Abstract
Background
Caries and periodontitis are important human diseases associated with formation of multi-species biofilms. The involved bacteria are intensively studied to understand the molecular basis of the interactions in such biofilms. This study established a basic in vitro single and mixed-species culture model for oral bacteria combining three complimentary methods. The setup allows a rapid screening for effects in the mutual species interaction. Furthermore, it is easy to handle, inexpensive, and reproducible.
Methods
Streptococcus mitis, S. salivarius and S. sanguinis, typical inhabitants of the healthy oral cavity, S. mutans as main carriogenic species, and Porphyromonas gingivalis, Fusobacterium nucleatum, Parvimonas micra, S. intermedius and Aggregatibacter actinomycetemcomitans as periodontitis-associated bacteria, were investigated for their biofilm forming ability. Different liquid growth media were evaluated. Safranin-staining allowed monitoring of biofilm formation under the chosen conditions. Viable counts and microscopy permitted investigation of biofilm behavior in mixed-species and transwell setups.
Findings
S. mitis, F. nucleatum, P. gingivalis and P. micra failed to form biofilm structures. S. mutans, S. sanguinis, S. intermedius and S. salivarius established abundant biofilm masses in CDM/sucrose. A. actinomycetemcomitans formed patchy monolayers. For in depth analysis S. mitis, S. mutans and A. actinomycetemcomitans were chosen, because i) they are representatives of the physiological-, cariogenic and periodontitis-associated bacterial flora, respectively and ii) their difference in their biofilm forming ability. Microscopic analysis confirmed the results of safranin staining. Investigation of two species combinations of S. mitis with either S. mutans or A. actinomycetemcomitans revealed bacterial interactions influencing biofilm mass, biofilm structure and cell viability.
Conclusions
This setup shows safranin staining, microscopic analysis and viable counts together are crucial for basic examination and evaluation of biofilms. Our experiment generated meaningful results, exemplified by the noted S. mitis influence, and allows a fast decision about the most important bacterial interactions which should be investigated in depth.
Introduction
Caries and Periodontitis are extremely frequent human diseases with high socioeconomic impact. They are associated with several potentially severe complications due to bacterial invasion of neighbouring anatomical structures or haematogenous spreading and purulent infections at distant sites. If not managed by appropriate therapies, both diseases are chronically progressive. Their pathogenesis is explained by a locally disturbed microecology within the bacterial biofilms covering the surfaces of the teeth and the subgingival sulci.
Bacterial biofilms are able to form and spread on the surfaces of the teeth in healthy oral cavities. Such biofilms display typical structural features such as i) a chemically conditioned support, ii) pioneer bacteria firmly adhering to the support's surface, iii) microcolony formation and production of macromolecular extracellular substances, iv) attachment of secondary colonizer binding to the growing biofilm, v) a predefined maximum thickness due to a balance between biofilm production and detachment (i.e. maturation) processes. These biofilms may contain up to several hundred bacterial species. These bacterial consortia are inconsistent between individual sites in one oral cavity and even more between diverse oral cavities [1]–[4].
Because of the extensive species variation between human individuals the concept of specific indicator bacteria for physiological and pathological biofilms in oral cavities is currently modified [5]–[9]. However, it is generally accepted that the cell number of the involved bacteria changes in a species-dependent manner during disease development. Simultaneously, individually differing species disappear below detection level while new species are temporally or constantly detectable during disease development [10], [11].
The involvement of many species and their constant qualitative and quantitative variations makes it extremely complicated to setup in vitro biofilms that truly reflect the natural situation. However, establishing of in vitro biofilms is still necessary to investigate substances suitable for the suppression of caries or periodontitis.
While the epidemiology of the microflora in healthy and diseased oral cavities has greatly been promoted by the introduction of advanced microscopic and molecular techniques, in vitro experiments have to rely on classical culture methods. It is currently impossible to grow representative in vitro multi-species biofilms resembling those encountered during caries or periodontitis. However, mixing a few important species to mimic biofilms encountered during complete health, transition to disease, developing disease, and finally, in deep lesions appears to be feasible. Such studies were performed in many laboratories [e.g. 12]–[20]. Depending on the scientific question in these laboratories different experimental setups were developed. Disparate setups and methods predominantly comprised 1) the species used in the studies, 2) the incubation conditions (static or flow, aerobic or anaerobic), which are very important and should mimic the environmental and physical parameters of the in vivo biofilm niche, 3) the documentation of biofilm formation, biofilm mass and biofilm maturation, and 4) the quantification of the individual species contained in the biofilms. Thus, a setup which could be used in many labs and which proves useful for many different scientific questions would be beneficial to basically investigate and understand biofilm formation and bacterial interaction in biofilm structures.
