) PCR amplification of virion DNA (V) and CV-1 infected DNA (CV1) with primers specific for HSV-1 gC (lanes 1–3) and tk (lanes 3–6). PCR reactions in lanes 1 and 4 contained 0.1 ng of virion DNA (V). Reactions in lanes 2 and 5 contained 0.1 ng of virion DNA and 0.1 ng of HSV CV-1 DNA (V+CV1). Lanes 3 and 6 PCR reactions contained 0.1 ng of HSV CV-1 DNA (CV1). DNAs were denatured with 0.3 N NaOH and followed by neutralization with Tris (pH 7.5) prior to 35 cycles of PCR amplification by using puReTaq Ready-To-Go PCR beads (Amersham). Amplified DNA fragments were subjected to agarose gel (2%) electrophoresis, stained with ethidium bromide, and photographed under an ultraviolet transilluminator.
Refractoriness occurs after approximately 20 days in culture
The neuron is known to have less DNA repair activity than dividing cells, as mentioned previously. It is possible that viral DNA inside of neuronal-like cells accumulated lesions that rendered the DNA a poorer substrate for PCR and that the longer the DNA persisted in the neuronal-like PC12 culture, the more damage it accumulated. Thus, it was of interest to determine if the refractoriness of HSV DNA to PCR was a function of the time in which the HSV DNA was maintained in tissue culture.
To test this hypothesis, DNA was isolated from either virions or NGF-differentiated PC12 cells maintained in culture for different times after infection, ranging from the day of infection to more than 30 days. The amount of HSV-1 DNA in each sample was quantified by Southern blot hybridization using the Bam B/E probe. The Bam B/E probe was chosen instead of Bam SP for easier quantification because two distinct bands (9 and 10 kb) were detected by the B/E probe instead of three not so focused bands (due to repeat sequences) by the SP probe. The results are shown in Figure 3A. Equimolar amounts of viral DNA (1 ng of viral DNA, as determined by Southern blot analysis) from each source and time point were subjected to PCR and the relative ability to be amplified was determined.
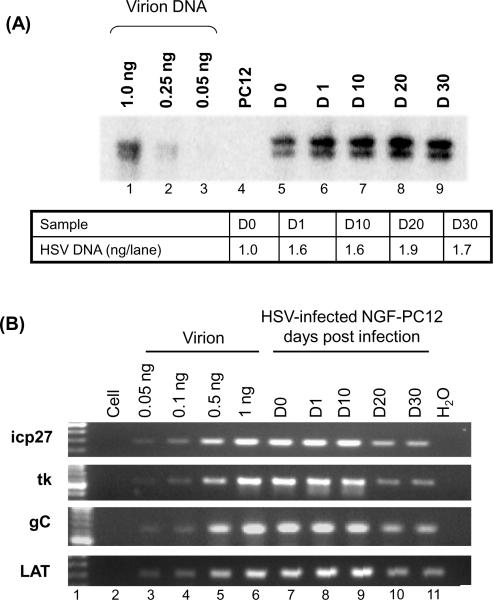
Figure 3.
HSV-1 DNA derived from quiescently infected PC12 cells becomes more refractory to PCR amplification as a function of time in culture. (A) Southern blot analysis of DNA from quiescently infected NGF-differentiated PC12 cells. Purified virion DNA (1, 0.25, and 0.05 ng) and 1.5 ng HSV DNA (quantified by Southern blot hybridization) from infected (MOI of 20) NGF-differentiated PC12 cells at day 0 (D0), day 1 (D1), day 10 (D 10), day 20 (D20), and day 30 (D30) post infection were digested with BamHI, resolved by electrophoresis through 1% agarose, transferred to nylon membranes, and hybridized to a 32P-labeled BamHI B/E fragment. The autoradiographic image was produced by a Bio-Rad phosphoimager. The density of viral DNA was calculated against the virion DNA standard curve. (B) PCR amplification of DNA derived from HSV-1 strain 17+ (MOI of 20) infected NGF-differentiated PC12 cells. Based on southern blot analysis (Panel A), 1 ng of viral DNA for day 0 (D0), 1 (D1), 10 (D10), 20 (D20) and 30 (D30) was used for PCR. purified virion DNA 0.05, 0.1, 0.5, and 1 ng was used as a positive control. Each sample contained 2.8 μg of cellular DNA to ensure that each reaction occurred under similar conditions. The PuReTaq Ready-To-Go PCR beads were used with the four primers shown, as representative of the HSV-1 genome. The products were resolved in 1.5% agarose gels.
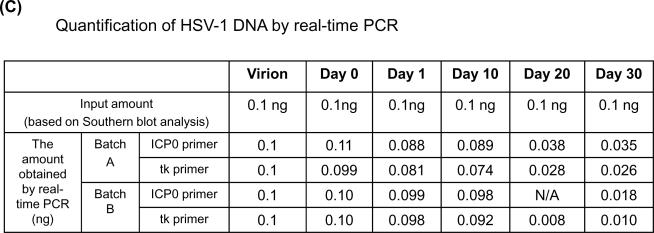
(C) Real-time PCR quantification of HSV-1 DNA derived from HSV-1 infected NGF-differentiated PC12 cells. 1 ng of viral DNA, based on the Southern blot analysis shown in Panel A, for day 0 (D0), 1 (D1), 10 (D10), 20 (D20), and 30 (D30) each was subjected to real-time PCR amplification. Two different sets of primers, ICP0 and tk, were used to quantify the HSV DNA derived from two independent batches (A and B) of infected cultures. Note that batch A is the same set of DNA shown in panel B. The amount of viral DNA obtained by real-time PCR was based on the standard curve generated by serial dilutions of virion DNA. Each sample contained 2.8 μg of cellular DNA to ensure that each reaction occurred under similar conditions.
