Abstract
Oligo/polynucleotide-based gene targeting strategies provide new options for achieving sequence-specific modification of genomic DNA and have implications for the development of new therapies and transgenic animal models. One such gene modification strategy, small fragment homologous replacement (SFHR), was evaluated qualitatively and quantitatively in human lymphoblasts that contain a single base substitution in the hypoxanthine-guanine phosphoribosyl transferase (HPRT1) gene. Because HPRT1 mutant cells are readily discernable from those expressing the wild type (wt) gene through growth in selective media, it was possible to identify and isolate cells that have been corrected by SFHR. Transfection of HPRT1 mutant cells with polynucleotide small DNA fragments (SDFs) comprising wild type HPRT1 (wtHPRT1) sequences resulted in clones of cells that grew in hypoxanthine-aminopterin-thymidine (HAT) medium. Initial studies quantifying the efficiency of correction in 3 separate experiments indicate frequencies ranging from 0.1% to 2%. Sequence analysis of DNA and RNA showed correction of the HPRT1 mutation. Random integration was not indicated after transfection of the mutant cells with an SDF comprised of green fluorescent protein (GFP) sequences that are not found in human genomic DNA. Random integration was also not detected following Southern blot hybridization analysis of an individual corrected cell clone.
Introduction
The development of genetic treatments for inherited disorders has been the focus for biomedical research for over 3 decades. While there have been significant advances in understanding the potential of various gene therapy strategies, researchers have also gained insight into their limitations. Although cDNA-based genetic therapy strategies employed currently have shown some success, these strategies have encountered problems such as insertional mutagenesis, which have limited their universal application to resolve pathologies associated with inherited diseases (Cavazzana-Calvo et al., 2004). To circumvent limitations inherent in cDNA-based strategies, alternative oligo/polynucleotide-based gene targeting approaches have been developed (Yanez and Porter, 1998; Richardson et al., 2002; Gruenert et al., 2003). One such approach using polynucleotides, small fragment homologous replacement (SFHR), has been shown to modify endogenous genomic loci responsible for cystic fibrosis (CF) (Kunzelmann et al., 1996; Goncz et al., 1998, 2001, 2002; Bruscia et al., 2002; Sangiuolo et al., 2002), Duchenne muscular dystrophy (DMD) (Kapsa et al., 2001, 2002; Todaro et al., 2007), spinal muscular (SMA) (Sangiuolo et al., 2005), severe combined immune deficiency (SCID) (Zayed et al., 2006), sickle cell disease (SCD) (Goncz et al., 2002, 2006, McNab et al., 2007), and β-thalassemia (Colosimo et al., 2007) as well as reporter/selectable genes in plasmids (Thorpe et al., 2000, 2002; Colosimo et al., 2001; Tsuchiya et al., 2005a, 2005b, 2005c). The mechanism(s) underlying SFHR are not well understood and, given the disease associated genomic targets, it has been difficult to quantify and optimize the efficiency of SFHR-mediated modification.
The present study evaluates SFHR-mediated modification and characterizes SFHR efficiency, following the transfection of human lymphoblasts with single- or double-stranded small DNA fragments (SDFs) designed to correct mutations in the hypoxanthine-guanine phosphoribosyl transferase (HPRT1) gene. While mutations in the HPRT1 genes are associated with Lesch-Nyhan syndrome (Nyhan and Wong, 1996; Nicklas et al., 2000), the HPRT1 gene has also been effectively used to measure the mutagenic potential of environmental agents and chemical carcinogens (Albertini et al., 1996). The HPRT1 gene product is a phosphoribosylation enzyme involved in the nucleotide salvage pathway. Phosphoribosylation of purine analogues such as 6-thioguanine (6-TG) by HPRT results in inhibition of cell proliferation and subsequent cell death, allowing cells containing mutant HPRT1 to be readily separated from the wild type cells by their ability to proliferate in the presence of TG. Conversely, cells with wtHPRT1 proliferate in medium containing a cocktail of hypoxanthine/aminopterin/thymidine (HAT), while cells with mutant HPRT1 perish.
In the studies reported here, transfection of mutant lymphoblasts with a wild type SDF (wtSDF), a minimum frequency that ranged from 0.1% to 2% of HAT resistant (HATr) was observed. This preliminary finding is several orders of magnitude higher than was observed previously using a similar gene targeting strategies (Hunger-Bertling et al., 1990; Kenner et al., 2002, 2004) and may reflect that apparently small differences can have a significant effect on gene targeting efficiencies. Since previous studies have indicated that homologous recombination is inhibited by wt p53 (Mekeel et al., 1997; Lu et al., 2003; Yoon et al., 2004; Saintigny et al., 2005; Gatz and Wiesmuller, 2006), the fact that these cells express wt p53 suggests that SFHR may be independent of the recombinatorial pathway, which is regulated by p53.
Materials and Methods
Cells and culture conditions
Human male lymphoblasts, TK6 (Liber and Thilly, 1982) and LT1-1B1 (Branda et al., 2001), were used for these experiments. The LT1-1B1 was derived after treatment of the Epstein Barr virus immortalized lymphoblastoid cell line, TK6 with the mutagen ethyl methanesulfonate, and contains a G>C transversion at base 152 in exon 3 of the HPRT1I gene and that inactivates the HPRT protein. Cultures were grown in RPMI1640 medium supplemented with 20% fetal bovine serum (FBS; UCSF Cell Culture Facility, CA) that contained either 10 μM 6-TG or HAT (hypoxanthine: 100 μM, aminopterin: 0,4 μM, thymidine: 16 μM; Sigma-Aldrich, St Louis, MO) or without any selection agent at 37°C under humidified conditions in 5% CO2. Successful conversion of HPRT1− > HPRT1+ was determined by proliferation in HAT medium, PCR screening of DNA and RNA, Southern blot hybridization, RFLP analysis, and sequencing.
