Abstract
Purpose
Proapoptotic BH3-only proteins (Bim, Bad, Bid, Puma, and Noxa) initiate apoptosis by binding to regulatory sites on antiapoptotic Bcl-2 proteins, directly neutralizing their cytoprotective function. Expression of these proteins in colon cancer patients may account for differences in recurrence and survival rates.
Experimental Design
Archival tumor-node-metastasis stage II and III primary colon carcinomas from patients treated in 5-fluorouracil – based adjuvant therapy trials were studied. Immunohistochemical analysis of Bim, Puma, and Noxa proteins was done using tissue microarrays (n = 431). Immunoscores were determined and correlated with clinicopathologic variables and disease-free survival (DFS) and overall survival (OS) rates.
Results
Elevated expression of proapoptotic Bim (hazard ratio, 0.65; 95% confidence interval, 0.44–0.97; P = 0.033) and Puma (hazard ratio, 0.59; 95% confidence interval, 0.37–0.93; P = 0.022), but not Noxa, proteins in the tumor cytoplasm was significantly associated with more favorable OS in a univariate analysis, and elevated Bim expression was also associated with better DFS (P = 0.023). Patient age, tumor stage, and histologic grade were also prognostic. Multivariate Cox analysis showed that Bim (DFS, P = 0.030; OS, P = 0.045) and Puma (OS, P = 0.037) expression were independent predictors of OS after adjustment for histologic grade, tumor stage, age, and treatment. Furthermore, the combined variable of Bim and Puma was highly discriminant for both DFS (P = 0.0034) and OS (P = 0.0011).
Conclusions
The proapoptotic BH3-only proteins Bim and Puma can provide prognostic information for stage II and III colon cancer patients receiving 5-fluorouracil – based adjuvant chemotherapy. Furthermore, our results support BH3-only proteins as molecular targets of novel anticancer drugs.
Colorectal cancer is the second leading cause of cancer-related death in the United States and the fourth leading cause worldwide (1). Clinical and pathologic staging information are the only data currently used to determine prognosis and to select patients to receive adjuvant chemotherapy. However, considerable stage-independent variability in patient outcome creates a need for prognostic markers to identify high-risk patients who may benefit from adjuvant chemotherapy and to spare others unnecessary toxicity. The ability of tumor cell populations to expand in number is determined by the rates of cell proliferation and apoptosis. Evasion of apoptosis is a hallmark of human cancers that leads to cancer development, progression, and treatment resistance (2). In response to intracellular damage signals, including those evoked by cancer therapy, the fate of a cell is determined by the Bcl-2 protein family that includes two types of proapoptotic molecules and prosurvival Bcl-2 proteins (Bcl-2, Bcl-xL Bcl-w, Mcl-1, and A1; ref. 3). The first type is BH3-only proteins (Bad, Bid, Bim, Puma, and Noxa) that are sensors of cellular damage and can engage and inactivate prosurvival Bcl-2 family members. They do so by inserting their BH3 domain into a groove on the prosurvival proteins to negate their cytoprotective function (4, 5). This interaction allows the second type of proapoptotic molecules (i.e., Bax and Bak) to be activated, which commits the cell to apoptosis by permeabilizing the outer mitochondrial membrane to enable cytochrome c release, which promotes caspase activation (6). Studies indicate that BH3-only proteins bind promiscuously or selectively to prosurvival Bcl-2 proteins (6–8). Bim (Bcl-2–interacting mediator of cell death) and Puma (p53 up-regulated modulator of apoptosis) have been shown to target all prosurvival protein and, accordingly, are more potent inducers of apoptosis in vitro than are Bad and Noxa that target only a subset (7).
The balance between BH3-only and prosurvival Bcl-2 proteins regulates tissue homeostasis and studies in animal models indicate that loss of proapoptotic genes can be oncogenic, as can up-regulation of prosurvival Bcl-2 family members (9, 10). Most tumors, including colorectal cancers, overexpress prosurvival Bcl-2 proteins (11) and also have defects in the p53 pathway (12). Puma and Noxa are transcriptional targets of p53 and play a role in p53-induced apoptosis (13, 14). A further understanding of the role of BH3-only proteins in tumorigenesis, including their expression levels and mechanisms of inactivation, is needed. To date, only sparse data are available about the expression of BH3-only proteins in normal and malignant human epithelia and their clinical significance remains unknown.
BH3-only proteins trigger apoptosis in response to signals elicited by activated oncogenes, DNA damage, chemotherapy drugs, and irradiation (3). Studies in cells from knockout mice confirm that BH3-only proteins have a dominant role in the response to diverse cytotoxic stimuli, including chemotherapeutic drugs (8). BH3-only proteins are activated, or their expression is induced, by conventional anticancer drugs, thus explaining, in part, how chemotherapy can regress tumors in patients (3, 15). Further insights into the mechanisms by which anticancer drugs trigger apoptosis are critical to improving treatment efficacy. Given the importance of BH3-only proteins in neutralizing antiapoptotic Bcl-2 proteins, small-molecule inhibitors (i.e., BH3 mimetics) have been developed as a novel class of drugs that can occupy the BH3-binding site on Bcl-2 or Bcl-xL thus suppressing their prosurvival function (16, 17). These agents have been shown to promote apoptosis of tumor cells in culture and to suppress tumor growth in mouse models (16).
We determined the expression and prognostic impact of the BH3-only proteins Bim, Puma, and Noxa in a large series of stage II and III human colon cancers from patients treated in an adjuvant chemotherapy trial. Given the role of BH3-only proteins in apoptosis induction and the observation that they can disable antiapoptotic Bcl-2 proteins, we tested the hypothesis that differences in the expression of Bim, Puma, or Noxa proteins may account for differences in clinical outcome among colon cancer patients. Our findings indicate that expression of proapoptotic BH3-only proteins Bim and Puma have potential utility as prognostic biomarkers in patients with colon cancer.
Materials and Methods
Patient population
Surgically resected primary colon carcinomas were analyzed from patients who participated in one of three 5-fluorouracil (5-FU)-based adjuvant chemotherapy trials conducted by the North Central Cancer Treatment Group (study numbers: 79-46-04, 89-46-51, and 91-46-53). Details of these completed, randomized, 5-FU–based adjuvant therapy trials have been previously reported (18, 19). Treatment consisted of 5-FU and leucovorin + standard dose or high-dose levamisole for resectable adenocarcinoma of the colon (91-46-53). Another study evaluated portal venous 5-FU versus observation (79-46-04), and the third evaluated 5-FU + levamisole versus 5-FU + levamisole + leucovorin (89-46-51). Paraffin-embedded tumor tissue blocks were available from a nonrandom subset of study participants and included tumor-node-metastasis (TNM) stage II (n = 85) and III (n = 346) colon cancers (total n = 431). Tumor histologic grade was determined as defined by the American Joint Committee on Cancer Prognostic Factors Consensus Conference (grade 1, well differentiated; grade 2, moderately differentiated; grade 3, poorly differentiated; grade 4, undifferentiated; ref. 20). Tumor site was defined relative to the splenic flexure and tumors located at the splenic flexure were included in the distal category. All patients were censored at 5 y after randomization for disease-free survival (DFS). Patients were followed for a median of 8 y after study randomization for overall survival (OS) data. The current analysis was in accordance with the original informed consent documents.