The results, shown in Figure 3B, show a clear trend. Regardless of the primer pair used, viral DNA isolated from NGF-differentiated PC12 cells after 1 day and up to 10 days of infection was similar to virion-derived DNA in terms of susceptibility to PCR, whereas the same amount of viral DNA (as determined by southern blot, see Figure 3A) isolated from cells 20 and 30 days after infection was refractory to PCR (see Figure 3B).
Real-time PCR analysis suggested that viral DNA 20 and 30 days after infection of PC12 cells is underestimated by approximately 3- to 10-fold compared to the same mole amounts from virions (Figure 3C). It was noted that the degree of refractoriness and the time required for this phenomenon to occur vary from batch to batch of the PC12 cultures. Nevertheless, this refractoriness was consistently observed from more than 5 batches of cultures and always occurred after 20 days postinfection.
LM-PCR results support a culture time dependent refractoriness of viral DNA
The suitability of the DNA template for primer binding, polymerase elongation, and completion of elongation can be monitored by LM-PCR, a method used to map the sites of DNA adducts that prevent polymerization during the primer extension (Dammann and Pfeifer, 1997; Hu et al, 2002; Pfeifer and Dammann, 1999; Tu et al, 1996; Zheng et al, 2001). In this technique, as illustrated in Figure 4A, substrate DNA is first digested by the restriction endonuclease of interest and then subjected to the primer extension. If the DNA template is suitable for the primer extension (for example, when no obstacle stops the extension reaction), the full-length first strand will be synthesized. However, if lesions exist that interfere with the primer extension reaction at the end of primer extension, there will either be no product (no priming) or truncated products due to the incomplete extensions. After primer extension, the products are ligated with the linker sequence at the 3'end and then subjected to the PCR reaction with the sequence-specific primer and linker primer. The PCR product is then analyzed by electrophoresis and Southern blot hybridization with both 3' and 5' internal probes respectively, as detailed in Material and Methods.
Figure 4.
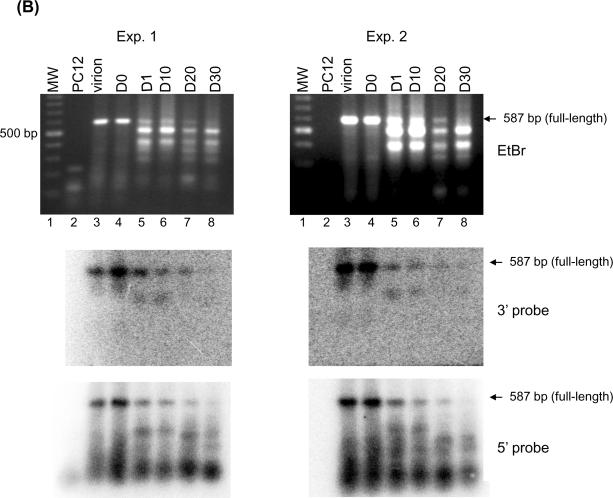
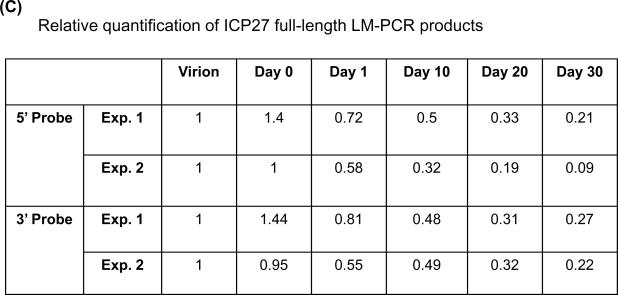
LM-PCR of HSV DNA from quiescently infected cells results in smaller than full length amplification products. (A) Schematic diagram of the LM-PCR procedure. (B) Southern blot hybridization of amplification products by LM-PCR. Total DNA was isolated from HSV-1 infected NGF-differentiated PC12 cells at indicated days post infection. Viral DNA was quantified by Southern blot hybridization as shown in Figure 3B. 100 ng of viral DNA from each sample was subjected to LM-PCR as outlined in panel A and detailed in Materials and Methods using primers specific for HSV-1 gene ICP27. The products were resolved on 1.5% agarose gel, stained with ethidium bromide, photographed under an ultraviolet transilluminator, and transferred to a nylon membrane. The membrane was then hybridized with 32P-labeled 25 bp internal oligo probes specific for either the 5' and 3' ends of the 587 bp product. Autoradiographs derived from the hybridization with each probe were generated and quantified using a Bio-Rad phosphoimager. The results are shown for two independent experiments. The locations of full-length 587 bp PCR products are indicated by arrows based on the molecular weight marker (MW). (C) The quantification results were normalized by data obtained from virion DNA and are summarized in panel C.
