Abstract
Objective
Genetic susceptibility to systemic lupus erythematosus (SLE) is well established, with the HLA class II DRB1 and DQB1 loci demonstrating the strongest association. However, HLA may also influence SLE through novel biologic mechanisms in addition to genetic transmission of risk alleles. Evidence for increased maternal–offspring HLA class II compatibility in SLE and differences in maternal versus paternal transmission rates (parent-of-origin effects) and nontransmission rates (noninherited maternal antigen [NIMA] effects) in other autoimmune diseases have been reported. Thus, we investigated maternal–offspring HLA compatibility, parent-of-origin effects, and NIMA effects at DRB1 in SLE.
Methods
The cohort comprised 707 SLE families and 188 independent healthy maternal–offspring pairs (total of 2,497 individuals). Family-based association tests were conducted to compare transmitted versus nontransmitted alleles (transmission disequilibrium test) and both maternally versus paternally transmitted (parent-of-origin) and nontransmitted alleles (using the chi-square test of heterogeneity). Analyses were stratified according to the sex of the offspring. Maternally affected offspring DRB1 compatibility in SLE families was compared with paternally affected offspring compatibility and with independent control maternal–offspring pairs (using Fisher’s test) and was restricted to male and nulligravid female offspring with SLE.
Results
As expected, DRB1 was associated with SLE (P < 1 × 10−4). However, mothers of children with SLE had similar transmission and nontransmission frequencies for DRB1 alleles when compared with fathers, including those for the known SLE risk alleles HLA–DRB1*0301, *1501, and *0801. No association between maternal–offspring compatibility and SLE was observed.
Conclusion
Maternal–offspring HLA compatibility, parent-of-origin effects, and NIMA effects at DRB1 are unlikely to play a role in SLE.
Systemic lupus erythematosus (SLE) is a systemic autoimmune disease characterized by autoantibodies to nuclear and cell surface antigens. Although the etiology of SLE remains unknown, evidence for genetic susceptibility is well established. The HLA class II alleles DRB1*1501, *0301, and *0801 and the HLA class I alleles A*01 and B*08 in the major histocompatibility complex (MHC) region are consistently associated with SLE.
HLA loci may also influence SLE through additional inherited or noninherited mechanisms. Differences in maternal and paternal transmission rates, or parent-of-origin effects, have not been previously examined in SLE. One potential mechanism influencing disease susceptibility is “genomic imprinting” due to epigenetic modification of the genome. This modification results in unequal transcription of parental alleles and subsequent allele expression, depending on whether alleles were transmitted from the mother or from the father.
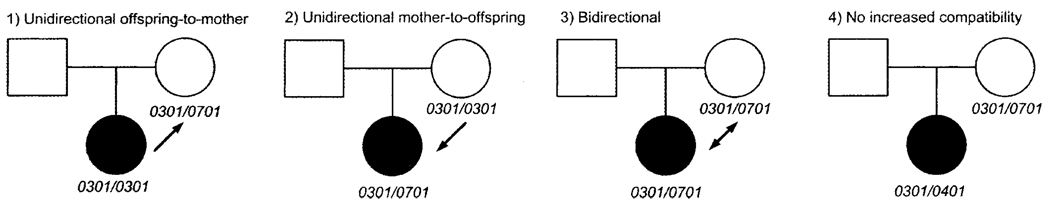
Increased HLA compatibility between a mother and her offspring is hypothesized to contribute to the risk of autoimmune disease. Maternal–offspring effects can present as excess HLA compatibility between the mother and affected offspring (Figure 1). In mice, HLA similarity between mother and fetus has been shown to promote the persistence of maternal cells in the offspring (maternal microchimerism) following pregnancy (1).
Figure 1.
Maternal–offspring HLA compatibility relationships.
The developing immune system of the fetus is also directly exposed to noninherited maternal antigens (NIMAs) in utero (2). Exposure to NIMAs can have a lifelong influence on the immune system and has been theorized to tolerize or predispose to autoimmune reactions. A tolerogenic effect may explain the longer survival of renal transplants from sibling donors expressing NIMAs versus noninherited paternal HLA. Decreased B cell responses to HLA class I NIMAs in humans have been reported (3). Recent data suggest that fetuses may also develop T cell tolerance to NIMAs in utero through tolerogenic fetal regulatory T cells that are maintained throughout life (4).