SDF design and production
The wtHPRT1 SDF (wtSDF) includes the wt-sequence for exon 3 (as well as the adjacent intron sequences; Table 1). The SDFs were generated by PCR amplification of cloned genomic DNA segment encompassing the region of homology defined SDF. The template for SDF preparation was derived from normal (TK6) cell DNA and cloned into the pCRII-TOPO plasmid (Invitrogen, Carlsbad, CA) for sequence confirmation for further SDF production. The primer pair used to generate the 579-bp SDF were: HPX3S579 (5′-CCTTATGAAACATGAGG-3′, sense) and HPX3A579 (5′-CCTACTGTTGCCACTAAAAAGAA-3′, antisense). The PCR amplification conditions were: initial denaturation, 95°C/2 minutes followed by 25 cycles: denaturation, 95°C/30 seconds, annealing, 65°C/30 seconds, extension, 72°C/1 minutes with a 7-minutes extension in the final cycle. The PCR mixture (100 μL) contained: 200 μM dNTPs, 200 nM each primer, 1X Taq Polymerase Buffer (50 mM KCl, 10 mM Tris-HCl, pH 8.3), 2 mM MgCl2 and 0.5 units of Taq polymerase (Applied Biosystems, Foster City, CA). The resultant wtSDFs were transfected either as double stranded DNA (dsDNA) or as heat denatured complementary single stranded DNA (ssDNA).
Table 1.
The Coding Sequences for Exons 2–4 of the HPRT Gene
| Exon | cDNA | Size (bp) | Coding sequence |
|---|---|---|---|
| 2 | 28–134 | 107 | att agt gat gat gaa cca ggt tat gac ctt gat tta ttt tgc ata cct aat cat tat gct gag gat ttg gaa agg gtg ttt att cct cat gga cta att atg gac ag |
| 3 | 135–318 | 184 | g act gaa cgt ctt gct c C a gat gtg atg aag gag atg gga ggc cat cac att gta gcc ctc tgt gtg ctc aag ggg ggc tat aaa ttc ttt gct gac ctg ctg gat tac atc aaa gca ctg aat aga aat agt gat aga tcc att cct atg act gta gat ttt atc aga ctg aag agc tat tgt |
| 4 | 319–384 | 66 | aat gac cag tca aca ggg gac ata aaa gta att ggt gga gat gat ctc tca act tta act gga aag |
The highlighted base indicates the LT1-1B1 G>C mutation in exon 3 at base 152 of the cDNA.
Transfection
Electroporation was carried out using the Amaxa nucleofection system (Amaxa Biosystems, Gaithersburg, MD). The Amaxa system was evaluated for the appropriate Nucleofection Program/Buffer combination to give optimal transfection/viability. Briefly, SDFs (106 and 107 SDFs/cell) were added to 106 cells suspended in 100 μL of Nucleofection Buffer T. The cell-SDF mixture was then electroporated with Program G16 in 1 cm transfection cuvettes. After electroporation, cells were transferred to dishes for selection. The survival of the cells after electroporation was ~80% and the transfection efficiency based on a GFP reporter plasmid was ~75% (data not shown).
Post-transfection cell selection
While the cells can be expected to produce HPRT+ enzyme in at least one doubling after transfection, DNA replication may be involved in SFHR-mediated modification (unpublished observations, B Bedayat and DC Gruenert), so the cells were grown for 48 hours without selection and then plated in T-25 flasks or in 12-well plates (Falcon, BD Biosciences, San Jose, CA) in HAT. Nontransfected control cells grown in HAT were used to assay spontaneous reversion of LT1-1B1 cells to HAT positive (HAT+) cells. To quantify the frequency of SFHR-mediated modification, cells were grown at densities of 105,104, 103, 102, and 10 cells per well in HAT medium in 12-well plates. The medium was changed everyday for 5 days and then every other day for up to 21 days. Plates were then harvested and analyzed.
Analysis of DNA and mRNA
In addition to the phenotypic selection for HATr, the DNA and RNA were analyzed to assess SFHR-mediated HPRT1 correction. DNA and RNA were isolated using the QIAGEN (Hilden, Germany) DNeasy Blood & Tissue Kit and the RNeasy Mini Kit, respectively. Since corrected cells regain an HPRT1-specific XhoI (CTCGAG)/TaqI (TCGA) cleavage site, restriction fragment length polymorphism (RFLP) analysis was used to characterize conversion to wtHPRT1. RFLP analysis was carried out after PCR, reverse transcriptase PCR (RT-PCR), Southern blot hybridization, and then confirmed by sequencing.
Genomic DNA analysis
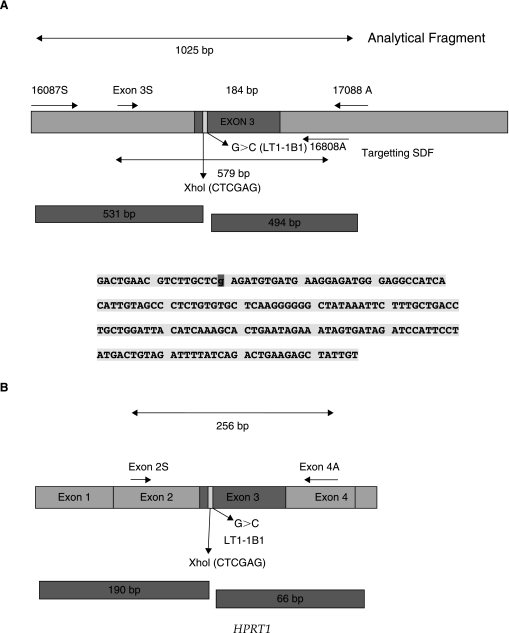
PCR and RFLP analysis was performed on the DNA from HAT+ cells. The genomic DNA was gel purified and then PCR amplified. The primer pair was outside the region of homology defined by the SDF to minimize, if not eliminate, the potential for amplification of unincorporated SDFs (Maurisse et al., 2006). Primers 16087S (5′-GCCCAGTTTCTACAGTCTCTCTTA-3′, sense) and 17088A (5′-TGACTCCCCACAAACCGATGTAGCTCAAGA-3′, anti-sense) flank the SDF region (Fig. 1A).