Thus, viral DNA was isolated from either virions or NGF-differentiated PC12 cells at various times after infection, as described before, and used as a substrate in LM-PCR with ICP27 primers. To detect the lesions, if any, we chose restriction endonuclease HphI in combination with ICP27 forward primer to synthesize a 587 bp fragment of the ICP27 gene in this method. The results obtained using DNA isolated from two independent batches of NGF-differentiated PC12 cultures are shown in Figure 4B. Clearly, full-length, virus-specific 587 nt products, as expected, were synthesized using DNA derived from virions and NGF-differentiated cells isolated on day 0 and isolated up to day 10 post infection. This full-length product was confirmed (Panel B) and quantified (panel C) by Southern blot hybridization using both 5' and 3' internal probes, respectively. As expected, the 3' internal probe only hybridized to the full-length products or to those less than 60 bp from full-length, as shown in the middle panels of Figure 4B, although many species of DNA fragments were revealed by ethidium bromide staining (top panels of Figure 4B, top panels). Interestingly, variable degrees of “smear” were revealed among samples tested by the 5' internal probes from the same membranes (Figure 4B, bottom panels). By day 20 post infection, little full-length product (less than 30% of the product from the virions) was generated. The shorter products detected with the 5' probe that become more prominent at later time point samples are suggestive of increasing likelihood of obstruction on the viral DNA substrate.
Taken together, the results are consistent with those observed using conventional PCR and demonstrate a time in culture dependent accumulation of refractoriness to polymerase-mediated synthesis from the viral template.
Alkaline gel electrophoresis of viral DNA derived from quiescently infected PC12 cells
The refractoriness to PCR exhibited by viral DNA from quiescently infected cells may be due to genomic damage. The DNA lesions most frequently found in cells are abasic lesions. It has been estimated that most tissues contain approximately 100,000 abasic lesions per cell, whereas more than 200,000 abasic lesions per cell were detected in the DNA isolated from brain tissue (Nakamura and Swenberg, 1999). It has been shown that virion DNA contains nicks and gaps (Ecker and Hyman, 1981; Frenkel and Roizman, 1972; Hyman et al, 1977; Kieff et al, 1971; Wilkie, 1973). Breaks and abasic site interruptions in even single strands of the viral genome would be expected to interfere with the processiveness of a PCR amplification. Alkaline denaturing techniques such as the single-cell COMET assay (Miyamae et al, 1997) or alkaline agarose gel analysis (Sutherland et al, 1999; Sutherland et al, 2003) have been used to assess the genomic DNA damage. Abasic sites are completely hydrolyzed in high pH conditions (pH 12.6) and become single-stranded break (Miyamae et al, 1997). Double-stranded DNA with single-stranded breaks would migrate as an intact species in native gels but may be refractory to PCR amplification and would migrate as fragments through alkaline denaturing gels.
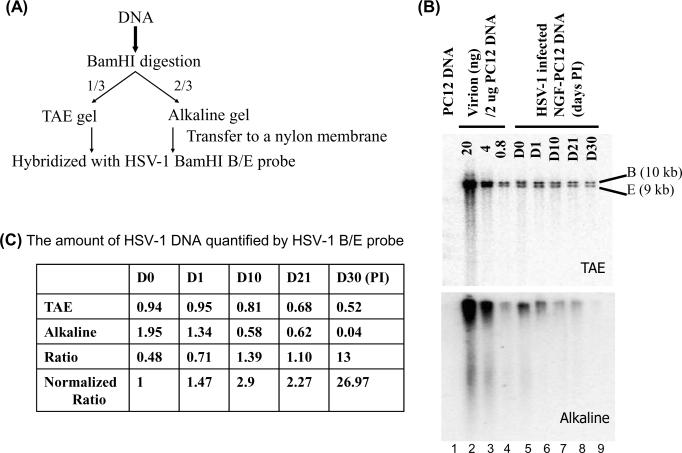
Thus, the possibility that viral DNA derived from quiescently infected PC12 cells accumulated strand breaks or abasic sites was determined by preparing DNA from virions and NGF-differentiated PC12 cells at different times after infection and quiescence and comparing their resolution through native and denaturing gels. Because native TAE gel will not reveal the single-stranded breaks unless two breaks are close to each other on the opposite strands, the intact HSV BamHI B/E fragments detected in the native TAE gel could serve as the loading control. In contrast, any nick or abasic lesion would result in the disappearance of an intact fragment and would migrate faster in the alkaline gel. The ratio of intact native TAE gel fragments to alkaline gel fragments detected indicates the number of alkaline-labile sites present in the fragments studied. This approach is outlined in Figure 5A.
Figure 5.
Native and alkaline gel analyses of HSV-1 DNA derived from quiescently infected NGF-PC12 cultures. (A) Outline of experimental procedure. (B) Southern blot analysis of HSV-1 DNA derived from infected NGF-PC12 cultures that was prepared and quantified as described in Figure 3. HSV-1 DNA (25 ng) from each sample was digested with restriction endonuclease BamHI. One-third of the digest was resolved on a TAE gel and the other two-thirds was resolved on an alkaline gel, transferred to nylon membranes, hybridized with 32P-labeled HSV BamHI B/E probe, scanned, and quantified using the Bio-Rad phosphoimager. (C) The ratio of the amount of HSV-1 BamHI B/E DNA detected in the native and alkaline gels. The quantity of HSV-1 DNA in indicated samples shown in panel B was quantified using the virion DNA standard. The ratio of quantified HSV DNA between the TAE gel and alkaline gel is calculated. At bottom, the ratio values were normalized so that the D0 sample ratio is set to 1.
