Abstract
Purpose
Proapoptotic BH3-only proteins Bad and Bid initiate apoptosis by binding to regulatory sites on prosurvival Bcl-2 proteins to directly neutralize their function. We determined if expression of these proteins in colon cancers may account for differences in patient survival.
Experimental Design
Tumor-node-metastasis stages II and III primary colon carcinomas from patients treated in 5-fluorouracil-based adjuvant therapy trials were studied. Immunohistochemical analysis of Bad and Bid proteins was done in tumors (n = 379) and adjacent normal mucosa. Expression was correlated with clinicopathologic variables, disease-free survival rates (DFS), and overall survival (OS) rates.
Results
High expression of the Bad protein [hazard ratio (HR), 0.64; 95% confidence interval (95% CI), 0.43–0.96; P = 0.031] in the cytoplasm of tumor cells was significantly associated with more favorable OS in a univariate analysis. The combined Bad and Bid variable was prognostic for DFS (P = 0.027) and OS (P = 0.006). Stage and histologic grade, but not DNA mismatch repair status, were also prognostic for OS. Multivariate Cox analysis showed that high expression of Bad (HR, 0.64; 95% CI, 0.43–0.97; P = 0.027) and Bid (HR, 0.68; 95% CI, 0.49–0.97; P = 0.034) were independent predictors of OS after adjustment for stage, grade, age, treatment, and study. The combined variable of Bad + Bid was independently associated with DFS (P = 0.020) and OS (P = 0.004).
Conclusion
Proapoptotic Bad and Bid proteins are independent prognostic variables in colon cancer patients receiving adjuvant treatment. If validated, Bad and Bid expression may assist in risk stratification and selection of patients to receive adjuvant chemotherapy.
Colorectal cancer is the second leading cause of cancer-related death in the United States and the fourth leading cause worldwide (1). Considerable stage-independent variability in patient survival is observed and underscores the need for additional prognostic markers. Apoptotic regulatory proteins are potentially important prognostic or predictive markers because impaired apoptosis is a critical event in tumor development and progression/metastasis and also renders the tumor cell resistant to cytotoxic chemotherapy (2). Whether a cell undergoes apoptosis in response to cellular stress, including chemotherapy, is determined largely by interactions between three factions of the Bcl-2 protein family (3). Two factions promote apoptosis and include the BH3-only proteins that sense intracellular damage and can trigger apoptosis by inserting their BH3 domain into a groove on the prosurvival Bcl-2 proteins to inactivate them (3, 4). The second faction are proapoptotic Bax and Bak proteins that when activated can permeabilize the outer mitochondrial membrane, enabling release of cytochrome c, which promotes activation of caspases (5). Activation of Bax and Bak is opposed by prosurvival Bcl-2 proteins (Bcl-2, Bcl-xL, Bcl-w, Mcl-1, and A1).
The BH3-only proteins Bid, Bim, and Puma bind to all prosurvival Bcl-2 proteins, whereas Bad and Noxa display selectivity (6). Cytotoxicity assays in fibroblasts show that Bid, Bim, and Puma are potent inducers of apoptosis, whereas Bad and Noxa alone are weaker, yet Bad in conjunction with Noxa kills potently (6). The interplay between BH3-only proteins and other Bcl-2 family members continues to evolve. Bid, similar to Bim and Puma, has been shown to directly activate Bax-Bak to release cytochrome c (7). Prosurvival Bcl-2 proteins (Bcl-2, Bcl-xL, and Mcl-1) sequester these “activator” BH3-only molecules into stable complexes, thus preventing the activation of Bax-Bak. The remaining “inactivator” BH3-only molecules, including Bad, serve to inactivate prosurvival Bcl-2 proteins. Bad displaces Bid, as well as Bim or Puma, from Bcl-2- Bcl-xL to activate Bax-Bak. In unstressed cells, Bad is phosphorylated by several protein kinases; however, in response to apoptotic stimuli, Bad is rapidly dephosphorylated and migrates to the mitochondria where it induces cell death (8). Bid and Bad proteins have been shown to directly affect the sensitivity of cancer cells to chemotherapeutic agents as shown in studies in cells from knockout mice (4). BH3 mimetic agents have been developed as a novel class of anticancer drugs. The BH3 mimetic ABT-737 has been shown to function like Bad to bind and inhibit prosurvival Bcl-2 family proteins but does not directly activate Bax and Bak (9). Prosurvival Bcl-2 proteins are commonly expressed in many types of human cancer and in most instances are associated with worse outcome and resistance to chemotherapy (4). Although the oncogenic potential of prosurvival Bcl-2 family members is well established (10, 11), experimental studies suggest that loss of a proapoptotic BH3 protein can also be oncogenic.