FIG. 1.
(A) Schematic of the target region encompassing HPRT1 Exon 3 and the PCR strategy for generating the 579-bp targeting SDF. The strategy for the assessment of genomic DNA targeting as well as the RFLP analysis of the 1025-bp diagnostic product is also depicted. The sequence represents Exon 3 and the LT1-1B1 cell line 152G>C (R51P) mutation. The mutation is indicated as (“g” = 152 G > C). (B) Schematic representation of the RT-PCR and RFLP analysis of HPRT1 mRNA. The XhoI digestion for the 256-bp amplicon will result in a 190- and 66-bp fragment.
RT/PCR
RNA was isolated and treated with DNase I to remove any possible DNA/SDF contamination (Maurisse et al., 2006). The DNase-treated RNA was reverse transcribed and then PCR amplified with primers in exons 2 and 4 (Ex2S, 5′-TGCTGAGGATTTGGAAAGGGTG-3′/Ex4A, 5′-GTCCCC TGTTGACTGGTCATTACA-3′), thereby crossing intron/exon boundaries (Fig. 1B; Table 1). Briefly, a pellet of 1 × 104 cells was resuspended in 5 μL of first-strand cDNA synthesis buffer (50 mM Tris-HCl pH 8.55, 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 500 μM of each of the dNTPs, 0.1 μg/μL BSA, 10 ng/μL oligo(dT)12–18 primer, 1 U/μL RNasin, 2.5 U/μL M-MuLV-RT, and 2.5% NP-40) and incubated at 37°C for 1 hours. The first-strand cDNA was then used as a template for PCR with the TaqI buffer (2.75 mM MgCl2, 60 mM KCl, 15 mM Tris-HCl, pH8.55), dNTPs at 400 uM each, 0.5 μL Ex2S (HPRT1 base pair −60 to −41) and Ex4A (HPRT1 base pair 721 to 702) (100 ng/μL), and 0.5 μL Taq polymerase). The conditions for PCR were as follows: 94°C for 5 minutes; followed by: denaturation at 94°C for 1 minutes, annealing at 55°C for 1 minutes and polymerization at 72°C for 2 minutes for 30 cycles with a final extension at 72°C for 7 minutes. Amplicons were subjected to digestion with XhoI and banded on 1% agarose gels.
Southern blot hybridization
Genomic DNA was isolated as described above and digested with TaqI. The DNA was then separated by electrophoresis on a 0.8% agarose gel and transferred to a Hybond N+ membrane (Amersham-Phamacia Biotech, Piscataway, NJ) for hybridization with an SDF-derived, 309-bp 32P-labeled probe as described previously (Chang and Kan, 1981). After an overnight hybridization at 65°C, the filters were washed once at room temperature for 20 minutes 2× SSC and then for at least 1 hour in fresh 2× SSC plus 1% SDS, and finally in 0.1× SSC plus 0.1% SDS at 65°C for 20 minutes. The filter was then evaluated on a Typhoon Phosphorimager (Molecular Dynamics, Sunnyvale, CA).
DNA sequencing
PCR products were directly sequenced. In preparation for sequencing, PCR products were gel-purified and further purified with Geneclean (Q-biogene, Montreal, Canada). Aliquots of 0.5 to 2 μL of the first round RT-PCR product were transferred to a new 0.5 mL tube containing 48 to 49.5 μL of Taq buffer that includes primer B: HPRT1 base pair −36 to −17 and primer A: HPRT1 base pair 701 to 682) for a second 30 cycle round of PCR as described. The product was then sequenced with primers A (5′-CACTATATTGCCCAGGTTGGT-3′: sense) and B (5′-TCATATATTAAATATACTCACACAATAGCT-3′: anti-sense). Sequencing carried out at Biothec Core, Sunnyvale, CA on an ABI 3100 Sequencer.
Assessment of random integration
In a separate experiment designed to detect genomic integration of residual free SDF present in the LT1-1B1 cells, the cells were transfected with an SDF comprising a green fluorescent protein (GFP) (GFP-SDF) sequence not present in the genomic DNA of the LT1-1B1 cells (Table 2). DNA was isolated from the transfected cells at different times post-transfection with and without gel purification (Maurisse et al., 2006). Nested PCR, using primers within the sequence defined by the GFP-SDF (Table 2), was carried out to detect whether there was any GFP sequence associated with the genomic DNA by amplifying a GFP-specific sequence smaller than that of the SDF. To ensure that the gel purified DNA was not degraded, an additional PCR was carried out using primers HPRTF2F and HPRTF2R to amplify a 337-bp segment of exon 3 of the HPRT1 gene (Table 2). The PCR conditions for generating the GFP-SDF were: initial denaturation: 94°C for 120 seconds followed by 35 cycles of; denaturation: 94°C for 30 seconds; annealing: 49°C for 30 seconds; elongation: 68°C for 60 seconds with a final elongation: 68°C for 120 seconds. The conditions for generating the smaller fragment for detection of GFP sequences in the cell and the genome as well as to test the quality of the genomic DNA were: initial denaturaion: 94°C for 120 seconds followed by 35 cycles of; denaturation: 94°C for 30 seconds; annealing: 56°C for 30 seconds; elongation: 68°C for 30 seconds with a final elongation: 68°C for 120 seconds.
Table 2.
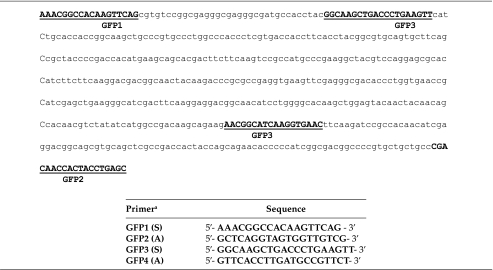
GFP-SDF: Generation and Analysis
GFP-SDF sequence (531-bp) with the primer placement. Lower panel indicates the primer sequence. Primer pair GFP1/GFP2 were used to generate the SDF and primer pair GFP3/GFP4 were used to detect GFP sequence.