DNA, isolated from virions or from HSV-1 infected, NGF-differentiated PC12 cells at different times after infection, was digested with restriction enzyme Bam HI. We expected to see less intensity from the alkaline gel after hybridization; therefore one-third of the digested product was loaded in TAE gel and two-thirds of the digested product was loaded in alkaline gel. The gels were resolved, blotted, hybridized with the probes generated using HSV BamHI B/E fragments, and scanned using a Bio-Rad phosphoimager to generate images (Figure 5 panel B) and to quantify each band. It was previously suggested that nicks and gaps are present in the virion DNA (Ecker and Hyman, 1981; Frenkel and Roizman, 1972; Hyman et al, 1977; Kieff et al, 1971; Wilkie, 1973). The nicks and gaps were represented by a slight smear below the B/E fragments in the alkaline gel. To determine the degree of fragmentation by alkaline treatment, we quantified the intact band intensity of B/E fragments. The amount of DNA in each lane was calculated based on the virion standard shown in the table in panel C. The ratio of the intensity of the image corresponding to each band between TAE gel and alkaline gel was calculated and normalized based on the ratio obtained from Day 0. The higher the normalized ratio, the more alkaline labile sites existed.
Strikingly, HSV-1 DNA derived from NGF-differentiated cells at later stages of infection was almost undetectable (highest normalized ratio, 26.97, as compared to D0, which is set as 1) when resolved through alkaline gels (comparing lane “D30” with lane D0). The loss of signal corresponding to the Bam H1 B/E fragments or the increase in normalized ratio values seemed to correlate with the length of time following HSV infection, with sample intensity declining progressively from day 1 to almost the level of detection at day 30. These data show that DNA derived from quiescently infected PC12 cells accumulates alkaline-labile lesions as a function of time in neuronal-like cultures and that the refractoriness to PCR may be due to this accumulation of DNA lesions.
Viral DNA derived from latently infected mice is refractory to PCR
Because viral DNA derived from NGF-differentiated PC12 cells in tissue culture becomes refractory to PCR (compared with a similar amount of viral DNA derived from virions), it was possible that the refractoriness was tissue culture specific and not indicative of a phenomenon occurring during HSV-1 latency in vivo. Thus, it was of interest to determine if viral DNA derived from latently infected animals becomes refractory to PCR.
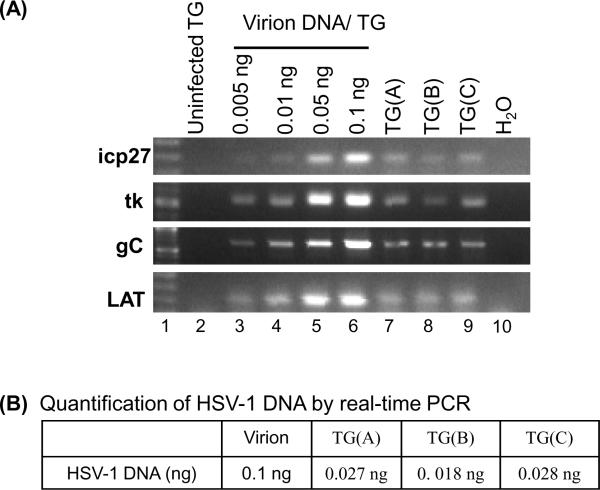
Similar amounts of viral DNA (as quantified by Southern blot analysis) isolated from virions and from the trigeminal ganglia of latently infected Balb/c mice 45 days after inoculation (see Methods) with either strain 17 or strain F were subjected to PCR. Despite the fact that the viral DNA derived from virions and murine ganglia migrates as a smear, indicating some degree of degradation, similar amounts could be obtained and used. The results of the comparative PCRs are shown in Figure 6A. As before, primers specific for 4 different regions of the viral genome were used. Compared to virion DNA, viral DNA derived from latently infected ganglia was much more refractory to PCR [compare Fig 6A lane “1ng virion” to latent HSV DNA, lanes “TG(A), TG(B) and TG(C)”]. Refractoriness to PCR was also shown using real-time PCR (panel B). Clearly, the amount of viral DNA derived from latently infected TG was consistently underestimated by PCR reaction (3- to 4-fold less compare to virion DNA).
Figure 6.
HSV-1 DNA from latently infected mice is refractory to PCR. DNA was isolated from TG and the amount of HSV-1 DNA in the TG DNA sample was quantified by Southern blot hybridization as shown in Figure 3. TG(A), TG(B), and TG(C) DNA samples were isolated from TGs of mice infected with 3 × 104, 3 × 105, and 4 × 107 PFU of HSV-1 strain F, respectively. HSV-1 DNA (1 ng) from each TG DNA sample were subjected to PCR with primers specific for HSV-1 ICP27, TK, gC, and Lat genes. PCR was also conducted using varying amounts of virion DNA reconstituted with 4.6 μg of uninfected TG DNA (lanes 3 – 6) and 4.6 μg of uninfected TG DNA (lane 2). PCR products derived from each reaction were analyzed on a 1% agarose gel, stained with ethidium bromide, and photographed. (B) Quantification of HSV-1 DNA by real-time PCR. Viral DNA (1 ng) of each sample and a range of concentrations of virion DNA standards were subjected to real-time PCR assay with primers specific for ICP0. The amount of viral DNA in each TG sample quantified by real-time PCR is shown.