In the present study, we intended to set up conditions for the investigation of bacterial biofilm formation and combined three complimentary methods (cfu, safranine staining, microscopy) as a basis for multi-species culture investigation. We used Streptococcus mitis, S. salivarius, S. mutans, S. sanguinis, S. intermedius, Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Fusobacterium nucleatum and Parvimonas micra as bacteria associated with a healthy oral cavity, caries, and periodontitis, respectively. Based on the experimental protocols used in this study, we could demonstrate in mixed species assays as well as in assays employing separating filter membranes between the partners, increased or decreased contribution of single species to biofilm formation and effects on viability exclusively in one co-incubation partner.
Results
Evaluation of basic parameters of biofilm formation
Investigation of biofilm structures and bacterial interaction required establishment of reliable biofilm setup protocols. For this purpose, different culture media were tested in a static biofilm setup to evaluate the best conditions for in vitro simulation of biofilm generation. It is assumed that not much liquid exchange occurs during periodontitis in vivo, thus static conditions best mimic this situation and also allow the action of potential signalling molecules in mixed species cultures. Six different media were examined for their effect on mono-species biofilm formation for a time period up to five days. Safranin staining was employed as an easy read-out approach. This method is used for the determination of biofilm mass, comprising bacterial cells and extrapolymeric substances. Typically, an OD492 nm with a value of more than 0.05 is required to indicate biofilm formation. Lower values are mostly caused by scattered bacteria in monolayers (data not shown). For comparison, also the growth curves of planktonic cells were recorded for each culture medium. The table S1 summarizes the results for all bacteria analyzed. However, the present study will only focus on the detailed results of S. mitis, S. mutans and A. actinomycetemcomitans as representatives of the physiological, cariogenic and periodontitis-associated oral microflora, respectively.
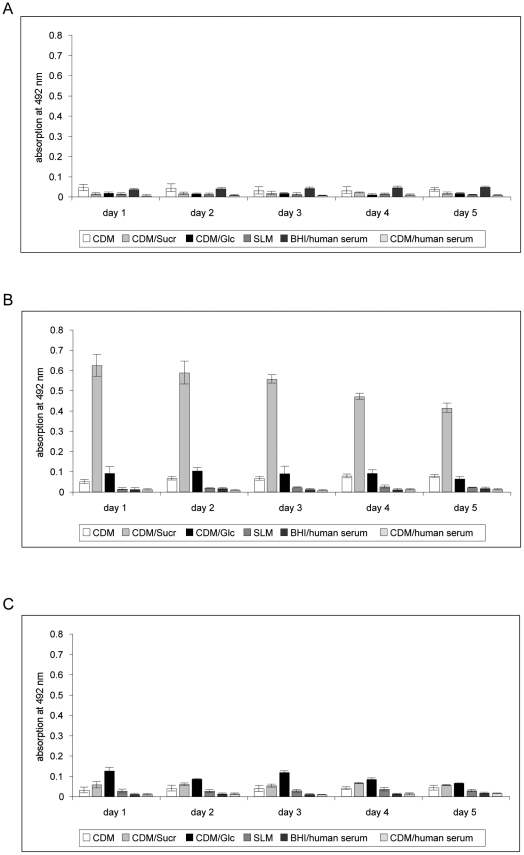
Monitoring the S. mitis, S. mutans and A. actinomycetemcomitans mono-species cultures for biofilm mass over a period of five days showed that biofilm formation of S. mutans occurred within the first 24 hours of incubation time in CDM without glucose (chemically defined medium; for details of composition please refer to reference 54), CDM supplemented with 50 mM glucose (CDM/glc) or sucrose (CDM/suc). The highest amounts of biofilm cell mass was formed in CDM/sucrose. The biofilm mass reached at this time point remained stable during the residual observation period. For A. actinomycetemcomitans, scattered patches of monolayers appeared after one day of incubation in CDM/glucose or CDM/sucrose. No multi-layered structures were observed in any tested medium. S. mitis bacteria failed to establish biofilms in all tested media (figure 1).