Of 431 patients, 404 were randomized to treatment arms and 27 received observation alone. Given that 22 patients received treatment that was later determined to be ineffective (i.e., portal venous 5-FU), there were a total of 382 patients who received effective treatment that included 5-FU. Accordingly and for purposes of the analyses, treatment was categorized as none or ineffective [n = 49; observation only (n = 27) + portal venous 5-FU (n = 22)] versus effective [i.e., consisting of modulated 5-FU (n = 382)].
Tissue microarrays
Tissue sections were cut from paraffin blocks and stained with H&E. H&E sections were examined by light microscopy to identify areas of invasive adenocarcinoma that were then marked on the H&E slide to facilitate tissue microarray construction. Tissue microarrays were made from paraffin blocks and used for subsequent immunohistochemical analysis of Bim, Puma, and Noxa. For tissue microarrays, the source block was cored and a 1-mm core was transferred to the recipient master block using a Beecher microarray instrument (Beecher Instruments). Six cores were arrayed per specimen and included three malignant and three normal colon tissue cores. Sections of normal liver, tonsil, and placenta were used as controls for immunostaining and navigation markers. Tissue microarrays allowed simultaneous evaluation of multiple cases on a single slide. This method virtually eliminates slide-to-slide variation, which is an important factor contributing to variability in immunohistochemical studies (21).
Immunohistochemistry for BH3-only proteins and p53
Immunohisistochemical staining for BH3-onlyproteins was done on an automated immunostaining system (Autostainer, DAKO). Paraffin sections were cut at 4 µm and mounted on silanized slides (SuperFrost Plus, Fisher Scientific). Heat-induced epitope retrieval using a steamer was done (citrate buffer, 40 min, 98–100°C). Slides were washed in PBS, placed into the Autostainer, and washed with hydrogen peroxide for 5 min. Subsequently, slides were placed into a wash buffer (TBS solution containing 0.05% Tween 20, pH 7.6; DAKO). Slides were incubated with the primary anti-Bim/Bod rabbit polyclonal antibody (NeoMarkers), diluted 1:200, for 60 min. For Puma, slides were incubated with the primary anti-Puma rabbit polyclonal antibody (Cell Signaling), diluted 1:50, for 60 min. Slides were incubated with the primary anti-Noxa rabbit polyclonal antibody (BioVision), diluted 1:50, for 60 min. All incubation steps were done at room temperature. For all of the antibodies, slides were rinsed in the wash buffer and a secondary antibody system was applied using Envision + Dual Link HRP (DAKO) for 15 min each. After rinsing in wash buffer, the 3,3′-diaminobenzidine chromagen was applied to the slides for 10 min.
Immunohistochemistry for p53 protein had been done in paraffin-embedded tissue sections as previously reported (18). Antigen retrieval was done by microwaving the slides in a 10 mmol/L citric acid buffer for 3 min × 3. Slides were incubated with the p53-D07 primary antibody recognizing both mutant and wild-type p53 (Novocastra Laboratories Ltd.) at a 1:50 dilution for 1 h at room temperature (18).
Immunohistochemical scoring
Bim, Puma, and Noxa staining localized to the tumor cell cytoplasm and was scored based on intensity (0/1/2/3) and percent tumor cell positivity [grouped into quartiles (0–4)]. Intensity and immunopercent were multiplied to yield an immunoscore. For purposes of our analysis, we dichotomized immunoscores as negative versus elevated for Bim, where elevated included any immunopositive case. Puma was dichotomized as negative/low (immunoscore, ≤2), versus elevated and Noxa was dichotomized as negative/low versus elevated where low was expanded to an immunoscore of ≤4. Puma and Noxa were dichotomized differently due to the small number of negative and/or low cases for these individual markers. We also examined the combined variable of Bim and Puma by adding their respective immunoscores, and their sum was dichotomized (≤4 or >4) for purposes of analysis. To standardize the scoring criteria, cases representative of the immunoscore categories were identified and reviewed to minimize subjectivity. All the specimens were analyzed by a pathologist (R.L.R.) without knowledge of any clinical information.
Slides were scanned by a cytotechnologist and imaging specialist using a slide scanner (Bacus Laboratories, Inc.). The BLISS system is capable of digitally capturing images at 480 × 752 pixel resolution and at × 40 magnification.
Nuclear p53 staining was regarded as positive when ≥10% of the malignant nuclei showed staining; all others were regarded as p53 negative (18).
Antibody specificity
To ensure the specificity of the antibodies selected against Bim, Puma, and Noxa (shown above), we did Western blotting in cultured HT29 and HCT116 human colon carcinoma cell lines. Protein samples were prepared in a lysis buffer per the procedure outlined above, normalized using detergent-compatible protein assay reagents (Bio-Rad), boiled in lithium dodecyl sulfate sample buffer (Invitrogen) containing 2-mercaptoethanol, resolved with a SDS-PAGE gel, and electrophoretically transferred onto a 0.22-µm polyvinylidene difluoride membrane (Bio-Rad). The membrane was blocked with 0.2% I-Block (Applied Biosystems) in PBS-T (PBS containing 0.1% Tween 20) and incubated with the primary antibody in PBS-T containing 0.2% I-Block overnight at 4°C. The membrane was then incubated with a secondary antibody conjugated to alkaline phosphatase in PBS-T containing 0.2% I-Block and then developed with CDP-Star substrate (Applied Biosystems).
DNA mismatch repair status
For the earlier studies (79-46-04 and 89-46-51), tumors were analyzed for microsatellite instability (MSI) using 11 dinucleotide microsatellite markers, as previously described (22, 23). In accordance with consensus definitions of the National Cancer Institute, tumors were classified into three groups: (a) microsatellite stable with no MSI at any of the loci examined, (b) low instability (MSI-L; <30% of the loci showing MSI), or (c) high instability (MSI-H; ≥30% of the loci showing MSI; refs. 23, 24). For a more recent study (91-46-53 study), defective mismatch repair (MMR) was defined by the presence of instability at the BAT 26 mononucleotide locus coupled with absent expression of a MMR protein, as previously described (22). MMR protein (hMLH1 and hMSH2) expression was analyzed by immunohistochemistry (n = 370). In cases with instability at BAT 26 but intact hMLH1 and hMSH2 expression, immunohisto-chemistry for hMSH6 and PMS2 proteins was also done as previously described (25). Slides were previously scored as either positive or negative for MMR proteins based on the presence (+) or absence (−) of tumor cell staining (26).