These results from latently infected mice are similar to those obtained using DNA from the quiescently infected PC12 cells (shown in Figure 3). Taken together, it is clear that viral DNA derived from latently infected mice is a poorer substrate for PCR than similar amount of viral DNA derived from virions.
Discussion
In this study, PCR analysis was used to show that viral DNA from quiescently infected PC12 cells is physically altered compared with that from virions used to establish the infection. Furthermore, it was shown that this change occurred in a time dependent fashion after infection. The physical alteration revealed by PCR analysis of quiescently infected PC12 cells was also determined to be present in viral DNA derived from latently infected mice. The physical alterations were also corroborated by analysis of the intactness of the viral genome using alkaline agarose gel electrophoresis, which suggested an accumulation of alkaline-labile lesions in HSV-1 DNA during quiescence.
We believe that the refractoriness of HSV-1 DNA to PCR is consistent with the accumulation of significant DNA damage. PCR involves processive polymerase synthesis along a template; it is influenced by both nucleotide composition and integrity. It thus can be taken as an indication of the intactness of a substrate. The viral DNA derived from latently infected cells could be mutated such that primers designed to hybridize to the virion-derived DNA no longer efficiently hybridized. However, LM-PCR results suggested that elongation was more affected than priming.
Because the appearance of refractoriness to PCR is a function of the length of time that the viral DNA is maintained in quiescent cultures, we believe that NGF-differentiated PC12 DNA is not inherently more refractory to PCR, thus making the result unlikely to be a technical artifact associated with the DNA extraction. However we can not rule out the possibility that long-term infected DNA is more sensitive to extraction. Also, previous work with these cultures showed that infectious virus was produced during the first several days of infection, after which it ceased (Su et al, 1999). Our detection of the accumulation of damage in the 20 to 30 day post infection time frame places the majority of damage introduction well beyond the time period of bulk DNA replication, supporting our belief that the damage was not a byproduct of an early technical event involving aspects of the NGF differentiation and infection protocols.
The results from our alkaline gel electrophoresis analysis clearly show that viral DNA from quiescently infected cells was, indeed, much more sensitive to alkaline fragmentation than was DNA from virions. Also, the degree to which the DNA was fragmented increased as a function of time in culture, supporting the results with PCR. The mobility of the HSV DNA restriction fragments through alkaline gels confirmed that viral DNA from long-term infected cultures does accumulate alkaline-labile lesions and that these lesions alone could account for refractoriness to PCR. However, since the kinetics of PCR refractoriness and alkaline degradation did not precisely match, it may be that the observed PCR effect was multifactorial.
Since alkaline lability could be caused by different types of DNA damage including nicked and abasic DNA, the accumulation of alkaline labile lesions could be due to several mechanisms. For instance, certain base modifications such as alkylation may produce highly labile glycosidic bonds resulting in higher hydrolytic depurination and greater alkaline sensitivity over time (Lawley and Brookes, 1963). It may be that HSV DNA synthesized and encapsidated during productive infection contains more of these base modifications than is typical in cellular DNA. The effect of alkylative (or oxidative) damage on PCR has been shown to be variable depending on the type of damage (Sikorsky et al, 2004). Alkaline lability could also be caused by nicks introduced into the DNA backbone. The LM-PCR data suggests that nicks may be a significant source of damage since nicked DNA, but not abasic site containing DNA, would be capable of the ligation step of this experiment. Ligatable DNA ends should be necessary to generate smaller than full-length LM-PCR products observed. The loss of full-length product, on the other hand, could occur due to the presence of abasic sites, nicks or other polymerase stopping lesions. The other side of this equation is the possibility of an impairment of DNA repair mechanisms in neuronal cells since damage accumulation would be a net accumulation of damage surviving DNA repair processes in infected cells. Preliminary evidence from our work suggests that damaged HSV-1 DNA was repaired less effectively than host DNA in quiescently infected NGF-differentiated cultures (Su, in progress).
It was also of interest to determine whether this phenomenon of PCR refractoriness occurred on host DNA. We noted that amplification of cellular (host) sequences derived from the long-term infected PC12 cells and PC12 cell DNA prepared shortly after plating in culture was similar (data not shown). This result suggests that the refractoriness to PCR amplification was specific for the viral DNA that had been maintained in the long-term cultures and was not a general property of the preparation or of sample handling. This observation suggests that viral DNA accumulated more lesions than did the host DNA. The damaged state of host TG DNA during latency in vivo was not examined in our studies and is complicated by the fact that only a subpopulation of cells in the tissue harbour latent virus, and pooled DNA preparations would contain a mixture of all DNA from neuronal and non-neuronal populations. As such, the damaged status of the host TG DNA remains undetermined.