Figure 1. Safranin-staining assay of mono-species-cultures in different media.
A), B), C) Results for S. mitis, S. mutans. A. actinomycetemcomitans, respectively. CDM - chemically defined medium, Sucr – sucrose, Glc – glucose, BHI – brain heart infusion, SLM- saliva-like medium.
Growth curves in CDM/sucrose revealed that S. mitis and S. mutans increased in their optical density. A parallel decrease in the medium pH within 24 hours was noted (7.7 to 4.77 and 5.8, respectively). Planktonic A. actinomycetemcomitans did not grow in this medium (figure S1). Nevertheless, determination of cfu/ml showed constant numbers of viable cells and a slight decrease of medium pH (7.7 to 7.47 within 24 h).
Next to the determination of biofilm mass and planktonic growth, viable cell counts and fluorescence microscopy were performed on the samples as outlined in the Materials/Methods section. In CDM/sucrose, the number of surface-adherent viable cells as measured by colony forming units per ml suspension (cfu/ml) decreased after day one of investigation for all three bacterial species. However, viable S. mutans and A. actinomycetemcomitans cells were retrievable at day five of incubation (table S2).
Thus, as the only medium supporting growth or at least viability of all three species while simultaneously allowing monolayer/biofilm growth for at least two species, CDM/sucrose was used for all subsequent experiments.
In this medium, biofilm formation of single bacterial species was also tested in fibronectin-coated wells. Although the biofilm mass was slightly different compared to uncoated plastic supports, all three species behaved similar concerning their biofilm forming ability or formation of monolayers, respectively (figure S2). Subsequently, the experiments were performed using uncoated supports.
Biofilm behavior of two-species cultures
To approach the natural situation and to obtain information about the species interactions, in the next step we employed co-cultivation of S. mitis with S. mutans and/or A. actinomycetemcomitans.
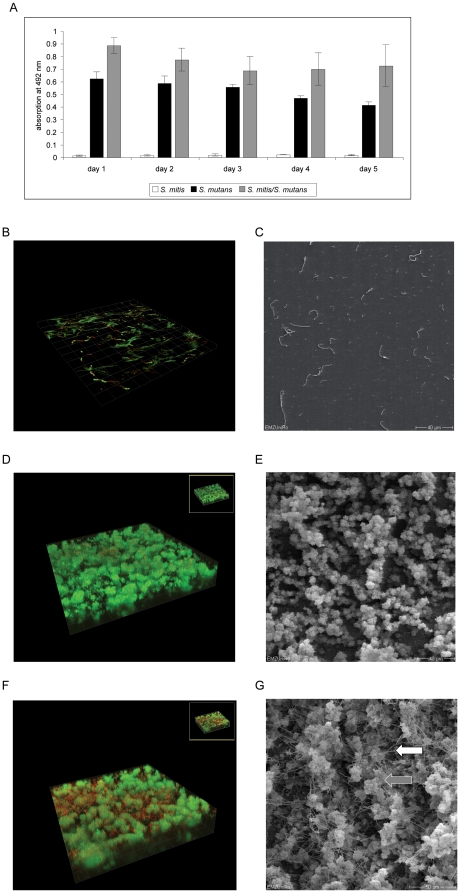
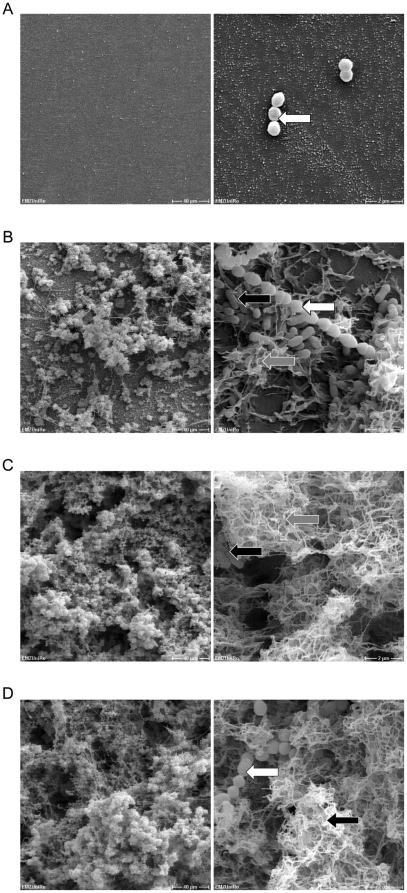
Safranin-assays revealed that the co-cultivation of S. mutans with S. mitis resulted in an increase of total biofilm mass compared to the S. mutans mono-species cultures (figure 2 A). Confocal laser scanning microscopy after live/dead-stain confirmed that S. mitis failed to form biofilms. In the two-species setting with S. mutans the integration of S. mitis within the biofilm structures was noted. Scanning electron microscopy confirmed this result (figure 2 B–F). Here, S. mitis chains could be found on and between the typical extracellular matrix structures synthesized by S. mutans (figure 2 E, grey arrow S. mutans extracellular matrix structure, white arrow S. mitis chain). Of note, within these mixed communities no colony forming units of S. mitis were detectable, whereas S. mutans numbers and viability were unchanged compared to single species settings (table S2). While incubating the two species, the culture fluid acidity reached pH 4.96 after 24 hours and decreased to pH 4.52 after 5 days.