S, sense; A, antisense.
Results
The G>C mutation at base152 of exon 3 in the HPRT1 gene results in conversion of an arginine to proline at codon (amino acid) 51 and renders the LT1-1B1 cells resistant to 6-TG (Branda et al., 2001). To ensure that mutant status of the LT1-1B1 cells was retained, the cells were grown in 10 μM 6-TG for at least 21 days prior to transfection. The cells were then transfected by Amaxa nucleofection to introduce wtHPRT1 ds- and ssSDFs into 106 cells at concentrations of 106 or 107 SDFs per cell.
Following transfection, the cells were allowed to recover in basal RPMI1640 medium for 10 minutes. They were then transferred to supplemented growth medium without HAT. After 48 hours the cells were transferred to complete medium containing HAT. After 21 days the cells were harvested and genotypically evaluated.
The frequency of SFHR-mediated conversion of the LT1-1B1 was determined by plating 105,104, 103, 102, and 10 cells per well in 10 wells of a 12-well plate immediately after transfection. The cells were cultured as described above in supplemented RPMI1640 medium for 48 hours after electroporation and then in HAT medium for 21 days. In 3 separate experiments, all the wells in the 105, 104, and 103 cells/well plates had cell growth after 3 weeks. In 2 experiments, plates containing 102 cells/well, all wells were positive for growth. In the 10 cells/well dish, the results of individual experiments showed 1, 0, and 2 wells with cells growing, respectively. In the experiment in which there were no cells at the 10 cells/well concentration, only 1 well of the 100 cells/well was positive. These results indicate that the frequency of SFHR-mediated modification ranged from a minimum of 0.1% to 2% (Table 3). No cell growth was detected in the control nontransfected cultures after HAT exposure at any of the cell densities.
Table 3.
SFHR-Mediated Correction of an HPRT Mutation in Human Lymphoblasts
| Experiments with correctiona | Minimal number of SFHR-corrected wellsb | Minimal SFHR correction efficiencyc |
|---|---|---|
| 1 | ND | ND |
| 2 | ND | ND |
| 3 | 2 wells of 10 with 10 cells/well | 0.02 |
| 4 | 1 well of 10 with 100 cells/well | 0.001 |
| 5 | 1 well of 10 with 10 cells/well | 0.01 |
3 of 5 positive experiments were analyzed for efficiency of correction.
SDF transfected cells were plated into 10 wells of a 12 well plate at densities of 1000, 100, or 10 cells per well.
The minimal frequency/efficiency was based on the fact that there had to be at least 1 corrected cell per well in 10 wells seeded with either 100 or 10 cells per well.
Abbreviation: ND, not detected.
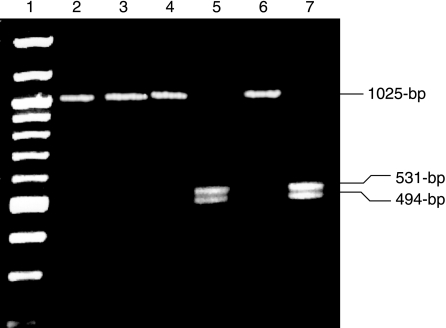
When the genomic DNA of the control TK6 cells and the wtSDF transfected LT1-1B1 cells that were able to grow in HAT was analyzed by PCR with a primer pair 16087S and 17088A (Fig. 1A), XhoI digestion of the resultant 1025-bp PCR product yielded products of 531- and 494-bp fragments for both the wt TK6 cells and the HAT+ LT1-1B1 cells transfected with wtSDF (Fig. 2). PCR amplification products from the mutant parental LT1-1B1 cell line were resistant to digestion by XhoI. Sequence analysis of the region homologous and adjacent to the ends of the SDF gave the expected sequence with either a C (mutant) or a G (wt) at base 152 (data not shown).
FIG. 2.
PCR/RFLP analysis of genomic DNA from transfected cells. The 1025-bp amplification product was either digested with XhoI (lanes 3, 5, and 7) or not digested (lanes 2, 4, and 6). LT1-1B1 cell DNA was isolated before transfection (lanes 2 and 3) and after transfection (lanes 4 and 5). Lanes 6 and 7 were derived from control TK6 cell DNA. Lane 1 is the 100-bp molecular weight marker.
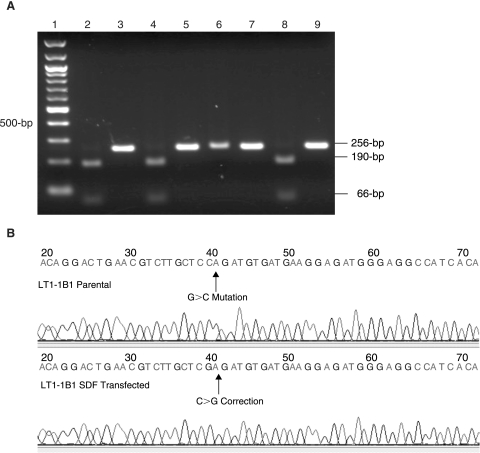
Analysis of the HPRT1 mRNA gave further confirmation of the SDF-mediated conversion of the mutant HPRT1 to the wt allele (Fig. 1B). XhoI digestion of the 256-bp RT-PCR product resulted in 66- and 190-bp fragments derived from the transfected HAT+ cells and the wt TK6 controls (Fig. 3A), while those derived from the untransfected parental LT1-1B1 line remained uncut. Direct sequencing of the HPRT1 mRNA-derived cDNA confirmed the presence of the wtHPRT1 sequence, that is, a C>G conversion, at the mutant locus (Fig. 3B).
FIG. 3.