We resolved HSV DNA restriction fragments through native and denaturing alkaline gels, following restriction endonuclease digestion to reduce the possibility of artifacts due to damage resulting from handling full length genomes. We found that HSV-1 DNA restriction fragments from viral DNA derived from quiescently infected NGF-differentiated PC12 cells, 30 days after infection, migrated through alkaline gels with a pattern consistent with significant fragmentation and nicking with almost no intact restriction fragments detectable. This despite the fact that nondenaturing gel electrophoresis showed the presence of significant amounts of viral DNA. It is clear that either some intact viral genomes remain, or the damaged genomes can be repaired, since all of our cultures could still reactivate infectious viruses (data not shown).
HSV DNA from virions has been reported to contain single-stranded nicks and gaps (Ecker and Hyman, 1981; Frenkel and Roizman, 1972; Hyman et al, 1977; Kieff et al, 1971; Wilkie, 1973). However, these nicks are generally believed to be repaired shortly after lytic infection. The host DNA repair pathway has been shown to play a crucial role in viral DNA replication during the period of productive infection (Iwahori et al, 2007; Lilley et al, 2005; Lilley et al, 2007; Muylaert and Elias, 2007; Shirata et al, 2005; Wilkinson and Weller, 2004; Wilkinson and Weller, 2005; Wilkinson and Weller, 2006). For example, HSV infection was found to elicit a cellular DNA damage response, with activation of the ataxia-telangiectasia-mutated (ATM) signal transduction pathway (Shirata et al, 2005) and thus resulted in the hypophosphorylation of transcription factor Sp1 (Shirata et al, 2005), while dismantling the ataxia telangiectasia-mutated and Rad3-related (ATR)-dependent DNA damage responses during the lytic cycle (Wilkinson and Weller, 2006).
Many cellular proteins that are involved in DNA recombination and repair are recruited to the sites of HSV DNA replication (Wilkinson and Weller, 2004). The activation of cellular DNA damage response has been sown to aid viral replication in non-neuronal cells (Lilley et al, 2005). Similarly, knockdown of host repair protein DNA ligase IV/XRCC4 greatly inhibits HSV replication (Muylaert and Elias, 2007). Interestingly, the activation of the DNA damage response was abrogated in neurons (Lilley et al, 2005), suggesting that virus might differentially manipulate the DNA repair machinery to favor lytic cycle or latency. However, little is known about how the host repair system maintains the integrity of the latent viral genome and its possible role in reactivation. Recent work implicated a new DNA damage response function for the HSV-1 ICP0 protein (Lilley et al). These studies showed how the ICP0 protein can target for degradation, cellular factors involved in the mobilization of DNA repair complexes to damaged cellular DNA. Such a function was suggested by the authors to possibly regulate transcriptional repression of viral genes and therefore, may modulate the course of latency and reactivation.
Interestingly, much of the work relating to HSV-1 modulation of the host DNA damage response has excluded direct examination of DNA. Therefore, while we believe that it may be premature to extrapolate the biological significance of the data presented here, we do think that in combination with the recent work of others, it necessitates a closer inspection of the viral DNA to see how perturbations of the cellular DNA damage response affect the maintenace of viral genomic integrity and how the regulation of DNA integrity may participate in transcriptional activity and ultimately viral replication. Although slower repair to damaged HSV-1 DNA could help explain the accumulation of lesions, it also remains possible that HSV-1 DNA is inherently more labile than host DNA for reasons involving DNA covalent modifications and chromatin effects. It could also be targeted for degradation via unknown mechanisms. Though additional studies are required to ascertain the biological ramifications of our findings, the implications of the accumulation of unrepaired lesions in latency and pathogenesis of HSV-1 could be significant.
Supplementary Material
Acknowledgements
This work was supported by National Institute of Health Grant NS 33768 and an appropriation from The Commonwealth of Pennsylvania through The Institute for Hepatitis and Virus Research.