Statistical analysis
Biomarker expression was dichotomized for DFS and OS. χ2 or Fisher’s exact tests were used to test for an association between prognostic markers for categorical variables. The Wilcoxon rank-sum test was used to test for an association between dichotomized markers and continuous variables of interest. OS (censored at 8 y) was calculated as the number of years from random assignment to the date of death (due to any cause) or last contact. DFS (censored at 5 y) was calculated as the number of years from random assignment to the date of disease recurrence or death. The distributions of OS and DFS were estimated using Kaplan-Meier methodology. Univariate and multivariate Cox proportional hazards models (27) were used to explore the association of clinical and biomarker expression factors with OS and DFS. Univariate models were stratified according to the patient’s original treatment study. Multivariate models were adjusted for treatment and study, but were not stratified by study, because the treatment effect was confounded with the study effect. The score and likelihood ratio test P values were used to test the significance of each covariate in the univariate and multivariate models, respectively. Multivariate models were adjusted for covariates including treatment study. The likelihood ratio test was also used to test for interactions in some limited multivariate models. Graphical and statistical methods were used to examine whether underlying model assumptions were satisfied (e.g., proportional hazards; ref. 28). Statistical tests were two sided, with P ≤ 0.05 considered significant. P values were not adjusted for multiple comparisons. Statistical analyses were done using Statistical Analysis System software (SAS Institute).
Results
Clinicopathologic features and tumor characteristics
Clininicopathologic features of the study patients and their resected colon carcinomas are shown in Table 1. The mean and median patient age were 62.9 (SD, 10.40) and 64 years (range, 26–89), respectively. The median duration of follow-up for patients who remain alive was 8.0 years. Of the 431 primary colon carcinomas studied, 85 (20%) were TNM stage II and 346 (80%) were stage III. DNA mismatch repair (MMR) status had been determined in 370 tumors and 36 (10%) cancers were found to have defective MMR (Table 1). Similar to previous reports (29–31), our data show that tumors with defective MMR were significantly more likely to be from females (P = 0.0078), located in the proximal colon (P < 0.0001), and to be poor/undifferentiated (P < 0.0001) compared with tumors with intact MMR.
Table 1.
Clinicopathologic features, tumor markers, and patient survival
| Variable | Total n | 5-y DFS (%) | HR (95% CI) | P* | 5-y OS (%) | HR (95% CI) | P* |
|---|---|---|---|---|---|---|---|
| Age (10-y increase) | 431 | NA | 1.09 (0.94–1.27) | 0.2445 | NA | 1.21 (1.03–1.41) | 0.0177 |
| Gender | |||||||
| Female | 203 (47%) | 63.5% | – | 0.5758 | 70.4% | – | 0.7029 |
| Male | 228 (53%) | 61.1% | 1.09 (0.80–1.49) | 70.6% | 1.06 (0.78–1.44) | ||
| TNM stage | |||||||
| II | 85 (20%) | 78.8% | – | 0.0013 | 84.7% | – | 0.0012 |
| III | 346 (80%) | 58.2% | 2.19 (1.34–3.59) | 67.0% | 2.16 (1.34–3.49) | ||
| T stage† | |||||||
| T1-T2 | 48 (13%) | 62.4% | – | 0.8091 | 75.0% | – | 0.3710 |
| T3-T4 | 334 (87%) | 64.5% | 0.94 (0.57–1.55) | 72.4% | 0.81 (0.51–1.29) | ||
| Grade‡ | |||||||
| 1, 2 | 291 (68%) | 64.1% | – | 0.1260 | 73.9% | – | 0.0411 |
| 3, 4 | 140 (32%) | 58.4% | 1.29 (0.93–1.77) | 63.6% | 1.39 (1.01–1.90) | ||
| Treatment§ | |||||||
| Ineffective/control | 49 (11%) | 46.9% | – | 0.0168 | 53.1% | – | 0.0163 |
| Effective | 382 (89%) | 64.2% | 0.60 (0.40–0.92) | 72.8% | 0.60 (0.40–0.92) | ||
| Tumor site | |||||||
| Distal | 215 (50%) | 61.2% | – | 0.5427 | 70.7% | – | 0.8470 |
| Proximal | 216 (50%) | 63.3% | 0.91 (0.67–1.24) | 70.4% | 0.97 (0.72–1.32) | ||
| Bim | |||||||
| Negative | 66 (18%) | 53.0% | – | 0.0225 | 66.7% | – | 0.0334 |
| Elevated | 295 (82%) | 64.5% | 0.62 (0.42–0.94) | 71.5% | 0.65 (0.44–0.97) | ||
| Puma | |||||||
| Negative/low§ | 37 (9%) | 51.4% | – | 0.1261 | 64.9% | – | 0.0220 |
| Elevated | 379 (91%) | 63.4% | 0.68 (0.42–1.12) | 71.0% | 0.59 (0.37–0.93) | ||
| Noxa | |||||||
| Negative/low§ | 146 (35%) | 67.8% | – | 0.1606 | 74.7% | – | 0.5442 |
| Elevated | 271 (65%) | 60.3% | 1.28 (0.91–1.80) | 69.0% | 1.11 (0.80–1.54) | ||
| Bim + Puma§ | |||||||
| Immunoscore sum ≤4 | 63 (18%) | 47.6% | – | 0.0007 | 60.3% | – | 0.0002 |
| Immunoscore sum >4 | 289 (82%) | 65.9% | 0.51 (0.34–0.76) | 73.0% | 0.48 (0.33–0.71) | ||
| MMR | |||||||
| Intact MMR | 334 (90%) | 62.1% | – | 0.1533 | 70.3% | – | 0.2155 |
| Defective MMR | 36 (10%) | 72.2% | 0.63 (0.33–1.20) | 80.6% | 0.68 (0.36–1.26) | ||
| p53† | |||||||
| Negative | 50 (36%) | 62.0% | – | 0.3150 | 74.0% | – | 0.4522 |
| Positive | 88 (64%) | 49.9% | 1.32 (0.77–2.28) | 59.1% | 1.22 (0.72–2.08) |
Abbreviation: NA, not available.
Score test P value from a Cox regression model after stratifying by study.
Missing cases.
Grade 1 and 2 (well/moderate); grade 3 and 4 (poor/undifferentiated).
See Materials and Methods.
BH3-only protein expression
Protein expression was analyzed by immunohistochemistry in tissue microarrays. Bim, Puma, and Noxa staining localized to the tumor cell cytoplasm (Fig. 1). Elevated Bim expression was observed in 295 (82%) cases and the remaining 66 (18%) tumors were negative for Bim (Table 1). Elevated Puma expression was observed in 379 (91%) cases and 37 (9%) tumors were negative/low for Puma (Table 1). We found that 271 (65%) tumors showed elevated staining for Noxa and 146 (35%) tumors showed negative/low expression (see Materials and Methods; Table 1). For the combined variable of Bim and Puma, 63 cases were negative/ low (immunoscore sum of ≤4) and 289 cases were elevated (score sum of >4; see Materials and Methods; Table 1).