(A) RT-PCR/RFLP analysis of mRNA-derived HPRT1 cDNA from LT1-1B1 cells transfected with ssSDFs (lanes 2 and 3), LT1-1B1 cells transfected with dsSDFs (lanes 4 and 5), mutant LT1-1B1 controls (lanes 6 and 7), and HPRT wt TK6 cells (lanes 8 and 9). The 256-bp product from LT1-1B1 cells transfected with ssSDFs (lanes 2 and 3) and from LT1-1B1 cells transfected with dsSDFs (lanes 4 and 5) were digested with XhoI (lanes 2 and 4). Assessment of mRNA from untransfected mutant LT1-1B1 controls (lanes 6 and 7) and HPRT wt TK6 cells (lanes 8 and 9) showed no XhoI digestion of the LT1-1B1 cell cDNA (lane 6) and the expected XhoI cleavage of the TK6 cDNA (lane 8). The RT-PCR products in lanes 3, 5, 7, and 9 are not digested by XhoI and serve as controls. (B) Sequence analysis of the mRNA-derived cDNA.
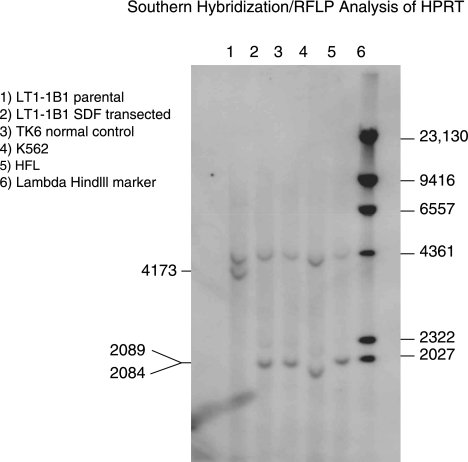
A clone from LT1-1B1 cells transfected with 106 ssSDFs/cell was analyzed by Southern blot hybridization. An individual cell was isolated from one of the wells of the 12-well dish that contained 10 cells/well and was grown for an additional 3 months as a single cell clone. Since the mutation in the LT1-1B1 cells eliminates the XhoI (CTCGAG) and TaqI (TCAG) restriction sites, it is possible to characterized wtHPRT1 sequence by RFLP analysis of the genomic DNA. DNA isolated from LT1-1B1 parental, SDF-transfected LT1-1B1, TK6, K562, and human fetal liver were digested with TaqI and analyzed by Southern blot hybridization (Fig. 4). Only the parental LT1-1B1 cells showed the absence of the HPRT1 specific TaqI restriction site that resulted in a single 4173-bp band. All the other cell lines including the SDF-transfected LT1-1B1 cells showed RFLP patterns consistent with wtHPRT1 sequence. There were also no other bands detected in the transfected clone, indicating that there was no spurious random integration of the SDF in this corrected clone.
FIG. 4.
Southern blot hybridization RFLP analysis of genomic HPRT1 digested with TaqI. The blot was hybridized with a 309-bp 32P-labeled probe. DNAs were as follows: lane 1, untransfected LT1-1B1; lane 2, ssSDF transfected LT1-1B1; lane 3, TK6 cells (normal lymphoblast control); lane 4, K562 (human erythroleukemia cell line, normal control); lane 5, HFL (human fetal liver normal control); and lane 6, lambda HindIII DNA marker. DNA from untransfected LT1-1B1 cells gave a 4173-bp band, while all the DNA derived from HPRT1wt cells and the SFHR corrected cells showed the product bands 2089- and 2084-bp resulting from TaqI digestion at the site of the corrected mutation.
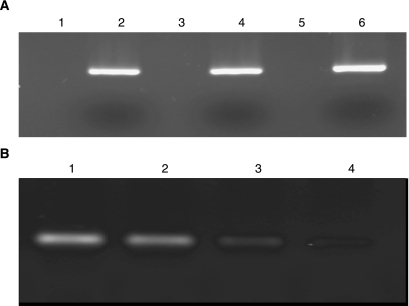
The question of random integration was further pursued by transfecting the cells with a 531-bp GFP-SDF that comprised sequences not present in the genomic DNA of the LT1-1B1 cells. PCR analysis of the genomic DNA isolated from cells at 1, 2, 3, and 4 weeks post-transfection showed that the fragment disappeared from the cells to barely detectable levels after 4 weeks (Fig. 5A). When gel purified DNA was analyzed to ensure that there was no unintegrated fragment associated with the genomic DNA, there were no GFP sequences detected even though there was no difficulty detecting HPRT1 gene sequences (Fig. 5B). This result is an additional indication that random integration by the SDFs is not a high probability event.
FIG. 5.
Nested PCR analysis of genomic DNA with and without gel purification following transfection with the GFP-SDF. (A) Lanes 1, 3, and 5 show the product of the GFP3/GFP4 amplification using gel purified genomic DNA as a template 1, 2, and 4 weeks post-transfection, respectively. Lack of amplification is in support of the absence of random integration. The quality of genomic DNA was ensured in an independent amplification using primers coding for a known genomic sequence. Lanes 2, 4, and 6 ascertain the gel purified genomic DNA quality with HPRT1 primers (5′-CACAGTTCACTCCAGCCTCA-3′, sense, 5′-CCAGCAGGTCAGCAAAGAAT-3′, antisense) at weeks 1, 2, and 4 post-transfection, respectively. (B) The GFP3/GFP4 PCR analysis of genomic DNA that was not gel purified indicates that the GFP-SDF disappears from the cells as a function of time. Lanes 1–4 indicates the presence of GFP sequences on weeks 1–4 post-transfection, respectively. At 4 weeks post-transfection GFP-SDF could barely be detected.