The aim of this study was to test the hypothesis that the DRB1 locus influences SLE through these novel biologic mechanisms in addition to genetic transmission of particular HLA risk alleles.
SUBJECTS AND METHODS
Subjects
The cohort consisted of 707 European American trio families (2,121 individuals) with 1 SLE-affected child and 2 parents and 188 Dutch healthy maternal–offspring pairs (376 individuals), for a total of 2,497 individuals. The child was female in 93% of the trio families (n = 661) and in 60% of the healthy maternal–offspring pairs (n = 111). All patients met the American College of Rheumatology revised criteria for the classification of SLE (5). Families were enrolled through the University of California, San Francisco (UCSF) (n = 314), the University of Minnesota (n = 233), and the Oklahoma Medical Research Foundation (n = 160), as previously described (6). The ages of the patients ranged from 15 years to 72 years (mean ± SD 45.4 ± 11.2 years), and the mean ± SD age at disease onset was 33.1 ± 13.2 years. Forty percent of the patients with SLE had renal disease or antibodies to double-stranded DNA.
Data were collected by questionnaire and chart review. Parity data were collected by questionnaire for patients recruited through the UCSF, of whom half were nulligravid (never pregnant at, or prior to, diagnosis). Independent controls consisted of healthy maternal–offspring pairs enrolled through Leiden University Medical Center, as previously described; these controls have also been used to study NIMAs in rheumatoid arthritis (RA) (7,8). DRB1 genotypes were generated using sequence-specific oligonucleotide–polymerase chain reaction methodology. Four-digit DRB1 resolution was available for the SLE trios, and 2-digit DRB1 resolution was available for the healthy maternal–offspring pairs.
Although healthy maternal–offspring pairs were Dutch and SLE families were North American, HLA–DRB1 allele frequencies derived from SLE families (nontransmitted alleles or controls) were statistically indistinguishable (P = 0.2) from those in Dutch control mothers, providing strong evidence that population frequencies for both groups were also very similar (data not shown). Furthermore, the family-based nature of all analyses used in the current study greatly reduced the impact of population stratification on the HLA compatibility, parent-of-origin, and NIMA analyses.
Statistical analysis
The transmission disequilibrium test (PLINK v1.02, http://pngu.mgh.harvard.edu/~purcell/plink) was used to examine differences between transmitted and nontransmitted DRB1 alleles in SLE-affected offspring. For compatibility analyses based on DRB1 genotypes, we defined maternal–offspring compatibility categorically, as follows: 1) unidirectional offspring-to-mother compatible, 2) unidirectional mother-to-offspring compatible, 3) bidirectional, and 4) no increased compatibility (Figure 1).
We created 2 × 2 contingency tables to test maternal–offspring DRB1 compatibility with Fisher’s exact test (R v2.6, http://cran.r-project.org), using paternal–offspring compatibility data as controls. The test was conducted using histocompatibility estimates based on 4-digit DRB1 resolution (data not shown) and 2-digit DRB1 resolution. These analyses were repeated with “any” DRB1 compatibility as the exposure (categories 1–3 above were combined). Similarly, we created 2 × 2 contingency tables to test maternal–offspring compatibility with Fisher’s exact test, using DRB1 compatibility from the independent healthy mother–offspring pairs as controls. Here, based on the availability of data, 2-digit DRB1 resolution was used to determine histocompatibility estimates for all analyses. Males and nulligravid females were examined as a separate subgroup. Furthermore, we restricted all of the above analyses to pairs in which offspring were carrying the SLE-associated DRB1*03 or DRB1*15 (for 2-digit DRB1 resolution) or DRB1*0301 or DRB1*1501 (for 4-digit DRB1 resolution) alleles (data not shown). Finally, we checked for an excess of DRB1 homozygotes among the mothers of patients with SLE by examining deviation from Hardy-Weinberg proportions in the pooled set of homozygous genotypes, using the chi-square goodness-of-fit test (PyPop v0.6; http://www.pypop.org).