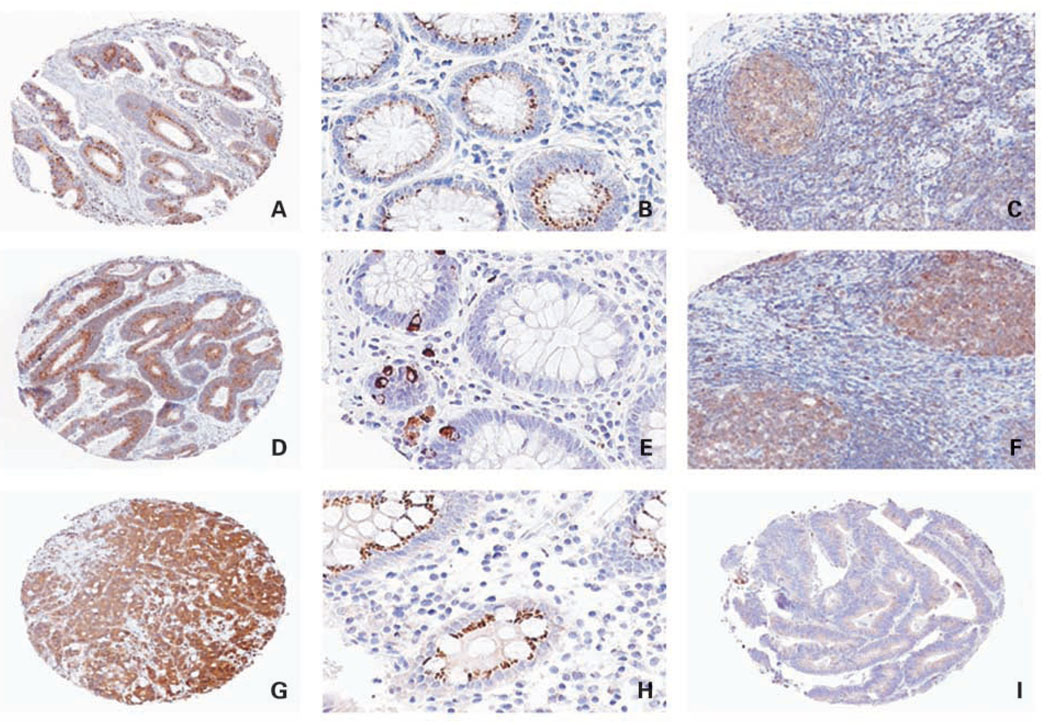
Fig. 1.
Immunohistochemical analysis of Bim, Puma, and Noxa proteins. A, colon carcinoma with elevated Bim expression in the tumor cell cytoplasm. Magnification, ×5. B, normal colonic mucosa shows Bim staining in colonic crypt epithelium. Magnification, ×20. C, tonsil shows Bim staining in lymphocytes within the germinal center (positive control). Magnification, ×10. D, colon carcinoma with elevated Puma staining in the tumor cell cytoplasm. Magnification, ×5. E, normal colonic mucosa shows cytoplasmic/perinuclear Puma staining in crypt epithelial cells. Magnification, ×20. F, tonsil serves as a positive control for Puma. Magnification, ×10. G, colon carcinoma (poorly differentiated) shows elevated Noxa staining in the tumor cell cytoplasm. Magnification, ×5. H, normal colonic mucosa with cytoplasmic Noxa staining in the crypt epithelium. Magnification, ×20.I, colon carcinoma with low-level Noxa expression. Magnification, ×5.
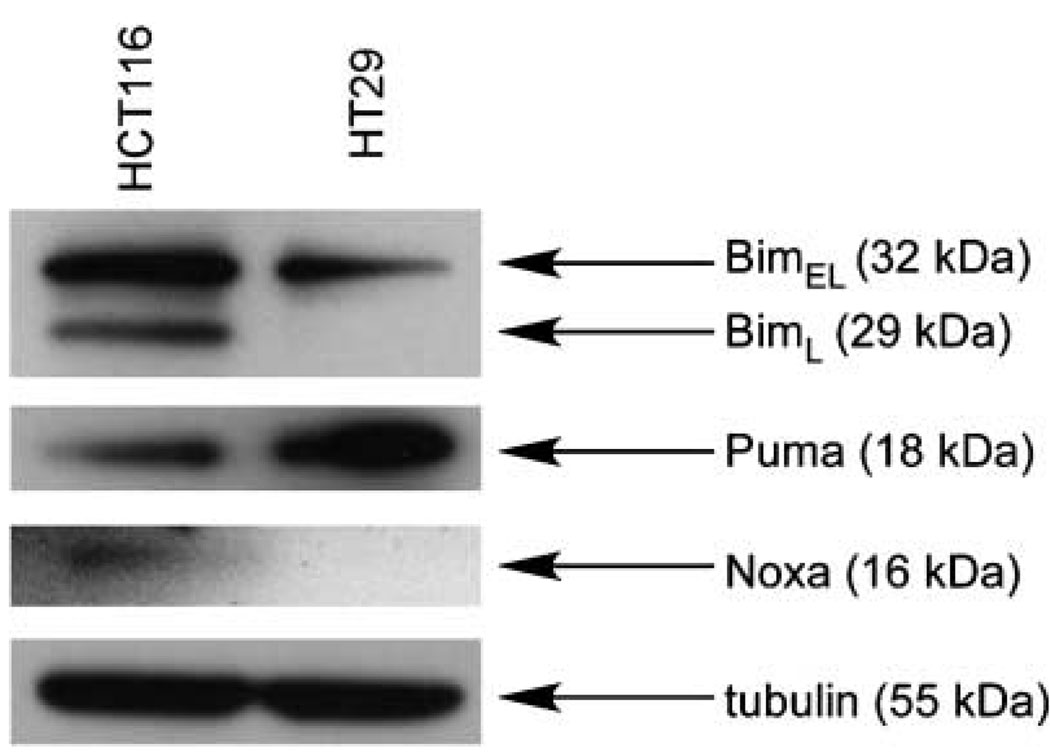
We also analyzed Bim, Puma, and Noxa protein expression in histologically normal-appearing colonic mucosa in a subset of 57 cases (Fig. 1). Within the normal mucosa, 23 (40%) showed staining for Bim, 25 (44%) were positive for Puma, and 41 (72%) were positive for Noxa. Bim and Noxa staining localized to the cytoplasm of colonic epithelial cells and no obvious gradient within the mucosa was appreciated. Puma staining was focally expressed in the cytoplasm and perinuclear regions of colonic epithelial cells, and some staining of mononuclear cells in the lamina propria was noted. Bim and Puma showed staining in germinal centers of tonsil (positive control), and Noxa showed an absence of staining in placenta (negative control). The specificity of the Bim, Puma, and Noxa antibodies was evaluated by Western blot assays done in cultured and untreated HT29 and HCT116 human colon carcinoma cell lines. Analysis of Bim, Puma, and Noxa expression revealed bands for each BH3 protein that corresponded to their known molecular weights (Fig. 2). The Bim antibody recognized two splice variants (BimEL and BimL), as shown in Fig. 2. Low-level or absent Noxa was detected in the cell lines examined, yet Noxa was readily detected in tissues by immunohistochemistry (Fig. 1). Within the tissue sections, additional determination of the antibody specificity was done using appropriate positive and negative tissue controls with the expected results obtained (Fig. 1). Therefore, the antibodies chosen for immunohistochemistry were specific for their target proteins.