Figure 2. Safranin-staining assay, Confocal laser scanning microscopy (CLSM) and Scanning electron microscopy (SEM) of S. mutans mono-species biofilms, S. mitis adherent cells and S. mutans/S. mitis two-species biofilms.
A) Safranin-staining assay of the mono- and two-species-biofilms of S. mitis and S. mutans. B) CLSM and C) SEM pictures of the S. mitis (mono-species) adherent cells. D) CLSM and E) SEM pictures of the S. mutans mono-species biofilm, F) CLSM and G) SEM pictures of the two-species combination S. mitis/S. mutans. For CLSM detection, cells were stained with Live/Dead dyes. Live cells are stained in green, dead cells light up in red. Grey arrow: S. mutans extracellular matrix structure; white arrow: S. mitis chain.
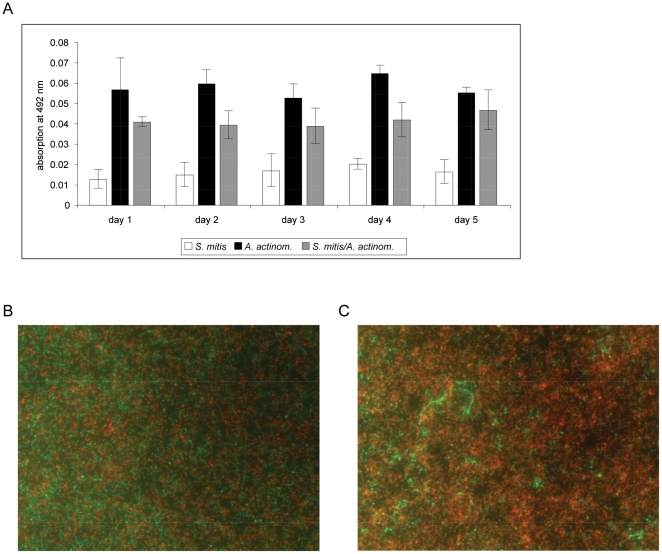
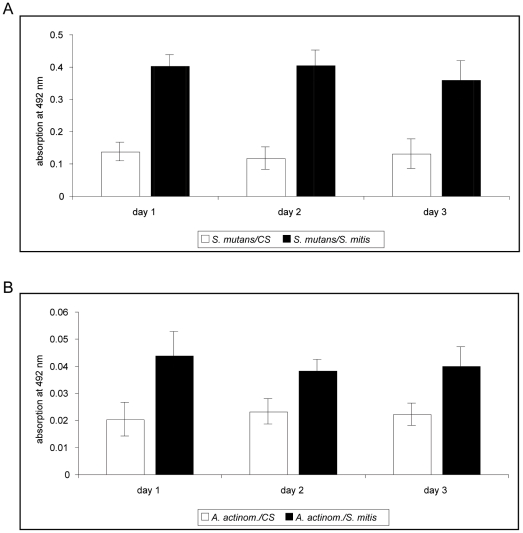
Combining S. mitis with A. actinomycetemcomitans resulted in safranin-stain values that were lower compared to the mono-species culture of A. actinomycetemcomitans (figure 3 A). Concomitantly, viable cell counts decreased for both S. mitis and A. actinomycetemcomitans as compared to single species incubations under the chosen conditions (table S2). Furthermore, the culture fluid pH reached values around 4.55 at day 1, which were lower than those of S. mitis and A. actinomycetemcomitans mono-species cultures (pH 4.77 and 7.47, respectively, day 1).