Given the importance of Bid and Bad proteins in the regulation and induction of cell death, we tested the hypothesis that differences in the expression of Bad and Bid proteins could account for differences in clinical outcome among colon cancer patients. Analysis of Bad and Bid protein expression was done in archival colon carcinomas from patients treated in 5-fluorouracil (5-FU)–based adjuvant chemotherapy trials. Our findings indicate that Bad and Bid expression are independent prognostic variables in patients with tumor-node-metastasis (TNM) stages II and III colon cancers.
Materials and Methods
Study population
Resected primary colon carcinomas were analyzed from participants in adjuvant chemotherapy trials conducted by the Mayo Clinic/North Central Cancer Treatment Group as reported previously (12, 13). Paraffin-embedded tumor blocks were available from a nonrandom subset of cases (n = 379). The current analysis was in accordance with the original informed consent documents. Of 379 patients, 349 were randomized to study treatment arms and 30 received observation alone. For purposes of the analysis, treatment was categorized as none or ineffective [n = 57; observation only (n = 30) + portal venous 5-FU (n = 27)] versus effective [5-FU + levamisole and/or leucovorin (n = 322); refs. 14, 15]. Eight stage II and 49 stage III patients received either control or ineffective treatments.
Immunohistochemistry
After deparaffinization, antigen retrieval was done in an EDTA (for Bad) or in a citrate buffer (for Bid) using a steamer (30–40 min, 98–100°C). Endogenous peroxidase activity was blocked, and slides were placed into a wash buffer [TBS solution containing 0.05% Tween 20 (pH 7.6); DAKO]. Subsequently, slides were incubated with a primary anti-Bad mouse monoclonal antibody (Santa Cruz Biotechnology), diluted 1:100, for 30 min. For Bid, slides were incubated with a primary anti-Bid rabbit polyclonal antibody (Cell Signaling), diluted 1:20, for 60 min. An automated immunostaining system (Autostainer; DAKO) was employed. Slides were rinsed in wash buffer and a secondary antibody was applied (Envision+ Dual-Link Horseradish Peroxidase; DAKO) for 15 min. The 3,3′-diaminobenzidine chromagen was then applied to the slides. As a negative control, the primary antibody was omitted, but all other steps were followed. A positive control for each antibody was used and consisted of prostate for Bad and tonsil for Bid.
Defective DNA mismatch repair (MMR) was defined as absent expression of an MMR protein by immunohistochemistry and instability at the BAT 26 locus as done and described previously (16–18).
Apoptotic cells had been analyzed previously by terminal nucleotidyl transferase–mediated dUTP nick end labeling (TUNEL) staining in a tumor subset using Apoptag Plus Peroxidase In situ Apoptosis Detection Kit (Serologicals) (18).
Immunohistochemical scoring
Immunointensity (0/1/2/3) was determined and categorized as high (3+) versus low (0–2+) staining. Immunopercent was determined and categorized as high (>50%) versus low (≤50% of immunopositive cells) at light microscopy (19). The combined variable of Bad + Bid was dichotomized for analysis, as high refers to either variable being high or both. All specimens were analyzed by a pathologist (R.L.R.) without knowledge of clinical information. To minimize subjectivity in the interpretation of immunohistochemical results, scoring criteria had been agreed upon before the data analysis.
Specificity of anti-Bad and anti-Bid antibodies
To ensure specificity of the antibodies against Bad and Bid proteins, we did Western blotting in cultured HT-29 and HCT116 human colon carcinoma cell lines.
Statistical analysis
Each slide was assigned a unique number that enabled blinding with respect to patient identity. χ2 or Fisher’s exact tests were used to test for an association between prognostic markers for categorical variables. The Wilcoxon rank-sum test was used to test for an association between dichotomized markers and continuous variables. McNemar’s test was used to test for an association between biomarkers in normal and tumor tissues from the same patients. Biomarker expression was dichotomized for disease-free survival (DFS) and overall survival (OS) censored at 5 or 8 years, respectively, and calculated as number of days from randomization to the date of disease recurrence for DFS or death or last contact for OS. The distributions of OS and DFS were estimated using Kaplan-Meier methodology. Univariate and multivariate Cox proportional hazards models (20) were used to explore the association of markers with OS and DFS. Univariate models were stratified according to the patient’s treatment study. Multivariate models were adjusted for covariates including treatment and study but were not stratified by study, because treatment effect was confounded with study effect. The score and likelihood ratio test P values were used to test the significance of each covariate in the univariate and multivariate models, respectively. The likelihood ratio test was also used to test for interactions in some limited multivariate models. Graphical and statistical methods were used to examine whether underlying model assumptions were satisfied (e.g., proportional hazards; ref. 21). Statistical tests were two sided, with P < 0.05 considered significant. Statistical analyses were done using SAS software (SAS Institute).