Fig. 2.
Western blot analysis of Bim, Puma, and Noxa expression in HCT116 and HT29 human colon carcinoma cell lines. Blot shows the specificity of the antibodies for Bim, Puma, and Noxa proteins in both cell lines, as indicated by bands corresponding to their known molecular weights. The Bim antibody recognized its known splice variants (BimEL and BimL).
Correlations between variables
We determined the relationship of Bim, Puma, and Noxa expression to clinicopathologic variables, p53 staining, and MMR status. We found that Noxa was correlated with older age (P = 0.0186) and proximal tumor site (P = 0.0174). Neither Bim nor Puma expression variables correlated with clinicopathologic features (data not shown). We found significant correlations between Bim, Puma, and Noxa in that tumors with elevated expression of Bim were significantly more likely to have elevated expression of Puma and Noxa (treated as a continuous variable) compared with Bim-negative tumors (P < 0.0001 and 0.0039, respectively).
We also determined the association of Bim, Puma, and Noxa staining with nuclear p53 expression, given that Puma and Noxa are transcriptionally regulated by p53 (13, 14, 32, 33). Noxa was correlated with p53 in that tumors with elevated Noxa expression were more likely to be positive for p53 than the negative/low tumors (P = 0.0394). Neither Puma nor Bim staining was significantly correlated with p53 (P = 0.4989 and 0.4501, respectively).
We also compared Bim, Puma, and Noxa in relation to MMR status. When markers were stratified by MMR status, tumors with defective MMR showed a trend toward a higher mean expression for Bim (treated as a continuous variable) compared with the tumors with intact MMR [4.8 (2.9) versus 3.7 (2.6), respectively; P = 0.0651]. Neither Puma nor Noxa was correlated with MMR status.
Tumor characteristics and patient survival
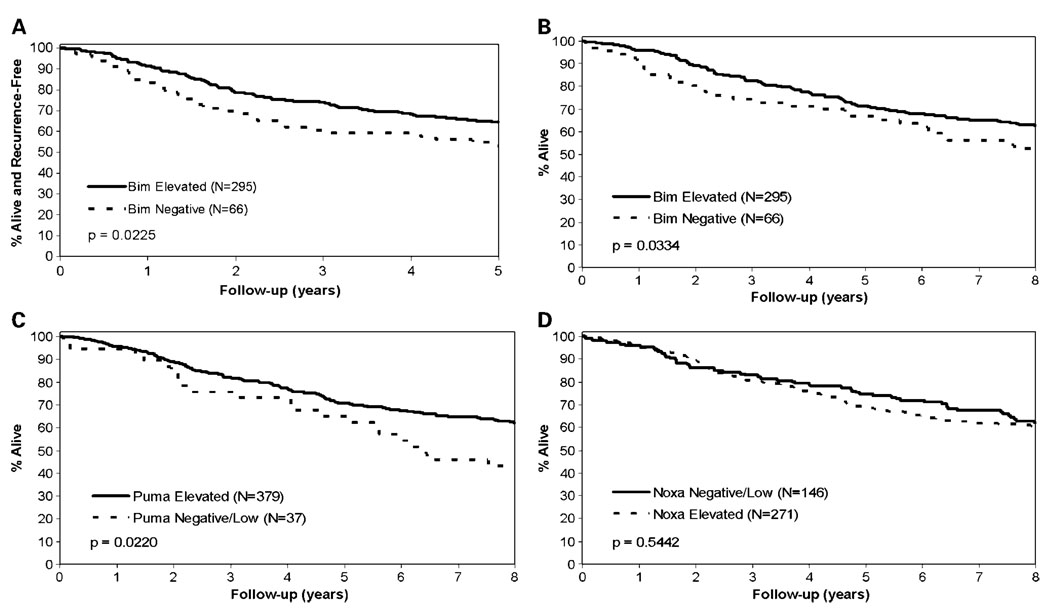
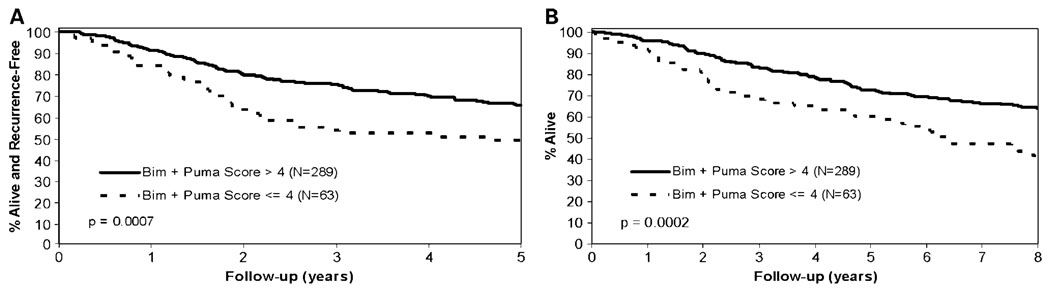
Patients with elevated Bim expression had significantly improved DFS and OS compared with patients with negatively expressed Bim tumors (5-year DFS: 64.5% versus 53%, P = 0.0225; OS: 71.5% versus 66.7%, P = 0.0334; Table 1; Fig. 3A and B). Patients with elevated expression of Puma were also found to have significantly improved OS compared with patients with negative/low expression. Specifically, 71.0% of patients whose tumors showed elevated Puma expression were alive at 5 years after randomization compared with 64.9% for patients with negative/low Puma expression (P = 0.0220; Table 1; Fig. 3C). In contrast, Noxa expression was not prognostic for either DFS or OS (Table 1; Fig. 3D). The combined variable of Bim and Puma was highly prognostic for both DFS and OS (P = 0.0007 and 0.0002, respectively; Table 1; Fig. 4A and B).
Fig. 3.
DFS (A) and OS (B) in patients with TNM stage II and III colon cancers by Bim expression. OS in patients with TNM stage II and III colon cancer by Puma (C) or Noxa (D) expression.
Fig. 4.
DFS (A) and OS (B) in patients with TNM stage II and III colon cancer by immunoscore for the combined variable of Bim and Puma.