Discussion
The assessment and verification of sequence-specific modification of genomic DNA has been complicated by the fact that the genomic targets to be modified generally involve endogenous genes that are not readily amenable to enrichment strategies. As a result, the quantification of sequence-specific modification is often based on indirect measurements. While SFHR has been shown to be effective at modifying genomic loci associated with various diseases (Kunzelmann et al., 1996; Goncz et al., 1998, 2006; Kapsa et al., 2001; Gruenert et al., 2003; Sangiuolo et al., 2005, 2008; Zayed et al., 2006; Colosimo et al., 2007; Todaro et al., 2007), quantification of its effectiveness at genomic modification have been limited to nonselectable systems and densitometric allele-specific hybridization analysis of PCR fragments or PCR–based quantification (Kunzelmann et al., 1996; Kapsa et al., 2001; Goncz et al., 2006; Todaro et al., 2007). These studies indicate that SFHR can induce sequence-specific gene modification at frequencies that generally range between 1% and 15%.
Studies have also used mutant heterologous reporter or selectable marker genes that are either episomal plasmids or randomly integrated into the host cell genome to assess the frequency of SFHR-mediated homologous replacement (Thorpe et al., 2000, 2002; Colosimo et al., 2001; Tsuchiya et al., 2005a, 2005b). The limitation with these approaches is that they do not accurately reflect the genomic context of an endogenous gene. The sequence-specific modification of an extrachromosomal target appears to involve enzymatic pathways that are distinct from those involved with modification of genomic chromatin (Yanez and Porter, 1999, 2002). Not only do the extrachromosomal target plasmids not have the same association with the nuclear matrix as the host genome, but they are also generally cDNA based and do not have the intron/exon structure associated with most eukaryotic genes. It is, therefore, more likely for new serendipitous mutations to be introduced while the original target mutation is corrected, and would lead to an underestimation of the frequency of correction.
The assessment of SFHR-mediated correction of mutations in the endogenous HPRT1 gene circumvents the issues associated with extrachromosomal targets and/or genomic cDNA targets. The studies presented here indicate that SFHR-mediated correction has occurred by genotypic and phenotypic criteria. The growth of the LT1-1B1 cells transfected with the wtHPRT1 SDF in HAT medium is a clear indicator that the inactivation induced by the G>C mutation in exon 3 has been reversed. Confirmation that this mutation was corrected and that the phenotypic correction was not the result of a secondary compensatory mutations was demonstrated by the RFLP analysis of both RT-PCR products and genomic DNA. While the data presented here indicate an efficiency that ranges between 0.1% and 2% that is consistent with the efficiencies observed by other groups studying SFHR (Gruenert et al., 2003), several studies evaluating SSOs or shorter DNA fragments in rodent cells (Kenner et al., 2002, 2004) or human lymphoblasts (Hunger-Bertling et al., 1990) have observed efficiencies of 10−4 to 10−5 and 6 × 10−6, respectively. The higher frequencies observed in this study may reflect differences in the cells used, the size of the correcting DNA fragments, and/or the methods of transfection. Further in-depth evaluation of the parameters influencing SFHR-mediated homologous exchange and homologous exchange in general are required to address these differences and provide insight into the elements responsible for the variations in the efficiencies observed.
While the isolation of a clone of corrected cells indicated that there was no evidence of random or Alu-directed integration (due to the Alu sequences in the SDF) into the genomic DNA, many more clones will need to be assessed whether, and at what level, random integration occurs. Studies using a GFP-SDF that has no genomic homologue, indicated that there was no random integration of this fragment up to 4 weeks after transfection (Fig. 5).
This study presents insights into SFHR-mediated modification of endogenous genes in chromatin and demonstrates that human cells defective in the endogenous HPRT1 gene can be effectively used for assessing the frequency. While the studies suggest that random integration is low at best, the possibility that it occurs below the level of detection of the analytical systems used cannot be dismissed. Future studies should provide additional insight into the mechanistic parameters that influence the SFHR efficacy and the possibility that low levels of random integration may occur.
Acknowledgments
We would like to thank Drs. Y.W. Kan, W.B. Browner, and R. Albertini for their input and support. We would also like to acknowledge the support of NIH Grants R01 GM75111 (D.C.G., J.A.N., H.P., and B.B.), NIH T32 DK007636 (A.A.), and R21 DK66403 (D.C.G., R.M.), as well as the California Pacific Medical Center Research Foundation.
References