Results
Clinicopathologic features and tumor characteristics
Clininicopathologic features of the study population are shown in Table 1. Mean (SD) and median (range) patient age were 63.0 (10.75) and 65 (26–89) years, respectively. Patient age was dichotomized at the median for survival analysis. The median duration of follow-up for patients who remain alive was 8.0 years. Of the 379 primary colon carcinomas studied, 72 (19%) were TNM stage II and 307 (81%) were stage III. DNA MMR status had been determined in 315 tumors and 37 (11.7%) cancers were found to have defective MMR (Table 1). These tumors were significantly more likely to be from females (P = 0.0049), to be located in the proximal colon (P < 0.0001), and to be poor/undifferentiated (P = 0.0030) compared with tumors with intact MMR as reported by ourselves and others (16, 19, 22–24).
Table 1.
Tumor markers, clinicopathologic features, and patient survival
| Parameter | Total n | 5-y DFS (%) | HR (95% CI) | P* | 5-y OS (%) | HR (95% CI) | P* |
|---|---|---|---|---|---|---|---|
| Bad expression† | |||||||
| Low | 282 (74.4) | 59.4 | — | 0.1175 | 65.5 | — | 0.0313 |
| High | 97 (25.6) | 67.9 | 0.73 (0.49–1.09) | 80.4 | 0.64 (0.43–0.96) | ||
| Bid expression† | |||||||
| Low | 112 (29.6) | 56.8 | — | 0.1421 | 63.1 | — | 0.0601 |
| High | 267 (70.4) | 63.6 | 0.77 (0.54–1.09) | 71.9 | 0.72 (0.51–1.02) | ||
| Bad + Bid† | |||||||
| Low | 88 (23.2) | 52.9 | 0.0267 | 58.6 | 0.0060 | ||
| High | 291 (76.8) | 64.2 | 0.66 (0.46–0.96) | 72.5 | 0.61 (0.42–0.87) | ||
| MMR (n = 315) | |||||||
| Intact | 278 (88.3) | 61.3 | — | 0.2123 | 69.3 | — | 0.1983 |
| Defective | 37 (11.7) | 70.3 | 0.67 (0.36–1.26) | 75.7 | 0.67 (0.36–1.24) | ||
| Age (y) | |||||||
| ≤65 | 199 (52.5) | 63.3 | — | 0.6662 | 71.9 | — | 0.3071 |
| >65 | 180 (47.5) | 59.8 | 0.93 (0.67–1.29) | 66.5 | 0.85 (0.61–1.17) | ||
| TNM stage | |||||||
| II | 72 (19) | 73.6 | — | 0.0597 | 79.2 | — | 0.0288 |
| III | 307 (81) | 58.8 | 1.59 (0.98–2.59) | 67.0 | 1.73 (1.05–2.85) | ||
| Histologic grade | |||||||
| 1, 2 (well/moderate) | 245 (64.4) | 63.9 | — | 0.0712 | 73.8 | — | 0.0119 |
| 3, 4 (poor/undifferentiated) | 134 (35.4) | 57.4 | 1.36 (0.97–1.90) | 61.2 | 1.52 (1.09–2.12) | ||
| Gender | |||||||
| Female | 190 (50.1) | 63.6 | — | 0.4360 | 70.5 | — | 0.4815 |
| Male | 189 (49.9) | 59.6 | 1.14 (0.82–1.58) | 68.1 | 1.12 (0.81–1.55) | ||
| Tumor site | |||||||
| Distal§ | 189 (49.9) | 59.3 | — | 0.2815 | 67.7 | — | 0.8314 |
| Proximal | 190 (50.1) | 64.0 | 0.84 (0.60–1.16) | 70.9 | 0.97 (0.70–1.34) | ||
Score test P from a Cox regression model after stratifying by study.
See Materials and Methods. For Bad + Bid, high refers to either or both variables showing high expression.
Distal to and including the splenic flexure.