For the parent-of-origin analyses, the frequencies of maternally transmitted versus paternally transmitted alleles were first derived from trio families using pedigree information and then compared using a chi-square contingency table test for heterogeneity (AFBAC v1.13; http://www.pypop.org/thomsonlab/afbac.html). The same test was used in the NIMA analyses to compare the frequencies of maternally transmitted versus paternally nontransmitted alleles. Both global and allele-specific analyses (when appropriate) were performed. Parent-of-origin and NIMA analyses were stratified by the sex of the offspring.
Statistical power was estimated for the parent-of-origin and NIMA analyses (Quanto v1.2.4; http://hydra.usc.edu/gxe/) assuming a 2-sided Type I error rate of 5% and using control frequencies derived from paternally transmitted and nontransmitted, respectively, DRB1*0301 and DRB1*1501 frequencies.
RESULTS
As expected, DRB1 was strongly associated with SLE (P < 1 × 10−4). DRB1*0301 exhibited the strongest association with SLE (odds ratio [OR] 2.2, 95% confidence interval [95% CI] 1.8–2.7, P = 9 × 10−14). DRB1*1501 was also associated with SLE (OR 1.4, 95% CI 1.1–1.7, P = 0.003). Results for the previously identified SLE-associated DRB1*0801 allele were not significant. DRB1 compatibility was not associated with SLE in the overall sample or in the subset restricted to males and nulligravid females (Table 1). Evidence for excess homozygosity at the DRB1 locus in mothers with SLE was not present.
Table 1.
Maternal–offspring HLA–DRB1 compatibility in SLE families compared with paternal–offspring compatibility (father controls) and independent healthy maternal–offspring pairs (healthy controls)*
| SLE, no. (%) |
Father controls | Healthy controls | |||||
|---|---|---|---|---|---|---|---|
| HLA–DRB1 compatibility | No. (%) | OR (95% CI) | P† | No. (%) | OR (95% CI) | P† | |
| Overall‡ | |||||||
| Unidirectional child-to-parent | 96 (13.6) | 93 (13.2) | 1.01 (0.73–1.41) | 0.94 | 17 (9.0) | 1.32 (0.91–2.98) | 0.10 |
| Unidirectional parent-to-child | 74 (10.5) | 72 (10.2) | 1.01 (0.70–1.46) | 0.99 | 22 (11.7) | 0.85 (0.56–1.68) | 0.89 |
| Bidirectional | 87 (12.3) | 99 (14.0) | 0.87 (0.62–1.20) | 0.38 | 21 (11.2) | 0.65 (0.69–2.08) | 0.61 |
| Not increased | 450 (63.6) | 443 (62.7) | Reference | 128 (68.1) | Reference | ||
| Male and nulligravid female SLE offspring§ | |||||||
| Unidirectional child-to-parent | 27 (13.9) | 25 (12.9) | 0.96 (0.50–1.82) | 0.88 | 17 (9.0) | 1.56 (0.78–3.21) | 0.19 |
| Unidirectional parent-to-child | 15 (7.7) | 19 (9.8) | 0.70 (0.31–1.53) | 0.36 | 22 (11.7) | 0.67 (0.31–1.43) | 0.29 |
| Bidirectional | 22 (11.3) | 35 (18.0) | 0.56 (0.29–1.04) | 0.06 | 21 (11.2) | 1.03 (0.51–2.08) | 0.99 |
| Not increased | 130 (67.0) | 115 (59.3) | Reference | 128 (68.1) | Reference | ||
Histocompatibility was estimated using 2-digit DRB1 typing resolution. SLE = systemic lupus erythematosus; OR = odds ratio; 95% CI = 95% confidence interval.
By Fisher’s exact test.
Comparing 707 maternally affected offspring pairs with 707 paternally affected offspring pairs and 188 independent healthy controls.
Comparing 194 maternally affected offspring pairs in which the affected offspring was either a male or a nulligravid female with 194 paternally affected offspring pairs and 188 independent healthy controls.