The favorable prognostic effect of Bim and Puma expression was similar in stage II and III patients, indicating that the improved survival for patients with elevated expression of Bim and Puma did not depend on tumor stage (interaction P = 0.75 and 0.78, respectively). However, the stage II patient cohort was relatively small, and thus, there was insufficient statistical power to show statistical significance within the stage II patient subset. The clinical utility of prognostic variables is especially relevant to stage II disease where adjuvant treatment is not standard of care, yet some patients receive a survival benefit (34). Similar results for OS and DFS were obtained for Bim and Puma expression when tumors with defective MMR were excluded from the analysis (data not shown), thus excluding bias that might arise from inclusion of tumors that arise through an alternative molecular pathway. MMR status (n = 370) was not significantly related to clinical outcome (Table 1). Clinicopathologic features, including tumor stage, histologic grade, and patient age, were prognostic (Table 1).
In a multivariate analysis, a Cox proportional hazards model was used to evaluate whether the BH3-only proteins Bim and Puma showed independent prognostic significance. We found that Bim was an independent marker for both DFS and OS (Table 2), and Puma was an independent prognostic marker for OS (Table 3), after adjustment for tumor stage, histologic grade, age, treatment status, and adjuvant study. Due to the strong correlation between Bim and Puma expression, we did not include them in the same multivariate model. Noxa was not included in these models as it was not significant in a univariate analysis. We also evaluated the prognostic effect of the combined variable of Bim and Puma. Importantly, we found that the combined variable was a strong independent prognostic marker for both DFS [hazard ratio (HR), 0.53; 95% confidence interval (95% CI), 0.36–0.79; P = 0.0034] and OS (Table 4). Adjustment for MMR status in the multivariate models yielded similar HRs for the included variables (Table 4).
Table 2.
Multivariate analysis of Bim immunoscore and DFS and OS
| Variable (n = 361) | DFS | OS | ||
|---|---|---|---|---|
| HR (95% CI) | P* | HR (95% CI) | P* | |
| Bim immunoscore (elevated vs negative) | 0.63 (0.42–0.94) | 0.0298 | 0.65 (0.44–0.98) | 0.0447 |
| Histologic grade (3 and 4 vs 1 and 2)† | 1.28 (0.91–1.82) | 0.1666 | 1.37 (0.98–1.93) | 0.0701 |
| TNM stage (III vs II) | 2.63 (1.48–4.67) | 0.0002 | 2.63 (1.52–4.58) | <0.0001 |
| Treatment (effective vs control/ineffective) | 0.77 (0.44–1.35) | 0.3709 | 0.70 (0.40–1.22) | 0.2163 |
| Age (10-y increase) | 1.07 (0.91–1.26) | 0.3831 | 1.21 (1.02–1.43) | 0.0211 |
Likelihood ratio P value also adjusted for adjuvant study.
Grade 1 and 2 (well/moderate); grade 3 and 4 (poor/undifferentiated).
Table 3.
Multivariate analysis of Puma immunoscore and DFS and OS
| Variable (n = 416) | DFS | OS | ||
|---|---|---|---|---|
| HR (95% CI) | P* | HR (95% CI) | P* | |
| Puma immunoscore (elevated vs negative/low) | 0.70 (0.43–1.15) | 0.1773 | 0.59 (0.37–0.94) | 0.0371 |
| Histologic grade (3 and 4 vs 1 and 2)† | 1.35 (0.97–1.87) | 0.0783 | 1.43 (1.04–1.97) | 0.0321 |
| TNM stage (III vs II) | 2.19 (1.34–3.59) | 0.0006 | 2.19 (1.35–3.54) | 0.0004 |
| Treatment (effective vs control/ineffective) | 0.65 (0.40–1.06) | 0.0918 | 0.65 (0.40–1.04) | 0.0815 |
| Age (10-y increase) | 1.08 (0.93–1.25) | 0.3131 | 1.19 (1.02–1.39) | 0.0237 |
Likelihood ratio P value adjusted for adjuvant study.
Grade 1 and 2 (well/moderate); grade 3 and 4 (poor/undifferentiated).
Table 4.
Multivariate analysis of Bim and Puma immunoscore sum and DFS and OS
| Variable (n = 304) | DFS | OS | ||
|---|---|---|---|---|
| HR (95% CI) | P* | HR (95% CI) | P* | |
| Bim and Puma immunoscore sum (>4 vs ≤4) | 0.58 (0.38–0.90) | 0.0184 | 0.53 (0.35–0.80) | 0.0039 |
| Histologic grade (3 and 4 vs 1 and 2)† | 1.64 (1.H–2.42) | 0.0139 | 1.67 (1.14–2.43) | 0.0092 |
| TNM stage (III vs II) | 2.44 (1.30–4.56) | 0.0018 | 2.67 (1.43–4.99) | 0.0005 |
| Treatment (effective vs control/ineffective) | 0.86 (0.37–1.96) | 0.7167 | 0.93 (0.41–2.14) | 0.8700 |
| Age (10-y increase) | 0.99 (0.81–1.20) | 0.9076 | 1.14 (0.93–1.39) | 0.1907 |
| Defective DNA MMR (vs intact) | 0.58 (0.30–1.14) | 0.0902 | 0.58 (0.31–1.10) | 0.0769 |
Likelihood ratio P value also adjusted for adjuvant study.
Grade 1 and 2 (well/moderate); grade 3 and 4 (poor/undifferentiated).
Discussion
Clinical and pathologic tumor staging insufficiently addresses tumor heterogeneity and, therefore, cannot account for interpatient variability in clinical outcome. Accordingly, prognostic markers are needed to identify colon cancer patients likely to benefit from adjuvant therapy, to guide clinical followup, and to provide a rational basis for use of molecularly targeted therapies. Abundant evidence supports the role of apoptotic regulatory proteins in tumor progression and metastasis (35) as well as in responsiveness to anticancer drugs (15). BH3-only proteins act as sensors of cellular stress and function to inactivate prosurvival Bcl-2 family members by inserting their BH3 domain into a groove on the Bcl-2 proteins to disable them (4, 5). We analyzed BH3 protein expression in tissue microarrays, which permitted their high-throughput evaluation. We found that elevated expression of Bim and Puma, but not Noxa, was significantly associated with improved patient survival rates in curatively resected stage II and III colon cancers from patients treated in 5-FU – based adjuvant therapy trials. Specifically, Bim and Puma expression were associated with a 35% and 41% reduction in the relative risk of death due to any cause, respectively. Our results are consistent with the finding that Bim and Puma are the most potent of the BH3 proteins as inducers of apoptosis because they can disable all of the prosurvival Bcl-2 proteins (7, 8). Multivariate Cox analysis revealed that both Bim and Puma expression were independent predictors of OS after adjustment for histologic grade, tumor stage, age, and treatment. Furthermore, the combined variable of Bim and Puma was highly discriminant for DFS and OS in both univariate and multivariate analyses. Interpretation of our study results is aided by rigorous data collection related to patient participation in adjuvant therapy trials and the availability of long-term survival data (median, 8 years). Our study findings are consistent with limited data in human melanomas where low Puma expression was associated with poor prognosis (36). A potential mechanism by which absent or reduced expression of BH3-only proteins can negatively affect prognosis is suggested by the observation that defects in apoptosis enable tumor cells to survive and grow intravascularly (35). Therefore, defective apoptosis can facilitate micrometastasis that seems to be responsible for shorter survival in colorectal cancer patients.