Bad and Bid expression
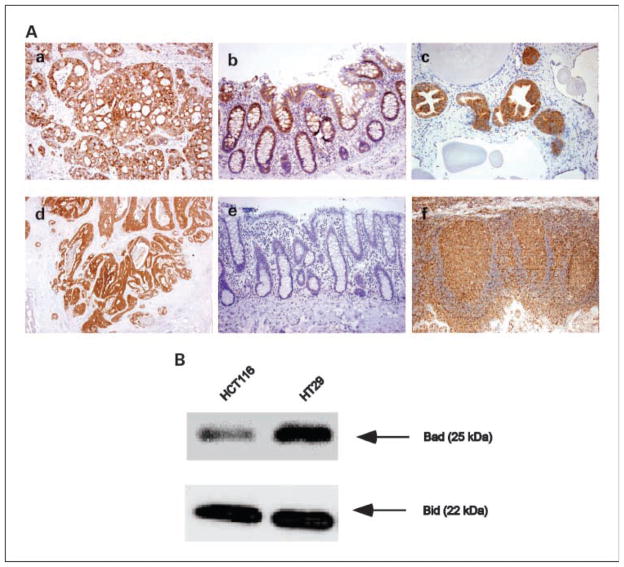
Bad and Bid staining localized to the cytoplasm of the tumor cells (Fig. 1A). High Bad expression was detected in 97 (25.6%) carcinomas and low expression was found in 282 (74.4%; Table 1). High Bid expression was found in 267 (70.4%) tumors and low Bid expression was detected in 112 (29.6%) cases. For a semiquantitative assessment of the level of protein expression, we used immunointensity for Bad and immunopercent for Bid. Adjacent histologically normal-appearing colonic mucosa was present in 234 of 379 (61.7%) tumor specimens (Fig. 1A). Bad and Bid expression (immunointensity) were significantly increased in tumor relative to normal colonic epithelium (both P < 0.0001).
Fig. 1.
A, immunohistochemical analysis of Bad and Bid proteins. a, colon carcinoma shows a high level of Bad expression in the tumor cell cytoplasm. b, normal colonic mucosa is positive for Bad staining. c, human prostate serves as a positive control for Bad staining. d, colon carcinoma shows Bid expression in a majority of tumor cells. e, normal colonic mucosa is negative for Bid staining. f, human tonsil serves as a positive control for Bid staining. Magnification, ×20 (a–e) and ×10 (f). B, Western blot of Bad and Bid expression in HCT116 and HT-29 human colon carcinoma cell lines. Blot shows the specificity of the antibodies for Bad and Bid proteins in both cell lines as indicated by a single band corresponding to their known molecular weights.
The specificity of the Bad and Bid antibodies was evaluated in HT-29 and HCT116 human colon carcinoma cell lines. In the Western blot assay, a single band corresponding to the known molecular weights of Bad or Bid proteins was detected (Fig. 1B). Within the tissue sections, determination of the antibody specificity was done using appropriate positive and negative tissue controls with the expected results obtained (Fig. 1A; refs. 14, 25).
Bad and Bid expression were significantly correlated with one another (P = 0.0322). High (versus low) Bid expression was associated with well/moderate versus poor/undifferentiated tumors (P = 0.0238). Bad expression did not correlate with clinicopathologic features. Bad or Bid expression did not differ based on adjuvant treatment status. Bid expression was strongly correlated with the tumor apoptotic index (n = 94). Specifically, tumors with high (versus low) Bid expression showed a higher median number of TUNEL stained tumor cells (P = 0.0076; Table 2). Bad expression was unrelated to tumor apoptotic indices.
Table 2.
Bid expression and apoptotic index
| Apoptotic index | Bid low (n = 33) | Bid high (n = 61) | P* |
|---|---|---|---|
| Mean (SD) | 0.8 (0.47) | 1.0 (0.44) | 0.0076 |
| Median | 0.7 | 0.9 | |
| Range | (0.0–1.9) | (0.3–2.6) |
Wilcoxon rank-sum test.