- ALBERTINI R.J. NICKLAS J.A. O'NEILL J.P. Future research directions for evaluating human genetic and cancer risk from environmental exposures. Environ. Health Perspect. 1996;104:503–510. doi: 10.1289/ehp.96104s3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRANDA R.F. O'NEILL J.P. BROOKS E.M. TROMBLEY L.M. NICKLAS J.A. The effect of folate deficiency on the cytotoxic and mutagenic responses to ethyl methanesulfonate in human lymphoblastoid cell lines that differ in p53 status. Mutat. Res. 2001;473:51–71. doi: 10.1016/s0027-5107(00)00138-x. [DOI] [PubMed] [Google Scholar]
- BRUSCIA E. SANGIUOLO F. SINIBALDI P. GONCZ K.K. NOVELLI G. GRUENERT D.C. Isolation of CF cell lines corrected at DeltaF508-CFTR locus by SFHR-mediated targeting. Gene Ther. 2002;9:683–685. doi: 10.1038/sj.gt.3301741. [DOI] [PubMed] [Google Scholar]
- CAVAZZANA-CALVO M. THRASHER A. MAVILIO F. The future of gene therapy. Nature. 2004;427:779–781. doi: 10.1038/427779a. [DOI] [PubMed] [Google Scholar]
- CHANG J.C. KAN Y.W. Antenatal diagnosis of sickle cell anaemia by direct analysis of the sickle mutation. Lancet. 1981;2:1127–1129. doi: 10.1016/s0140-6736(81)90584-5. [DOI] [PubMed] [Google Scholar]
- COLOSIMO A. GONCZ K.K. NOVELLI G. DALLAPICCOLA B. GRUENERT D.C. Targeted correction of a defective selectable marker gene in human epithelial cells by small DNA fragments. Mol. Ther. 2001;3:178–185. doi: 10.1006/mthe.2000.0242. [DOI] [PubMed] [Google Scholar]
- COLOSIMO A. GUIDA V. ANTONUCCI I. BONFINI T. STUPPIA L. DALLAPICCOLA B. Sequence-specific modification of a beta-thalassemia locus by small DNA fragments in human erythroid progenitor cells. Haematologica. 2007;92:129–130. doi: 10.3324/haematol.10560. [DOI] [PubMed] [Google Scholar]
- GATZ S.A. WIESMULLER L. p53 in recombination and repair. Cell Death Differ. 2006;13:1003–1016. doi: 10.1038/sj.cdd.4401903. [DOI] [PubMed] [Google Scholar]
- GONCZ K.K. COLOSIMO A. DALLAPICCOLA B. GAGNE L. HONG K. NOVELLI G. PAPAHADJOPOULOS D. SAWA T. SCHREIER H. WIENER-KRONISH J. XU Z. GRUENERT D.C. Expression of DeltaF508 CFTR in normal mouse lung after site-specific modification of CFTR sequences by SFHR. Gene Ther. 2001;8:961–965. doi: 10.1038/sj.gt.3301476. [DOI] [PubMed] [Google Scholar]
- GONCZ K.K. KUNZELMANN K. XU Z. GRUENERT D.C. Targeted replacement of normal and mutant CFTR sequences in human airway epithelial cells using DNA fragments. Hum. Mol. Genet. 1998;7:1913–1919. doi: 10.1093/hmg/7.12.1913. [DOI] [PubMed] [Google Scholar]
- GONCZ K.K. PROKOPISHYN N.L. ABDOLMOHAMMADI A. BEDAYAT B. MAURISSE R. DAVIS B.R. GRUENERT D.C. Small fragment homologous replacement-mediated modification of genomic beta-globin sequences in human hematopoietic stem/progenitor cells. Oligonucleotides. 2006;16:213–224. doi: 10.1089/oli.2006.16.213. [DOI] [PubMed] [Google Scholar]
- GONCZ K.K. PROKOPISHYN N.L. CHOW B.L. DAVIS B.R. GRUENERT D.C. Application of SFHR to gene therapy of monogenic disorders. Gene Therapy. 2002;9:691–694. doi: 10.1038/sj.gt.3301743. [DOI] [PubMed] [Google Scholar]
- GRUENERT D.C. BRUSCIA E. NOVELLI G. COLOSIMO A. DALLAPICCOLA B. SANGIUOLO F. GONCZ K.K. Sequence-specific modification of genomic DNA by small DNA fragments. J. Clin. Invest. 2003;112:637–641. doi: 10.1172/JCI19773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUNGER-BERTLING K. HARRER P. BERTLING W. Short DNA fragments induce site specific recombination in mammalian cells. Mol. Cell Biochem. 1990;92:107–116. doi: 10.1007/BF00218128. [DOI] [PubMed] [Google Scholar]
- KAPSA R. QUIGLEY A. LYNCH G.S. STEEPER K. KORNBERG A.J. GREGOREVIC P. AUSTIN L. BYRNE E. In vivo and in vitro correction of the mdx dystrophin gene nonsense mutation by short-fragment homologous replacement. Hum. Gene Ther. 2001;12:629–642. doi: 10.1089/104303401300057324. [DOI] [PubMed] [Google Scholar]
- KAPSA R.M. QUIGLEY A.F. VADOLAS J. STEEPER K. IOANNOU P.A. BYRNE E. KORNBERG A.J. Targeted gene correction in the mdx mouse using short DNA fragments: towards application with bone marrow-derived cells for autologous remodeling of dystrophic muscle. Gene Therapy. 2002;9:695–699. doi: 10.1038/sj.gt.3301737. [DOI] [PubMed] [Google Scholar]
- KENNER O. KNEISEL A. KLINGLER J. BARTELT B. SPEIT G. VOGEL W. KAUFMANN D. Targeted gene correction of hprt mutations by 45 base single-stranded oligonucleotides. Biochem. Biophys. Res. Commun. 2002;299:787–792. doi: 10.1016/s0006-291x(02)02749-3. [DOI] [PubMed] [Google Scholar]
- KENNER O. LUTOMSKA A. SPEIT G. VOGEL W. KAUFMANN D. Concurrent targeted exchange of three bases in mammalian hprt by oligonucleotides. Biochem. Biophys. Res. Commun. 2004;321:1017–1023. doi: 10.1016/j.bbrc.2004.07.062. [DOI] [PubMed] [Google Scholar]
- KUNZELMANN K. LEGENDRE J.Y. KNOELL D.L. ESCOBAR L.C. XU Z. GRUENERT D.C. Gene targeting of CFTR DNA in CF epithelial cells. Gene Therapy. 1996;3:859–867. [PubMed] [Google Scholar]
- LIBER H. THILLY W. Mutation assay at the thymidine kinase locus in diploid human lymphoblasts. Mutat. Res. 1982;94:467–482. doi: 10.1016/0027-5107(82)90308-6. [DOI] [PubMed] [Google Scholar]
- LU X. LOZANO G. DONEHOWER L.A. Activities of wildtype and mutant p53 in suppression of homologous recombination as measured by a retroviral vector system. Mutat. Res. 2003;522:69–83. doi: 10.1016/s0027-5107(02)00261-0. [DOI] [PubMed] [Google Scholar]