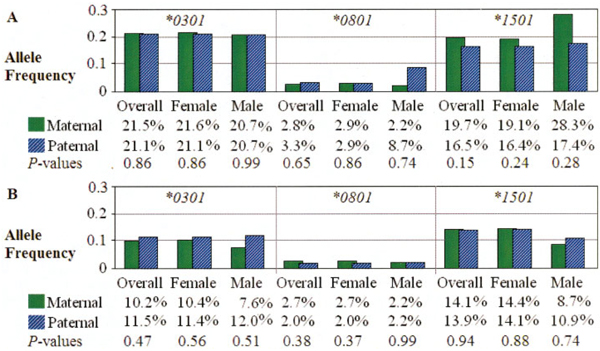
There was no evidence for parent-of-origin and noninherited maternal HLA (NIMA) effects in SLE (data not shown), even when specific SLE risk alleles (DRB1*1501, *0301, and *0801) were considered separately. Comparisons of paternally versus maternally transmitted and paternally versus maternally nontransmitted SLE risk DRB1 alleles in the overall data set, and for male and female patients analyzed separately, demonstrated that the frequencies did not differ (Figure 2).
Figure 2.
HLA–DRB1 susceptibility allele frequencies in systemic lupus erythematosus families. A, Results of tests of parent-of-origin effects (maternally versus paternally transmitted alleles). B, Results of tests of noninherited maternal antigen effects (maternally versus paternally nontransmitted alleles).
DISCUSSION
The MHC confers the strongest genetic effect in SLE known to date; associations are well established for class I and class II MHC loci, particularly for HLA–DRB1*0301– and HLA–DRB1*1501–associated haplotypes. HLA loci may also influence SLE susceptibility through additional inherited or noninherited mechanisms. These hypotheses were tested using a large, well-characterized data set of SLE and control families.
The current study is the largest study to date to examine maternal–offspring HLA compatibility in SLE. Several biologically based hypotheses have been proposed, in which increased compatibility could result in a small number of nonhost cells that could 1) cause dysregulation among host cells, 2) lead to the presentation of nonhost peptides by host cells to other host cells, 3) inactivate T lymphocytes upon interaction, or 4) undergo differentiation and become targets of a later immune response (9). Evidence for increased maternal–offspring HLA class II compatibility has been previously reported for both SLE and systemic sclerosis, suggesting that HLA class II loci may be involved through an undefined pathway dependent on maternal–offspring compatibility (10,11). A recent study demonstrated that maternal–offspring HLA compatibility does not influence the risk of type 1 diabetes mellitus (12).
Stevens et al reported evidence for increased bidirectional DRB1 compatibility for DRB1 allele groups in 30 pairs of mothers and their sons with SLE when compared with 76 independent, healthy mother–son pairs (OR 5.0, 95% CI 1.6–15.7, P = 0.006) (10). When the analyses were restricted to sons carrying DRB1*03 or DRB1*15/16, the results were stronger (OR 7.2, 95% CI 1.6–32.8, P = 0.01) and remained significant when non–European Americans were excluded. In contrast, our results indicate that maternal–offspring DRB1 compatibility does not influence SLE susceptibility. We observed some weak evidence for decreased bidirectional compatibility in male and nulligravid female maternal–offspring pairs compared with paternal–offspring pairs; however, this result did not reach statistical significance (OR 0.56, 95% CI 0.29–1.04, P = 0.06). Larger studies will be required to exclude the possibility of very modest DRB1 compatibility effects on the risk of SLE.
Several factors may have contributed to the disparity between the results of the current study and those reported by Stevens et al. The current study was larger and used both independent and family-based controls for all histocompatibility analyses. In addition, male and nulligravid (never pregnant at, or prior to, diagnosis) female patients were analyzed separately to account for the potential contribution of fetal microchimerism. In contrast, Stevens et al excluded a role for fetal microchimerism by including only mother–son pairs and used an independent control group for comparison. Whereas our study was limited to individuals of European ancestry, the previous study also included African American and Asian American individuals. Finally, SLE cases in our study were derived from trio families, whereas the study by Stevens et al included cases from families with multiple affected individuals. It is possible that 1 or more of these factors, or an undetermined difference in clinical phenotype represented by both groups, may help explain the observed differences. For example, disease differences attributed to familial SLE or subgroups defined by sex, race/ethnicity, or other clinical features such as the presence of particular autoantibodies and/or lupus nephritis may be relevant to studies of histocompatibility and SLE. Finally, to account for the potential contribution of sibling microchimerism, a future study could focus on analyzing male and nulligravid female patients conceived to mothers who had not previously been pregnant.