Tumor characteristics and patient survival rates
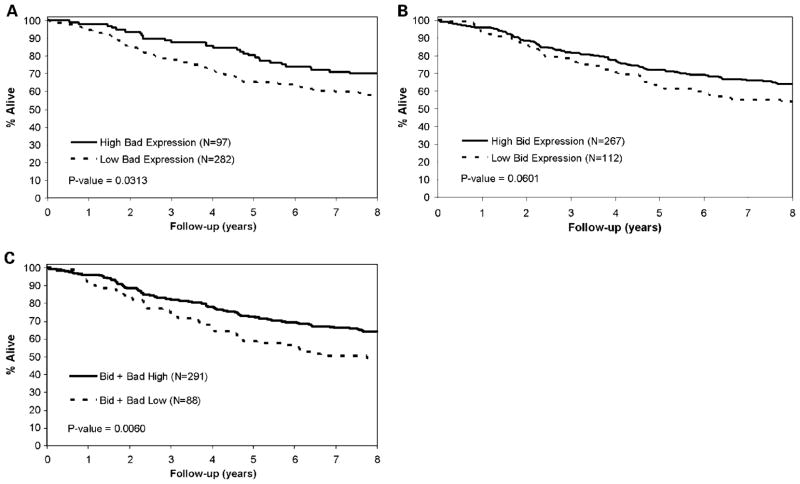
A significant association was observed between longer OS and high Bad expression (Table 1; Fig. 2A). Specifically, 80.4% of patients whose tumors showed high level Bad were alive at 5 years postsurgical resection compared with 65.5% for tumors with low Bad expression (P = 0.0313). Additionally, 71.9% of patients whose tumors showed high Bid expression were alive at 5 years postresection compared with 63.1% for tumors with low Bid levels (P = 0.0601; Table 1; Fig. 2B). We also examined the combined Bad + Bid variable. As shown in Table 1, high expression of the combined variable was significantly associated with better DFS (P = 0.0267) and OS (P = 0.0060) (Fig. 2C) rates. Calculation of hazard ratios [HR; with 95% confidence interval (95% CI)] is shown as estimates of relative risk of death due to colon cancer (Table 1). The favorable prognostic effect of Bad expression was observed in both stages II and III cancers as shown by similar HRs, although fewer stage II tumors were studied. Similar correlations with OS were obtained for Bad and Bid expression when tumors with defective MMR were excluded from the analysis (data not shown), thus excluding bias that might arise from inclusion of tumors that arise through an alternative molecular pathway.
Fig. 2.
A, OS in patients withTNM stages II and III colon carcinoma by Bad expression. B, OS in patients withTNM stages II and III colon carcinoma by Bid expression. C, OS in patients withTNM stages II and III colon carcinoma by the Bad + Bid combined variable.
In a multivariate analysis, a Cox proportional hazards model was used to evaluate whether the BH3-only proteins Bad and Bid showed independent prognostic significance. High levels of Bad and Bid expression were independent prognostic markers for better OS after adjustment for histologic grade, tumor stage, treatment, age, and study (Table 3). HR calculations indicated a reduced relative risk of death from colon cancer of 36% for high Bad and 28% for high Bid expression (Table 1). Furthermore, expression of the combined variable of Bad + Bid was an independent predictor of both DFS (HR, 0.64; 95% CI, 0.44–0.92; P = 0.0202) and OS (Table 4).
Table 3.
Multivariate analysis of OS (n = 379)
| Variable | HR (95% CI) | P* |
|---|---|---|
| Bad expression (high vs low) | 0.65 (0.43–0.98) | 0.0336 |
| Bid expression (high vs low) | 0.69 (0.49–0.98) | 0.0420 |
| Histologic grade (3, 4 vs 1, 2)† | 1.65 (1.18–2.31) | 0.0037 |
| TNM stage (III vs II) | 1.80 (1.09–2.96) | 0.0140 |
| Treatment (effective vs ineffective) | 0.62 (0.40–0.97) | 0.0429 |
| Age (≤65 vs >65 y) | 0.86 (0.62–1.19) | 0.3701 |
Likelihood ratio P, adjusted for histologic grade, tumor stage, treatment, age, and study.
1, 2 (well/moderate) and 3, 4 (poor/undifferentiated).
Table 4.
Multivariate analysis of OS (n = 379)
| Variable | HR (95% CI) | P* |
|---|---|---|
| Bad + Bid (high vs low) | 0.57 (0.40–0.82) | 0.0038 |
| Histologic grade (3, 4 vs 1, 2)† | 1.69 (1.21–2.36) | 0.0025 |
| TNM stage (III vs II) | 1.84 (1.12–3.03) | 0.0101 |
| Treatment (effective vs ineffective) | 0.61 (0.38–0.96) | 0.0354 |
| Age (≤65 vs >65 y) | 0.86 (0.62–1.20) | 0.3779 |
Likelihood ratio P, adjusted for histologic grade, tumor stage, treatment, age, and study.
1, 2 (well/moderate) and 3, 4 (poor/undifferentiated).