Figure 3. Safranin-staining assay and fluorescence microscopy of the A. actinomycetemcomitans/S. mitis two-species biofilms.
A) Result of Safranin-staining assay of the mono- and two-species biofilms. B) Fluorescence microscopy of A. actinomycetemcomitans and C) A. actinomycetemcomitans/S. mitis biofilms. For the assay the cells were stained with the Live/Dead dyes. Live cells are stained in green, dead cells light up in red. Magnification: 400×.
SEM and fluorescence microscopy revealed that the surface of the wells was mainly covered by A. actinomycetemcomitans cells with few S. mitis cells on top of their partner cells. Finally, as determined by live/dead stain and viable counts, cells of A. actinomycetemcomitans died faster in the presence of S. mitis compared to mono-species cultures (figure 3 B and C, and table S2, respectively).
Successive seeding strategy
The results from simultaneously seeded two-species cultures indicated the presence of bacterial interaction mechanisms. The aim of the next set of experiments was to evaluate the influence of timing of bacterial adherence on biofilm formation. Thus, we next employed a successive seeding strategy.
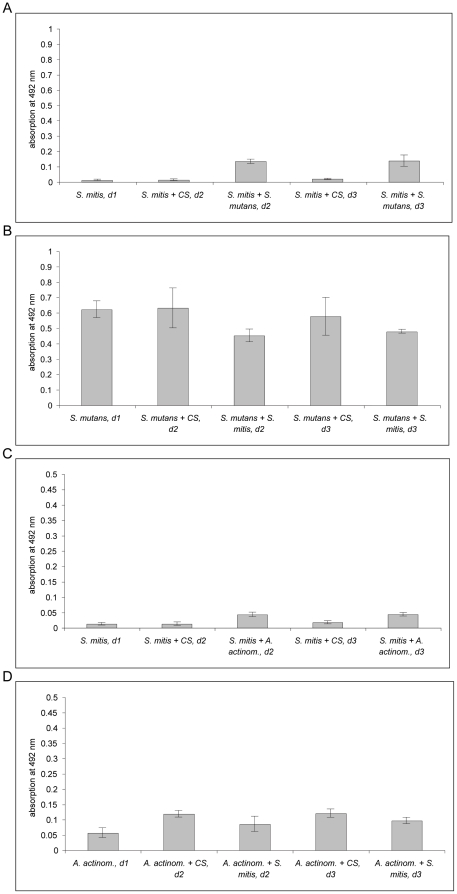
The data obtained from the safranin-assays showed that inoculation of S. mutans on top of S. mitis lead to biofilm formation (figure 4 A). However, biofilm mass was not as abundant as for the S. mutans mono-species culture (compare figure 2 A). In the reverse setup, when S. mutans was used as the primary colonizer, the results showed a decrease of biofilm mass compared to the S. mutans mono-species culture as well as after seeding with CDM/sucrose as control (S. mutans + CS, d2 and d3; figure 4 B). Nevertheless, in both cases the number of S. mutans colony forming units did not differ from those of simultaneously seeded cultures (table S3). For S. mitis no colony forming units were detectable in such successive seeding combinations.
Figure 4. Successive seeding of S. mitis and S. mutans or A. actinomycetemcomitans combinations in biofilm experiments and vice versa.
In both experiments, on day 0 S. mitis, S. mutans or A. actinomycetemcomitans was inoculated into separate wells. Following incubation of 24 hours, biofilm mass of the mono-species was determined by safranin-stain (d1 = day 1). In parallel, S. mutans or A. actinomycetemcomitans was inoculated to S. mitis or, in reverse order, S. mitis to S. mutans or A. actinomycetemcomitans. After further incubation for 24 or 48 hours, biofilm mass was again quantified by safranin-staining (d2 = day 2 and d3 = day 3). The graph shows the data obtained by safranin-staining for the combination S. mitis with S. mutans and vice versa (A and B) and S. mitis with A. actinomycetemcomitans and vice versa (C and D). For better optical discrimination, the grading of the y-axis is different in both graphs. CS – chemically defined medium with sucrose.