Parent-of-origin effects, potentially operating through imprinting, have been reported in multiple sclerosis with respect to the inheritance of HLA class II alleles (13). The results of a similar HLA study in type 1 diabetes mellitus were negative (12). Likewise, results from our study do not support a role for DRB1-associated parent-of-origin effects in SLE, even for the known risk alleles DRB1*1501, *0301, and *0801. Additional classic HLA loci were not the focus of the current study and should be included in future studies. Although strong linkage disequilibrium is present between DRB1, DQB1, and DQA1 loci, the association between particular alleles on haplotypes is not complete, and therefore, more may be learned by including additional HLA class II loci, as well as all HLA class I loci, in larger SLE studies.
There is evidence that HLA alleles may act as environmental risk factors. Exposure to HLA NIMAs may therefore shape the immune repertoire of the offspring and either predispose to or protect against future immune reactions. In addition to maternal–offspring cell trafficking and oral exposure through breast milk, NIMA effects may occur through maternal microchimerism. Both risk and protective NIMA effects have been reported for RA (8,14). NIMA effects do not appear to play a strong role in type 1 diabetes mellitus, although some evidence for an association has been reported (12,15). We tested the hypothesis that maternal histocompatibility antigens, specifically those for HLA–DRB1, may contribute to the risk of SLE. Our study did not reveal any evidence for NIMA effects in SLE at the DRB1 locus, even for established risk alleles. The current study had 80% power to detect a modest association (OR ≥1.5) for parent-of-origin or NIMA effects conferred by the SLE risk alleles DRB1*0301 and DRB1*1501. A role for other class I or class II NIMAs in the risk of SLE cannot be excluded.
In conclusion, the results of this large study of SLE families and healthy maternal–offspring pairs do not support a major role for DRB1 in disease susceptibility mediated through maternal–offspring compatibility, parent-of-origin effects, or NIMA effects. Future studies should examine additional classic HLA loci.
ACKNOWLEDGMENTS
We thank Benjamin Goldstein, Farren Briggs, Ira Tager, and Gary Artim.
This study was performed in part at the General Clinical Research Center, Moffitt Hospital, University of California, San Francisco, with funds provided by the NIH (grant 5-M01-RR-00079 from the National Center for Research Resources [NCRR], USPHS). Dr. Bronson’s work was supported by a Graduate Student Achievement Award from the American College of Rheumatology Research and Education Foundation and by the NIH (grant F31-AI-075609 from the National Institute of Allergy and Infectious Diseases [NIAID]). Dr. Harley is recipient of a Kirkland Scholar Award, and his work was supported by the Alliance for Lupus Research, the US Department of Veterans Affairs, and the NIH (grants P01-AR-049084, R01-AR-42460, and N01-AR-62277 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases [NIAMS]; grants R37-AI-24717 and P01-AI-083194 from NIAID; and grant P20-RR-020143 from NCRR). Dr. Moser’s work was supported by the Alliance for Lupus Research, the Lupus Foundation of Minnesota, and the NIH (grant R01-AR-043274-14 from NIAMS). Dr. Gaffney’s work was supported by the NIH (grant 5R01-A1-063274-06 from NIAID). Dr. Criswell is recipient of a Kirkland Scholar Award, and her work was supported by the NIH (grants R01-AR-22804, R01-AR-052300, and K24-AR-02175 from NIAMS).
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Barcellos had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Bronson, Barcellos.
Acquisition of data. Komorowski, May, Noble, Lane, Claas, Seldin, Kelly, Harley, Moser, Gaffney, Behrens, Criswell, Barcellos.
Analysis and interpretation of data. Bronson, Komorowski, Ramsay, Thomson, Seldin, Barcellos.