Discussion
Diverse intracellular damage signals, including those evoked by cancer chemotherapy, are transduced by the BH3-only proteins, including Bad and Bid, which inactivate prosurvival Bcl-2 family members and commit the cell to apoptosis. In this study, we found a high level of Bad expression in 97 (25.6%) cases that was significantly associated with better OS in patients with curatively resected stages II and III colon cancers treated in 5-FU-based adjuvant therapy trials. Specifically, cancers with high Bad expression showed a 36% reduction in 5-year OS rates compared with tumors with low Bad levels. Additionally, high Bid expression was detected in 267 (70.4%) cases and was associated with better OS that was of borderline significance in a univariate analysis. The combined variable of Bad + Bad was highly discriminant for both DFS and OS. Moreover, multivariate analysis revealed that both high levels of Bad and Bid expression were independent predictors of improved OS after adjustment for tumor stage, histologic grade, age, study, and treatment. The combined variable of Bad + Bad was independently associated with DFS and OS. The reliability of our study results is aided by the availability and rigorous collection of long-term (median, 8 years) survival data. Consistent with our findings in colon cancers, Krajewska et al. (15) found that higher levels of Bid expression were associated with longer recurrence-free survival in men with locally advanced prostate cancer. The interpretation of our data is further supported by the observation that tumor stage and grade were prognostic and have consistently been the most robust prognostic markers in colorectal cancer patients (26), including prior reports using these same studies (19, 22).
Within tumor cells, Bad functions to dimerize with Bcl-xL and with Bcl-2 and can neutralize the prosurvival function of Bcl-xL (27). Dimerization of Bad with Bcl-xL results in displacement of Bax from the Bcl-xL/Bax complex, thereby causing restoration of Bax-mediated apoptosis. Bad is regulated by its phosphorylation status and when phosphoryated at serine residues is unable to dimerize with Bcl-xL. The explanation for the higher frequency of Bid relative to Bad expression found in our study is unclear. In humans, a mutation in the BH3 domain of the Bad gene was reported in only 2 of 47 colon adenocarcinomas analyzed and was shown to strongly reduce the binding of Bad to Bcl-2 and Bcl-xL (28). Further studies to analyze other genetic mechanisms or potential epigenetic silencing of the Bad gene may resolve this issue. The proapoptotic Bid protein requires proteolytic processing typically by a caspase (29, 30) or granzyme B (31). Bid can be activated by members of the tumor necrosis factor (TNF) family, including the novel anticancer drug TRAIL/Apo2L (32). Therefore, Bid connects the TNF family death receptor apoptotic pathway with the mitochondrial pathway (29, 30, 32). Although no data are available on the frequency of Bid mutations in colorectal cancers, inactivating mutations in the Bid gene were detected in only 1 of 67 advanced gastric cancers studied (33).
Given that Bad and Bid are proapoptotic proteins, we analyzed their expression in relationship to tumor cell apoptotic indices. We found that Bid, but not Bad, expression was significantly correlated with increased tumor cell apoptosis as determined by TUNEL staining. This finding is consistent with evidence showing that Bid as well as the BH3-only proteins Bim and Puma engage all prosurvival proteins that constrain them and are thus highly potent inducers of apoptosis compared with the selective BH3 proteins Bad and Noxa (6). Furthermore, Bad has been reported to be phosphorylated in human tumors and this alteration appears to prevent its proapoptotic function (8). Studies in BH3 knockout mice have confirmed the importance of Bad and Bid proteins in tumorigenesis and in apoptotic susceptibility. Spontaneous tumorigenesis was observed in a cohort of aged Bad−/− mice, with diffuse large B-cell lymphomas being the most frequent tumor observed, accompanied by hematopoietic malignancies (34). Furthermore, Bad-deficient mice succumbed to radiation-induced thymic lymphomas significantly earlier than their wild-type littermates (34). Mice lacking Bid spontaneously develop a clonal malignancy that resembles human chronic myelomonocytic leukemia (35). Bid also plays a role in the DNA damage response that is mediated through Bid phosphorylation and is independent of its proapoptotic role (36). Together, these data support a tumor suppressor role of these BH3-only genes. In this regard, our finding that low or absent expression of Bad and Bid proteins is associated with worse clinical outcome is consistent with a tumor suppressor role for these proteins in human colon cancers.