References
- Abner CW, McKinnon PJ. The DNA double-strand break response in the nervous system. DNA Repair (Amst) 2004;3:1141–7. doi: 10.1016/j.dnarep.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Batterson W, Furlong D, Roizman B. Molecular genetics of herpes simplex virus. VIII. further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks PJ. DNA repair in neural cells: basic science and clinical implications. Mutat Res. 2002;509:93–108. doi: 10.1016/s0027-5107(02)00222-1. [DOI] [PubMed] [Google Scholar]
- Cline SD, Hanawalt PC. Who's on first in the cellular response to DNA damage? Nat Rev Mol Cell Biol. 2003;4:361–72. doi: 10.1038/nrm1101. [DOI] [PubMed] [Google Scholar]
- Dammann R, Pfeifer GP. Lack of gene- and strand-specific DNA repair in RNA polymerase III-transcribed human tRNA genes. Mol Cell Biol. 1997;17:219–29. doi: 10.1128/mcb.17.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker JR, Hyman RW. Analysis of interruptions in the phosphodiester backbone of herpes simplex virus DNA. Virology. 1981;110:213–6. doi: 10.1016/0042-6822(81)90024-6. [DOI] [PubMed] [Google Scholar]
- Francisconi S, Codenotti M, Ferrari-Toninelli G, Uberti D, Memo M. Preservation of DNA integrity and neuronal degeneration. Brain Res Brain Res Rev. 2005;48:347–51. doi: 10.1016/j.brainresrev.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Fraser NW, Lawrence WC, Wroblewska Z, Gilden DH, Koprowski H. Herpes simplex type 1 DNA in human brain tissue. Proc Natl Acad Sci U S A. 1981;78:6461–5. doi: 10.1073/pnas.78.10.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel N, Roizman B. Separation of the herpesvirus deoxyribonucleic acid duplex into unique fragments and intact strand on sedimentation in alkaline gradients. J Virol. 1972;10:565–72. doi: 10.1128/jvi.10.4.565-572.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci U S A. 1976;73:2424–8. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Feng Z, Chasin LA, Tang MS. Transcription-coupled and transcription-independent repair of cyclobutane pyrimidine dimers in the dihydrofolate reductase gene. J Biol Chem. 2002;277:38305–10. doi: 10.1074/jbc.M206375200. [DOI] [PubMed] [Google Scholar]
- Hyman RW, Oakes JE, Kudler L. In vitro repair of the preexisting nicks and gaps in herpes simplex virus DNA. Virology. 1977;76:286–94. doi: 10.1016/0042-6822(77)90303-8. [DOI] [PubMed] [Google Scholar]
- Iwahori S, Shirata N, Kawaguchi Y, Weller SK, Sato Y, Kudoh A, Nakayama S, Isomura H, Tsurumi T. Enhanced phosphorylation of transcription factor sp1 in response to herpes simplex virus type 1 infection is dependent on the ataxia telangiectasia-mutated protein. J Virol. 2007;81:9653–64. doi: 10.1128/JVI.00568-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L, Linn S. A reduced rate of bulky DNA adduct removal is coincident with differentiation of human neuroblastoma cells induced by nerve growth factor. Mol Cell Biol. 1988;8:3964–8. doi: 10.1128/mcb.8.9.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy PG, Clements GB. Reactivation of latent herpes simplex virus from dissociated identified dorsal root ganglion cells in culture. J Gen Virol. 1983;64(Pt 7):1629–35. doi: 10.1099/0022-1317-64-7-1629. [DOI] [PubMed] [Google Scholar]
- Kieff ED, Bachenheimer SL, Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971;8:125–32. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley PD, Brookes P. Further Studies on the Alkylation of Nucleic Acids and Their Constituent Nucleotides. Biochem J. 1963;89:127–38. doi: 10.1042/bj0890127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley CE, Carson CT, Muotri AR, Gage FH, Weitzman MD. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc Natl Acad Sci U S A. 2005;102:5844–9. doi: 10.1073/pnas.0501916102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley CE, Chaurushiya MS, Boutell C, Landry S, Suh J, Panier S, Everett RD, Stewart GS, Durocher D, Weitzman MD. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. EMBO J. 2010;29:943–55. doi: 10.1038/emboj.2009.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley CE, Schwartz RA, Weitzman MD. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 2007;15:119–26. doi: 10.1016/j.tim.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Maggioncalda J, Mehta A, Su YH, Fraser NW, Block TM. Correlation between herpes simplex virus type 1 rate of reactivation from latent infection and the number of infected neurons in trigeminal ganglia. Virology. 1996;225:72–81. doi: 10.1006/viro.1996.0576. [DOI] [PubMed] [Google Scholar]
- McMurray CT. To die or not to die: DNA repair in neurons. Mutat Res. 2005;577:260–74. doi: 10.1016/j.mrfmmm.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Mitchell BM, Bloom DC, Cohrs RJ, Gilden DH, Kennedy PG. Herpes simplex virus-1 and varicella-zoster virus latency in ganglia. J Neurovirol. 2003;9:194–204. doi: 10.1080/13550280390194000. [DOI] [PubMed] [Google Scholar]
- Miyamae Y, Iwasaki K, Kinae N, Tsuda S, Murakami M, Tanaka M, Sasaki YF. Detection of DNA lesions induced by chemical mutagens using the single-cell gel electrophoresis (comet) assay. 2. Relationship between DNA migration and alkaline condition. Mutat Res. 1997;393:107–13. doi: 10.1016/s1383-5718(97)00091-0. [DOI] [PubMed] [Google Scholar]
- Moxley MJ, Block TM, Liu HC, Fraser NW, Perng GC, Wechsler SL, Su YH. Herpes simplex virus type 1 infection prevents detachment of nerve growth factor-differentiated PC12 cells in culture. J Gen Virol. 2002;83:1591–600. doi: 10.1099/0022-1317-83-7-1591. [DOI] [PubMed] [Google Scholar]