Our finding that loss of Bim and/or Puma expression is associated with worse patient survival is consistent with a tumor suppressor role for these BH3-only proteins (10, 37–39). Bim deficiency was shown to facilitate epithelial tumor growth in a mouse xenograft model (40), and loss of even one Bim allele was shown to accelerate tumorigenesis in Eu-myc mice (38). Moreover, biallelic deletion of the Bim gene was found in a proportion of human mantle cell lymphomas consistent with a tumor suppressor role for human Bim (10, 37). Bim is silenced by promoter methylation in certain other B-cell lymphomas (10); however, its methylation status and that of other BH3-only proteins in nonhematologic malignancies is unknown. Similarly, down-regulation of Puma or its deletion has been shown to accelerate Myc-induced lymphomagenesis (41). Taken together, our data support a tumor suppressor role for Bim and Puma in colon cancers in that loss or reduced expression of either protein product can contribute to tumor growth and progression. In this regard, we show that absent or low-level Bim and Puma are associated with significantly shorter patient survival.
The BH3 proteins are regulated by both transcriptional and posttranslational mechanisms (42). The Bim protein can be induced downstream of Akt by a forkhead transcription factor (43) and is subject to posttranslational regulation. We found that Noxa expression in colon cancers was significantly associated with nuclear p53 expression. Both Noxa and Puma are regulated at the transcriptional level by p53 and contribute to p53-mediated apoptosis (12–14). Noxa and Puma also have a role in several p53-independent responses (44) and Puma can be regulated by transcription factors, such as p73 and E2F1 (32, 33). A reduction or loss of BH3-only protein expression can confer resistance to anticancer therapies. Deletion of Puma was shown to inhibit p53-mediated apoptosis in a colon carcinoma cell line (12). Furthermore, infection with a Puma adenovirus markedly increased sensitivity to 5-FU in esophageal cancer cells (45). Suppression of Noxa expression by RNA interference inhibited apoptosis in mouse fibroblasts (46), and Bim was shown to determine the sensitivity of carcinoma cells to paclitaxel (40) and to proteasome inhibitors (47).
In our study of primary colon cancers, the majority of patients received 5-FU–based adjuvant therapy among three clinical trials conducted over different times. In our multivariate models, the independent prognostic effect of Bim and/or Puma remained statistically significant after adjustment for treatment. In an exploratory analysis, treatment was found to be beneficial for patients with increased expression of Bim and/or Puma, although this benefit did not always reach statistical significance. Unfortunately, our numbers were too small to explore the effect of treatment for patients with low expression of Bim and/or Puma, and we could not appropriately test for the interaction between the markers and treatment to find evidence of the markers being predictive. However, we regard this issue as important for future study. Our findings are therapeutically relevant in that the BH3-only proteins represent novel targets for which BH3 mimetic agents have been developed. In this regard, the BH3 mimetics ABT-737 (Abbott) and GX015-070 (GeminX) have been shown to reverse apoptosis resistance in vitro and ABT-737 treatment induced the regression of solid tumor xenografts (16, 17, 48). The clinical appeal of BH3 mimetics is that most tumors, including colorectal cancers, overexpress prosurvival Bcl-2 proteins (11) and have defects in the p53 pathway that preclude induction of the Puma and Noxa (12).
In conclusion, we show for the first time that the proapoptotic BH3-only proteins Bim and Puma are independent prognostic variables in stage II and III colon cancer patients receiving 5-FU–based adjuvant chemotherapy. These data support experimental evidence that these BH3 proteins serve a tumor suppressor role. If validated, analysis of Bim and Puma, or the combination of these markers, may assist in the risk stratification of colon cancer patients and may be useful in therapeutic decision making.
Acknowledgments
We thank Dr. John Reed (Burnham Institute, La Jolla, CA) for helpful discussion and Jonelle Morales for capable secretarial assistance.
Grant support: National Cancer Institute grant CA104683-02 (F.A. Sinicrope) and Mayo Clinic Cancer Center core grant CA15083.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sattler M, Liang H, Nettesheim D, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 5.Liu X, Dai S, Zhu Y, Marrack P, Kappler JW. The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity. 2003;19:341–352. doi: 10.1016/s1074-7613(03)00234-6. [DOI] [PubMed] [Google Scholar]
- 6.Kuwana T, Bouchier-Hayes L, Chipuk JE, et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Willis SN, Wei A, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 8.Willis SN, Fletcher JI, Kaufmann T, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 9.Kitada S, Pedersen IM, Schimmer AD, Reed JC. Dysregulation of apoptosis genes in hematopoietic malignancies. Oncogene. 2002;21:3459–3474. doi: 10.1038/sj.onc.1205327. [DOI] [PubMed] [Google Scholar]
- 10.Mestre-Escorihuela C, Rubio-Moscardo F, Richter JA, et al. Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood. 2007;109:271–280. doi: 10.1182/blood-2006-06-026500. [DOI] [PubMed] [Google Scholar]
- 11.Sinicrope FA, Hart J, Michelassi F, Lee JJ. Prognostic value of bcl-2 oncoprotein expression in stage II colon carcinoma. Clin Cancer Res. 1995;1:1103–1110. [PubMed] [Google Scholar]
- 12.Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L. PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc Natl Acad Sci USA. 2003;100:1931–1936. doi: 10.1073/pnas.2627984100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oda E, Ohki R, Murasawa H, et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 14.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 15.Reed JC. Proapoptotic multidomain Bcl-2/Bax-family proteins: mechanisms, physiological roles, and therapeutic opportunities. Cell Death Differ. 2006;13:1378–1386. doi: 10.1038/sj.cdd.4401975. [DOI] [PubMed] [Google Scholar]
- 16.Oltersdorf T, Elmore SW, Shoemaker AR, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 17.Trudel S, Li ZH, Rauw J, Tiedemann RE, Wen XY, Stewart AK. Preclinical studies of the pan- Bcl inhibitor obatoclax (GX015-070) in multiple myeloma. Blood. 2007;109:5430–5438. doi: 10.1182/blood-2006-10-047951. [DOI] [PubMed] [Google Scholar]