BH3-only protein expression is highly relevant to chemo-sensitivity. Knockdown of Bad and Bid have been shown to confer resistance to anticancer drugs, including 5-FU (37, 38). Furthermore, overexpression of a Bid mutant in 293 T cells impaired the proapoptotic function of Bid in response to 5-FU (33). Accordingly, Bad and Bid expression may be important determinants of the efficacy of 5-FU in vivo, in that low or absent marker expression may hinder a cell’s ability to respond to chemotherapy. Unfortunately, our study does not permit an evaluation of the predictive utility of Bad and Bid proteins. The majority of patients in our study received 5-FU-based adjuvant therapy, and due to the small number of untreated patients, we could not appropriately test for the interaction between marker expression and treatment. We did adjust for treatment effect in the multivariate model and Bad and Bid expression remained as independent prognostic variables after taking into account differences in treatment.
In conclusion, proapoptotic Bad and Bid proteins are independent prognostic variables for stages II and III colon cancer patients. These data also indirectly support the rationale for targeting prosurvival Bcl-2 proteins using newly developed BH3 mimetic agents to enhance tumor cell apoptosis (39, 40) that have entered phase I testing. Validation of our study results in an independent data set is planned, and evaluation of these markers in colon cancer patients treated with current standard adjuvant chemotherapy (FOLFOX) is warranted. If validated, our results suggest that Bad and Bid expression may be able to risk stratify colon cancer patients and potentially to aid in the selection of patients to receive adjuvant therapy.
Acknowledgments
Grant support: National Cancer Institute grant CA104683-02 (F.A. Sinicrope) and Mayo Clinic Cancer Center core grant CA15083.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Bedi A, Pasricha PJ, Akhtar AJ, et al. Inhibition of apoptosis during development of colorectal cancer. Cancer Res. 1995;55:1811–6. [PubMed] [Google Scholar]
- 3.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–37. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willis SN, Fletcher JI, Kaufmann T, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–9. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 5.Reed JC. Proapoptotic multidomain Bcl-2/Bax-family proteins: mechanisms, physiological roles, and therapeutic opportunities. Cell Death Differ. 2006;13:1378–86. doi: 10.1038/sj.cdd.4401975. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Willis SN, Wei A, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 7.Kim H, Rafiuddin-Shah M, Tu HC, et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol. 2006;8:1348–58. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 8.Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BADin response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–28. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 9.Walensky LD, Kung AL, Escher I, et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–70. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonnell TJ, Korsmeyer SJ. Progression from lymphoid hyperplasia to high-grade malignant lymphoma in mice transgenic for the t(14;18) Nature. 1991;349:254–6. doi: 10.1038/349254a0. [DOI] [PubMed] [Google Scholar]
- 11.Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci U S A. 2004;101:6164–9. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allegra CJ, Parr AL, Wold LE, et al. Investigation of the prognostic and predictive value of thymidylate synthase, p53, and Ki-67 in patients with locally advanced colon cancer. JClin Oncol. 2002;20:1735–43. doi: 10.1200/JCO.2002.07.080. [DOI] [PubMed] [Google Scholar]
- 13.O’Connell MJ, Sargent DJ, Windschitl HE, et al. Randomized clinical trial of high-dose levamisole combined with 5-fluorouracil and leucovorin as surgical adjuvant therapy for high-risk colon cancer. Clin Colorectal Cancer. 2006;6:133–9. doi: 10.3816/ccc.2006.n.030. [DOI] [PubMed] [Google Scholar]
- 14.Kitada S, Krajewska M, Zhang X, et al. Expression and location of pro-apoptotic Bcl-2 family protein BAD in normal human tissues and tumor cell lines. Am J Pathol. 1998;152:51–61. [PMC free article] [PubMed] [Google Scholar]
- 15.Krajewska M, Krajewski S, Banares S, et al. Elevated expression of inhibitor of apoptosis proteins in prostate cancer. Clin Cancer Res. 2003;9:4914–25. [PubMed] [Google Scholar]
- 16.Thibodeau SN, French AJ, Cunningham JM, et al. Microsatellite instability in colorectal cancer: different mutator phenotypes and the principal involvement of hMLH1. Cancer Res. 1998;58:1713–8. [PubMed] [Google Scholar]