- MAURISSE R. FICHOU Y. DE SEMIR D. CHEUNG J. FEREC C. GRUENERT D.C. Gel purification of genomic DNA removes contaminating small DNA fragments interfering with polymerase chain reaction analysis of small fragment homologous replacement. Oligonucleotides. 2006;16:375–386. doi: 10.1089/oli.2006.16.375. [DOI] [PubMed] [Google Scholar]
- McNAB G.L. AHMAD A. MISTRY D. STOCKLEY R.A. Modification of gene expression and increase in alpha1-antitrypsin (alpha1-AT) secretion after homologous recombination in alpha1-AT-deficient monocytes. Hum Gene Ther. 2007;18:1171–1177. doi: 10.1089/hum.2007.073. [DOI] [PubMed] [Google Scholar]
- MEKEEL K.L. TANG W. KACHNIC L.A. LUO C.M. DEFRANK J.S. POWELL S.N. Inactivation of p53 results in high rates of homologous recombination. Oncogene. 1997;14:1847–1857. doi: 10.1038/sj.onc.1201143. [DOI] [PubMed] [Google Scholar]
- O'NEILL J.P. JINNAH H.A. HARRIS J.C. NYHAN W.L. Lesch-Nyhan Syndrome geneReviews. 2009. www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=gene&part=lns www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=gene&part=lns
- NYHAN W. WONG D. New approaches to understanding Lesch-Nyhan disease. N. Engl. J. Med. 1996;334:1602–1604. doi: 10.1056/NEJM199606133342411. [DOI] [PubMed] [Google Scholar]
- RICHARDSON P.D. KREN B.T. STEER C.J. Gene repair in the new age of gene therapy. Hepatology. 2002;35:512–518. doi: 10.1053/jhep.2002.32421. [DOI] [PubMed] [Google Scholar]
- SAINTIGNY Y. BERTRAND P. LOPEZ B.S. [Direct role of p53 on homologous recombination] Med. Sci. (Paris) 2005;21:43–48. doi: 10.1051/medsci/200521143. [DOI] [PubMed] [Google Scholar]
- SANGIUOLO F. BRUSCIA E. SERAFINO A. NARDONE A.M. BONIFAZI E. LAIS M. GRUENERT D.C. NOVELLI G. In vitro correction of cystic fibrosis epithelial cell lines by small fragment homologous replacement (SFHR) technique. BMC Med. Genet. 2002;3:8. doi: 10.1186/1471-2350-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANGIUOLO F. FILARETO A. SPITALIERI P. SCALDAFERRI M.L. MANGO R. BRUSCIA E. CITRO G. BRUNETTI E. DE FELICI M. NOVELLI G. In vitro restoration of functional SMN protein in human trophoblast cells affected by spinal muscular atrophy by small fragment homologous replacement. Hum. Gene Ther. 2005;16:869–880. doi: 10.1089/hum.2005.16.869. [DOI] [PubMed] [Google Scholar]
- SANGIUOLO F. SCALDAFERRI M.L. FILARETO A. SPITALIERI P. GUERRA L. FAVIA M. CAROPPO R. MANGO R. BRUSCIA E. GRUENERT D.C. Cftr gene targeting in mouse embryonic stem cells mediated by Small Fragment Homologous Replacement (SFHR) Front Biosci. 2008;13:2989–2999. doi: 10.2741/2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THORPE P. STEVENSON B. GOHIL A. PORTEOUS D.J. Towards CFTR gene correction: a comparison of two key strategies. Pediatr. Pulmonal. Suppl. 2000;20:241. [Google Scholar]
- THORPE P. STEVENSON B.J. PORTEOUS D.J. Optimising gene repair strategies in cell culture. Gene Ther. 2002;9:700–702. doi: 10.1038/sj.gt.3301750. [DOI] [PubMed] [Google Scholar]
- TODARO M. QUIGLEY A. KITA M. CHIN J. LOWES K. KORNBERG A.J. COOK M.J. KAPSA R. Effective detection of corrected dystrophin loci in mdx mouse myogenic precursors. Hum. Mutat. 2007;28:816–823. doi: 10.1002/humu.20494. [DOI] [PubMed] [Google Scholar]
- TSUCHIYA H. HARASHIMA H. KAMIYA H. Factors affecting SFHR gene correction efficiency with single-stranded DNA fragment. Biochem. Biophys. Res. Commun. 2005a;336:1194–1200. doi: 10.1016/j.bbrc.2005.08.258. [DOI] [PubMed] [Google Scholar]
- TSUCHIYA H. HARASHIMA H. KAMIYA H. Increased SFHR gene correction efficiency with sense single-stranded DNA. J. Gene Med. 2005b;7:486–493. doi: 10.1002/jgm.673. [DOI] [PubMed] [Google Scholar]
- TSUCHIYA H. SAWAMURA T. HARASHIMA H. KAMIYA H. Correction of frameshift mutations with single-stranded and double-stranded DNA fragments prepared from phagemid/plasmid DNAs. Biol. Pharm. Bull. 2005c;28:1958–1962. doi: 10.1248/bpb.28.1958. [DOI] [PubMed] [Google Scholar]
- YANEZ R.J. PORTER A.C. Gene targeting is enhanced in human cells overexpressing hRAD51. Gene Ther. 1999;6:1282–1290. doi: 10.1038/sj.gt.3300945. [DOI] [PubMed] [Google Scholar]
- YANEZ R.J. PORTER A.C. Differential effects of Rad52p overexpression on gene targeting and extrachromosomal homologous recombination in a human cell line. Nucleic Acids Res. 2002;30:740–748. doi: 10.1093/nar/30.3.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANEZ R.J. PORTER A.C.G. Therapeutic gene targeting. Gene Ther. 1998;5:149–159. doi: 10.1038/sj.gt.3300601. [DOI] [PubMed] [Google Scholar]
- YOON D. WANG Y. STAPLEFORD K. WIESMULLER L. CHEN J. P53 inhibits strand exchange and replication fork regression promoted by human Rad51. J. Mol. Biol. 2004;336:639–654. doi: 10.1016/j.jmb.2003.12.050. [DOI] [PubMed] [Google Scholar]
- ZAYED H. MCIVOR R.S. WIEST D.L. BLAZAR B.R. In vitro functional correction of the mutation responsible for murine severe combined immune deficiency by small fragment homologous replacement. Hum. Gene Ther. 2006;17:158–166. doi: 10.1089/hum.2006.17.158. [DOI] [PubMed] [Google Scholar]