- 18.Allegra CJ, Parr AL, Wold LE, et al. Investigation of the prognostic and predictive value of thymidylate synthase, p53, and Ki-67 in patients with locally advanced colon cancer. J Clin Oncol. 2002;20:1735–1743. doi: 10.1200/JCO.2002.07.080. [DOI] [PubMed] [Google Scholar]
- 19.O’Connell MJ, Sargent DJ, Windschitl HE, et al. Randomized clinical trial of high-dose levamisole combined with 5-fluorouracil and leucovorin as surgical adjuvant therapy for high-risk colon cancer. Clin Colorectal Cancer. 2006;6:133–139. doi: 10.3816/ccc.2006.n.030. [DOI] [PubMed] [Google Scholar]
- 20.Compton C, Fenoglio-Preiser CM, Pettigrew N, Fielding LP. American Joint Committee on Cancer Prognostic Factors Consensus Conference: Colorectal Working Group. Cancer. 2000;88:1739–1757. doi: 10.1002/(sici)1097-0142(20000401)88:7<1739::aid-cncr30>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 21.Jourdan F, Sebbagh N, Comperat E, et al. Tissue microarray technology: validation in colorectal carcinoma and analysis of p53, hMLH1, and hMSH2 immunohistochemical expression. Virchows Arch. 2003;443:115–121. doi: 10.1007/s00428-003-0833-z. [DOI] [PubMed] [Google Scholar]
- 22.Thibodeau SN, French AJ, Cunningham JM, et al. Microsatellite instability in colorectal cancer: different mutator phenotypes and the principal involvement of hMLH1. Cancer Res. 1998;58:1713–1718. [PubMed] [Google Scholar]
- 23.Halling KC, French AJ, McDonnell SK, et al. Microsatellite instability and 8p allelic imbalance in stage B2 and C colorectal cancers. J Natl Cancer Inst. 1999;91:1295–1303. doi: 10.1093/jnci/91.15.1295. [DOI] [PubMed] [Google Scholar]
- 24.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 25.Parc YR, Halling KC, Wang L, et al. HMSH6 alterations in patients with microsatellite instability-low colorectal cancer. Cancer Res. 2000;60:2225–2231. [PubMed] [Google Scholar]
- 26.Garrity MM, Burgart LJ, Mahoney MR, et al. Prognostic value of proliferation, apoptosis, defective DNA mismatch repair, and p53 overexpression in patients with resected Dukes’ B2 or C colon cancer: a North Central Cancer Treatment Group Study. J Clin Oncol. 2004;22:1572–1582. doi: 10.1200/JCO.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 27.Cox DR. Regression models and life tables. J R Stat Soc. 1972;34:187–202. [Google Scholar]
- 28.Grambsch PMT, Terry M. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;8:515–526. [Google Scholar]
- 29.Sinicrope FA, Rego RL, Foster N, et al. Microsatellite instability accounts for tumor site-related differences in clinicopathologic variables and prognosis in human colon cancers. Am J Gastroenterol. 2006;101:2818–2825. doi: 10.1111/j.1572-0241.2006.00845.x. [DOI] [PubMed] [Google Scholar]
- 30.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 31.Lothe RA, Peltomaki P, Meling GI, et al. Genomic instability in colorectal cancer: relationship to clinicopathological variables and family history. Cancer Res. 1993;53:5849–5852. [PubMed] [Google Scholar]
- 32.Jeffers JR, Parganas E, Lee Y, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 33.Villunger A, Michalak EM, Coultas L, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 34.Quasar Collaborative Group. Gray R, Barnwell J, McConkey C, Hills RK, Williams NS, Kerr DJ. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370:2020–2029. doi: 10.1016/S0140-6736(07)61866-2. [DOI] [PubMed] [Google Scholar]
- 35.Wong CW, Lee A, Shientag L, et al. Apoptosis: an early event in metastatic inefficiency. Cancer Res. 2001;61:333–328. [PubMed] [Google Scholar]
- 36.Karst AM, Dai DL, Martinka M, Li G. PUMA expression is significantly reduced in human cutaneous melanomas. Oncogene. 2005;24:1111–1116. doi: 10.1038/sj.onc.1208374. [DOI] [PubMed] [Google Scholar]
- 37.Tagawa H, Karnan S, Suzuki R, et al. Genome-wide array-based CGH for mantle cell lymphoma: identification of homozygous deletions of the proapoptotic gene BIM. Oncogene. 2005;24:1348–1358. doi: 10.1038/sj.onc.1208300. [DOI] [PubMed] [Google Scholar]
- 38.Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci U S A. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erlacher M, Labi V, Manzl C, et al. Puma cooperates with Bim, the rate-limiting BH3-only protein in cell death during lymphocyte development, in apoptosis induction. J Exp Med. 2006;203:2939–2951. doi: 10.1084/jem.20061552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan TT, Degenhardt K, Nelson DA, et al. Key roles of BIM-driven apoptosis in epithelial tumors and rational chemotherapy. Cancer Cell. 2005;7:227–238. doi: 10.1016/j.ccr.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 41.Hemann MT, Zilfou JT, Zhao Z, Burgess DJ, Hannon GJ, Lowe SW. Suppression of tumorigenesis by the p53target PUMA. Proc Natl Acad Sci U S A. 2004;101:9333–9338. doi: 10.1073/pnas.0403286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puthalakath H, Strasser A. Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ. 2002;9:505–512. doi: 10.1038/sj.cdd.4400998. [DOI] [PubMed] [Google Scholar]
- 43.Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol. 2000;10:1201–1204. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- 44.Armstrong JL, Veal GJ, Redfern CP, et al. Role of Noxa in p53-independent fenretinide-induced apoptosis of neuroectodermal tumours. Apoptosis. 2007;12:613–622. doi: 10.1007/s10495-006-0020-1. [DOI] [PubMed] [Google Scholar]
- 45.Wang H, Qian H, Yu J, et al. Administration of PUMA adenovirus increases the sensitivity of esophageal cancer cells to anticancer drugs. Cancer Biol Ther. 2006;5:380–385. doi: 10.4161/cbt.5.4.2477. [DOI] [PubMed] [Google Scholar]
- 46.Schuler M, Maurer U, Goldstein JC, et al. p53 triggers apoptosis in oncogene-expressing fibroblasts by the induction of Noxa and mitochondrial Bax translocation. Cell Death Differ. 2003;10:451–460. doi: 10.1038/sj.cdd.4401180. [DOI] [PubMed] [Google Scholar]
- 47.Nikrad M, Johnson T, Puthalalath H, Coultas L, Adams J, Kraft AS. The proteasome inhibitor bortezomib sensitizes cells to killing by death receptor ligand TRAIL via BH3-only proteins Bik and Bim. Mol Cancer Ther. 2005;4:443–449. doi: 10.1158/1535-7163.MCT-04-0260. [DOI] [PubMed] [Google Scholar]
- 48.Huang SS, Sinicrope FA. The BH3 mimetic ABT-737 potentiates TRAIL-mediated apoptotic signaling by unsequestering Bim and Bak in human pancreatic cancer cells. Cancer Res. 2008;68:2944–2951. doi: 10.1158/0008-5472.CAN-07-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]