In order to visually complement the results from the S. mitis/S. mutans successive seeding assays we performed scanning electron microscopy (SEM). As shown in figure 5 A–D, SEM pictures were consistent with the data obtained from safranin-stain. The inoculation of S. mutans to S. mitis led to biofilm formation, dominated by S. mutans and its extracellular matrix structures (figure 5 B, black arrow S. mutans, white arrow S. mitis, grey arrow S. mutans extracellular matrix structure). In the reverse order, the inoculation of S. mitis to S. mutans showed again the integration of S. mitis into the S. mutans biofilm, which also consisted of abundant extracellular matrix (figure 5 D).
Figure 5. SEM analysis of biofilms obtained by successive seeding of S. mitis and S. mutans.
A) S. mitis mono-culture. B) S. mitis as first colonizer, S. mutans as second species. C) S. mutans mono-culture. D) S. mutans as first colonizer, S. mitis inoculated as second species. Magnification 500× and 10 000×. White arrows: S. mitis; Grey arrows: S. mutans extracellular matrix structure; Black arrows: S. mutans.
Next, the same experimental setup was used to study the S. mitis/A. actinomycetemcomitans interactions. According to safranin-staining, A. actinomycetemcomitans was able to attach to the surface when S. mitis was inoculated as first bacterium, leading to monolayer formation (figure 4 C). The total mass was similar to that of A. actinomycetemcomitans mono-species culture (compare figure 3A). In the reverse seeding order, A. actinomycetemcomitans was enabled to form higher biofilm masses when seeded with CDM/sucrose compared to the mono-species culture. The inoculation of S. mitis led to a marginal decrease of biofilm mass compared to the mono-species control (A. actinomycetemcomitans + CS), but values were still higher as for the A. actinomycetemcomitans mono-species culture (figure 4 D).
Of note, viable cell counts for both bacteria decreased in this experiment, similar to the results obtained for the simultaneously seeded two-species culture (table S3). Fluorescence microscopic analysis of A. actinomycetemcomitans + CDM/sucrose and A. actinomycetemcomitans + S. mitis did not support the results obtained by safranin-stain. No obvious change in number of adherent cells and live/dead stain could be determined (data not shown).
Transwell experiments
We next employed transwell assays to investigate if a change in biofilm mass and/or viable counts as seen in simultaneous and successive seeding experiments was caused by direct cell-cell contact or by soluble substances secreted by the tested species. Therefore, experiments were repeated in the above-mentioned combinations, but the bacteria were separated by a membrane with 0.2 µm sized pores.
The data presented in figure 6 illustrate that S. mutans biofilm formation was stimulated in the presence of S. mitis (figure 6 A). Unfortunately, A. actinomycetemcomitans formed less dense mono-layers on the plastic of the transwell system compared to the 96-well polystyrene microtiter plates. However, adherence of this bacterium increased under the influence of S. mitis (figure 6 B).
Figure 6. Results of safranin-staining assay after transwell experiments.
S. mitis was inoculated in the upper compartment. A) and B) show the results for safranin-staining assays when S. mutans or A. actinomycetemcomitans, respectively, were inoculated in the lower compartment. For better optical discrimination, the grading of the y-axis is different in both graphs. CS – chemically defined medium with sucrose.
The corresponding colony counts for the combinations investigated in the transwell setup are shown in the table S4. Viable counts of S. mitis adherent cells decreased when S. mutans was seeded in the upper compartment. In turn, growth and viable counts of S. mutans were slightly enhanced by the presence of S. mitis in the upper compartment.
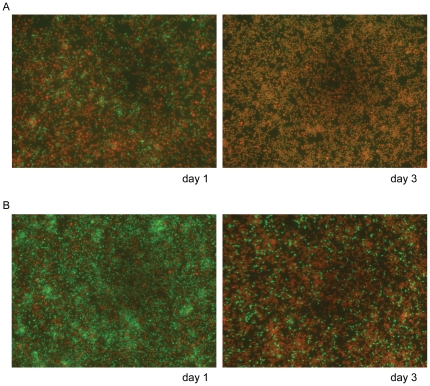
Viable counts of A. actinomycetemcomitans were reduced to zero when S. mitis was present in the upper compartment, whereas S. mitis viability was marginally affected (table S4). In contrast to the results from the viable counts, rod shaped and, according to their green stain, viable A. actinomycetemcomitans cells were detectable in presence of S. mitis, when using fluorescence microscopy after live/dead staining. Moreover, viable A. actinomycetemcomitans cells were still present at day three of co-incubation with S. mitis (figure 7).