- Muylaert I, Elias P. Knockdown of DNA ligase IV/XRCC4 by RNA interference inhibits herpes simplex virus type I DNA replication. J Biol Chem. 2007;282:10865–72. doi: 10.1074/jbc.M611834200. [DOI] [PubMed] [Google Scholar]
- Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–6. [PubMed] [Google Scholar]
- Nouspikel T, Hanawalt PC. Terminally differentiated human neurons repair transcribed genes but display attenuated global DNA repair and modulation of repair gene expression. Mol Cell Biol. 2000;20:1562–70. doi: 10.1128/mcb.20.5.1562-1570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouspikel T, Hanawalt PC. DNA repair in terminally differentiated cells. DNA Repair (Amst) 2002;1:59–75. doi: 10.1016/s1568-7864(01)00005-2. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Dammann R. Measuring the formation and repair of UV photoproducts by ligation-mediated PCR. Methods Mol Biol. 1999;113:213–26. doi: 10.1385/1-59259-675-4:213. [DOI] [PubMed] [Google Scholar]
- Renlund M, Chester MA, Lundblad A, Aula P, Raivio KO, Autio S, Koskela SL. Increased urinary excretion of free N-acetylneuraminic acid in thirteen patients with Salla disease. Eur J Biochem. 1979;101:245–50. doi: 10.1111/j.1432-1033.1979.tb04237.x. [DOI] [PubMed] [Google Scholar]
- Roizman B, Knipe DM, Whitley RJ. Herpes Simplex Viruses. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 2501–2601. [Google Scholar]
- Roizman B, Sears AE. An inquiry into the mechanisms of herpes simplex virus latency. Annu Rev Microbiol. 1987;41:543–71. doi: 10.1146/annurev.mi.41.100187.002551. [DOI] [PubMed] [Google Scholar]
- Shirata N, Kudoh A, Daikoku T, Tatsumi Y, Fujita M, Kiyono T, Sugaya Y, Isomura H, Ishizaki K, Tsurumi T. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J Biol Chem. 2005;280:30336–41. doi: 10.1074/jbc.M500976200. [DOI] [PubMed] [Google Scholar]
- Sikorsky JA, Primerano DA, Fenger TW, Denvir J. Effect of DNA damage on PCR amplification efficiency with the relative threshold cycle method. Biochem Biophys Res Commun. 2004;323:823–30. doi: 10.1016/j.bbrc.2004.08.168. [DOI] [PubMed] [Google Scholar]
- Su YH, Meegalla RL, Chowhan R, Cubitt C, Oakes JE, Lausch RN, Fraser NW, Block TM. Human corneal cells and other fibroblasts can stimulate the appearance of herpes simplex virus from quiescently infected PC12 cells. J Virol. 1999;73:4171–80. doi: 10.1128/jvi.73.5.4171-4180.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YH, Moxley M, Kejariwal R, Mehta A, Fraser NW, Block TM. The HSV 1 genome in quiescently infected NGF differentiated PC12 cells can not be stimulated by HSV superinfection. J Neurovirol. 2000;6:341–9. doi: 10.3109/13550280009030760. [DOI] [PubMed] [Google Scholar]
- Su YH, Moxley MJ, Ng AK, Lin J, Jordan R, Fraser NW, Block TM. Stability and circularization of herpes simplex virus type 1 genomes in quiescently infected PC12 cultures. J Gen Virol. 2002;83:2943–50. doi: 10.1099/0022-1317-83-12-2943. [DOI] [PubMed] [Google Scholar]
- Sutherland BM, Bennett PV, Sutherland JC. Methods in Molecular Biology: DNA Repair Protocols: Mammalian Systems. Humana Press Inc.; Totowa, NJ: 1999. DNA damage quantification by gel electrophoresis; pp. 251–271. [Google Scholar]
- Sutherland BM, Georgakilas AG, Bennett PV, Laval J, Sutherland JC. Quantifying clustered DNA damage induction and repair by gel electrophoresis, electronic imaging and number average length analysis. Mutat Res. 2003;531:93–107. doi: 10.1016/j.mrfmmm.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Tu Y, Tornaletti S, Pfeifer GP. DNA repair domains within a human gene: selective repair of sequences near the transcription initiation site. EMBO J. 1996;15:675–83. [PMC free article] [PubMed] [Google Scholar]
- Wagner EK, Bloom DC. Experimental investigation of herpes simplex virus latency. Clin Microbiol Rev. 1997;10:419–43. doi: 10.1128/cmr.10.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley RJ, Gnann J. The epidemiology and clinical manifestations of herpes simplex virus infections. In: Roizman B, Whitley RJ, editors. The Human Herpes Viruses. Raven Press; New York City, NY: 1993. pp. 69–105. [Google Scholar]
- Wilkie NM. The synthesis and substructure of herpesvirus DNA: the distribution of alkali-labile single strand interruptions in HSV-1 DNA. J Gen Virol. 1973;21:453–67. doi: 10.1099/0022-1317-21-3-453. [DOI] [PubMed] [Google Scholar]
- Wilkinson DE, Weller SK. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J Virol. 2004;78:4783–96. doi: 10.1128/JVI.78.9.4783-4796.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DE, Weller SK. Inhibition of the herpes simplex virus type 1 DNA polymerase induces hyperphosphorylation of replication protein A and its accumulation at S-phase-specific sites of DNA damage during infection. J Virol. 2005;79:7162–71. doi: 10.1128/JVI.79.11.7162-7171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DE, Weller SK. Herpes simplex virus type I disrupts the ATR-dependent DNA-damage response during lytic infection. J Cell Sci. 2006;119:2695–703. doi: 10.1242/jcs.02981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–9. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Pao A, Adair GM, Tang M. Cyclobutane pyrimidine dimers and bulky chemical DNA adducts are efficiently repaired in both strands of either a transcriptionally active or promoter-deleted APRT gene. J Biol Chem. 2001;276:16786–96. doi: 10.1074/jbc.M010973200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.