- 17.Parc YR, Halling KC, Wang L, et al. HMSH6 alterations in patients with microsatellite instability-low colorectal cancer. Cancer Res. 2000;60:2225–31. [PubMed] [Google Scholar]
- 18.Garrity MM, Burgart LJ, Mahoney MR, et al. Prognostic value of proliferation, apoptosis, defective DNA mismatch repair, and p53 overexpression in patients with resected Dukes’ B2 or C colon cancer: a North Central Cancer Treatment Group Study. J Clin Oncol. 2004;22:1572–82. doi: 10.1200/JCO.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 19.Sinicrope FA, Rego RL, Halling KC, et al. Thymidylate synthase expression in colon carcinomas with microsatellite instability. Clin Cancer Res. 2006;12:2738–44. doi: 10.1158/1078-0432.CCR-06-0178. [DOI] [PubMed] [Google Scholar]
- 20.Cox DR. Regression models and life tables. JR Stat Soc. 1972;34:187–202. [Google Scholar]
- 21.Grambsch PMT, Terry M. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;8:515–26. [Google Scholar]
- 22.Sinicrope FA, Rego RL, Halling KC, et al. Prognostic impact of microsatellite instability and DNA ploidy in human colon carcinoma patients. Gastroenterology. 2006;131:729–37. doi: 10.1053/j.gastro.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–9. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 24.Lothe RA, Peltomaki P, Meling GI, et al. Genomic instability in colorectal cancer: relationship to clinicopathological variables and family history. Cancer Res. 1993;53:5849–52. [PubMed] [Google Scholar]
- 25.Krajewska M, Zapata JM, Meinhold-Heerlein I, et al. Expression of Bcl-2 family member Bid in normal and malignant tissues. Neoplasia. 2002;4:129–40. doi: 10.1038/sj.neo.7900222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Compton CC. Colorectal carcinoma: diagnostic, prognostic, and molecular features. Mod Pathol. 2003;16:376–88. doi: 10.1097/01.MP.0000062859.46942.93. [DOI] [PubMed] [Google Scholar]
- 27.Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ. Bad, a heterodimeric partner for Bcl-xL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80:285–91. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 28.Lee JW, Soung YH, Kim SY, et al. Inactivating mutations of proapoptotic Bad gene in human colon cancers. Carcinogenesis. 2004;25:1371–6. doi: 10.1093/carcin/bgh145. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 30.Kim TH, Zhao Y, Barber MJ, Kuharsky DK, Yin XM. Bid-induced cytochrome c release is mediated by a pathway independent of mitochondrial permeability transition pore and Bax. J Biol Chem. 2000;275:39474–81. doi: 10.1074/jbc.M003370200. [DOI] [PubMed] [Google Scholar]
- 31.Waterhouse NJ, Sedelies KA, Browne KA, et al. A central role for Bid in granzyme B-induced apoptosis. J Biol Chem. 2005;280:4476–82. doi: 10.1074/jbc.M410985200. [DOI] [PubMed] [Google Scholar]
- 32.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–90. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 33.Lee JH, Soung YH, Lee JW, et al. Inactivating mutation of the proapoptotic gene BID in gastric cancer. J Pathol. 2004;202:439–45. doi: 10.1002/path.1532. [DOI] [PubMed] [Google Scholar]
- 34.Ranger AM, Zha J, Harada H, et al. Bad-deficient mice develop diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9324–9. doi: 10.1073/pnas.1533446100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zinkel SS, Ong CC, Ferguson DO, et al. Proapoptotic BID is required for myeloid homeostasis and tumor suppression. Genes Dev. 2003;17:229–39. doi: 10.1101/gad.1045603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zinkel SS, Hurov KE, Ong C, Abtahi FM, Gross A, Korsmeyer SJ. A role for proapoptotic BID in the DNA-damage response. Cell. 2005;122:579–91. doi: 10.1016/j.cell.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 37.Labi V, Erlacher M, Kiessling S, Villunger A. BH3-only proteins in cell death initiation, malignant disease and anticancer therapy. Cell Death Differ. 2006;13:1325–38. doi: 10.1038/sj.cdd.4401940. [DOI] [PubMed] [Google Scholar]
- 38.Sax JK, Fei P, Murphy ME, Bernhard E, Korsmeyer SJ, El-Deiry WS. BID regulation by p53 contributes to chemosensitivity. Nat Cell Biol. 2002;4:842–9. doi: 10.1038/ncb866. [DOI] [PubMed] [Google Scholar]
- 39.Oltersdorf T, Elmore SW, Shoemaker AR, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–81. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 40.Trudel S, Li ZH, Rauw J, Tiedemann RE, Wen XY, Stewart AK. Preclinical studies of the pan-Bcl inhibitor obatoclax (GX015-070) in multiple myeloma. Blood. 2007;109:5430–8. doi: 10.1182/blood-2006-10-047951. [DOI] [PubMed] [Google Scholar]