REFERENCES
- 1.Kaplan J, Land S. Influence of maternal-fetal histocompatibility and MHC zygosity on maternal microchimerism. J Immunol. 2005;174:7123–7128. doi: 10.4049/jimmunol.174.11.7123. [DOI] [PubMed] [Google Scholar]
- 2.Hall JM, Lingenfelter P, Adams SL, Lasser D, Hansen JA, Bean MA. Detection of maternal cells in human umbilical cord blood using fluorescence in situ hybridization. Blood. 1995;86:2829–2832. [PubMed] [Google Scholar]
- 3.Claas FH, Gijbels Y, van der Velden-de Munck J, van Rood JJ. Induction of B cell unresponsiveness to noninherited maternal HLA antigens during fetal life. Science. 1988;241:1815–1817. doi: 10.1126/science.3051377. [DOI] [PubMed] [Google Scholar]
- 4.Mold JE, Michaelsson J, Burt TD, Muench MO, Beckerman KP, Busch MP, et al. Maternal alloantigens promote the development of tolerogenic fetal regulatory T cells in utero. Science. 2008;322:1562–1565. doi: 10.1126/science.1164511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 6.Barcellos LF, May SL, Ramsay PP, Quach HL, Lane JA, Nititham J, et al. High-density SNP screening of the major histocompatibility complex in systemic lupus erythematosus demonstrates strong evidence for independent susceptibility regions. PLoS Genet. 2009;5:e1000696. doi: 10.1371/journal.pgen.1000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dankers MK, Roelen DL, Korfage N, de Lange P, Witvliet M, Sandkuijl L, et al. Differential immunogenicity of paternal HLA class I antigens in pregnant women. Hum Immunol. 2003;64:600–606. doi: 10.1016/s0198-8859(03)00058-2. [DOI] [PubMed] [Google Scholar]
- 8.Feitsma AL, Worthington J, van der Helm-van Mil AH, Plant D, Thomson W, Ursum J, et al. Protective effect of noninherited maternal HLA-DR antigens on rheumatoid arthritis development. Proc Natl Acad Sci U S A. 2007;104:19966–19970. doi: 10.1073/pnas.0710260104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson JL. Microchimerism and human autoimmune diseases. Lupus. 2002;11:651–654. doi: 10.1191/0961203302lu271oa. [DOI] [PubMed] [Google Scholar]
- 10.Stevens AM, Tsao BP, Hahn BH, Guthrie K, Lambert NC, Porter AJ, et al. Maternal HLA class II compatibility in men with systemic lupus erythematosus. Arthritis Rheum. 2005;52:2768–2773. doi: 10.1002/art.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Artlett CM, Welsh KI, Black CM, Jimenez SA. Fetal-maternal HLA compatibility confers susceptibility to systemic sclerosis. Immunogenetics. 1997;47:17–22. doi: 10.1007/s002510050321. [DOI] [PubMed] [Google Scholar]
- 12.Bronson PG, Ramsay PP, Thomson G, Barcellos LF. Analysis of maternal-offspring HLA compatibility, parent-of-origin and noninherited maternal effects for the classical HLA loci in type 1 diabetes. Diabetes Obes Metab. 2009;11 Suppl 1:74–83. doi: 10.1111/j.1463-1326.2008.01006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramagopalan SV, Herrera BM, Bell JT, Dyment DA, DeLuca GC, Lincoln MR, et al. Parental transmission of HLA-DRB1*15 in multiple sclerosis. Hum Genet. 2008;122:661–663. doi: 10.1007/s00439-007-0442-z. [DOI] [PubMed] [Google Scholar]
- 14.Guthrie KA, Tishkevich NR, Nelson JL. Non-inherited maternal human leukocyte antigen alleles in susceptibility to familial rheumatoid arthritis. Ann Rheum Dis. 2009;68:107–109. doi: 10.1136/ard.2008.092312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akesson K, Carlsson A, Ivarsson SA, Johansson C, Weidby BM, Ludvigsson J, et al. The non-inherited maternal HLA haplotype affects the risk for type 1 diabetes. Int J Immunogenet. 2009;36:1–8. doi: 10.1111/j.1744-313X.2008.00802.x. [DOI] [PubMed] [Google Scholar]