Figure 7. Fluorescence microscopy analysis of A. actinomycetemcomitans in the transwell system.
A) A. actinomycetemcomitans mono-species culture in CDM/sucrose at days 1/3. B) A. actinomycetemcomitans grown in the remote presence of S. mitis on days 1/3. For the assay the cells were stained with the Live/Dead dyes. Live cells are stained in green, dead cells light up in red. Magnification: 400× for every picture.
Biofilm susceptibility to degrading substances
To analyze the nature of extracellular substances involved in formation of biofilm masses, we investigated the effect of pronase, DNase and sodium metaperiodate on established biofilms/monolayers. The increase or decrease of the biofilm mass subsequent to exposure would indicate prominent functions of proteins, extracellular DNA (eDNA) or carbohydrates, respectively, in biofilm mass and structure.
For S. mutans mono-species biofilms, an increase of biofilm mass could be observed when DNase was added to the 2 and 3 days old biofilm, whereas pronase treatment had such effects only on 2-day-old biofilms. The addition of sodiummetaperiodate led to a decreased biofilm mass of 1-day-old S. mutans biofilms, whereas treatment at days 2 or 3 caused no change. The analysis of A. actinomycetemcomitans monolayers with these substances revealed a significant decrease of safranin staining intensity when pronase was added to 1, 2 or 3 day old monolayers while the other substances had no obvious effects (figure S3 A–C).
For S. mitis, no biofilm formation was observed after the addition of pronase, DNase or sodiummetaperiodate (data not shown).
Feasability of the basic setup for three-species communities
The next set of experiments was performed to prove the feasibility of the test system for three-species investigations. The mixture of A. actinomycetemcomitans, S. mitis, and S. mutans in a three-species culture revealed safranin values only marginally increased compared to the S. mutans mono-species culture (figure S4 A). In this combination only S. mutans was recovered from the established biofilm (figure S4 B). SEM and fluorescence microscopy visualized S. mitis and S. mutans bacteria in the biofilm, however, no A. actinomycetemcomitans was detected (figure S4 C and D).
Discussion
The aims of this study were i) the establishment of an in vitro setup for mixed-species cultures from which the biofilm behavior of oral bacteria could be studied and which is easy and inexpensive to handle, as well as reproducible in other laboratories, ii) the introduction of a combination of complementary methods to substantiate results, and iii) the demonstration of its usefulness for investigating bacteria from the oral cavity of healthy subjects and/or patients with periodontitis (S. mitis, S. mutans, A. actinomycetemcomitans).
Initially, we sought the best suited medium for biofilm formation of the chosen bacterial species. Brain heart infusion medium (BHI) supplemented with human serum or saliva-like medium (SLM) was studied because of its similarity to sulcus fluid. Both media preparations hardly supported biofilm formation. The included human serum constituents [17] or mucin could potentially interfere with the bacterial adhesion to the plastic surface. Either a sterical hindrance after adhesion to the bacterial cell envelope or changes in the electrical charge of the bacterial or plastic surface are explanations of these effects.
Next, a chemically defined medium without glucose (CDM) supplemented with sucrose was tested and found to be optimal for the biofilm formation of S. mutans, S. salivarius, S. sanguinis, S. intermedius and the monolayer formation of A. actinomycetemcomitans. One major advantage of this medium is that its composition is known in detail, allowing an easy modulation of the presence and/or concentration of amino acids, phosphate or sugar. Unfortunately, the composition of CDM is more remote from sulcus fluid than that of the complex BHI medium. This potential disadvantage could not be resolved by the addition of human serum, due to the negative influence of the supplement on biofilm formation. Yet, a sucrose (or glucose) supplement has been established as an important substrate for the synthesis of extracellular polysaccharides, which in turn are crucial components of streptococcal biofilms [21], [22]. Therefore, the combination of CDM and sucrose was chosen for the experiments.
Although more closely reflecting the natural situation, fibronectin coating of surfaces did not significantly influence the biofilm forming ability of the tested mono-species as compared to uncoated supports.
Introduced washing steps in biofilm formation experiments are critically discussed [23]. For biofilms established under flow conditions Gomez-Suarez et al. [23] described detachment of bacteria from substratum surfaces after air-bubble exposure. However, under the chosen conditions in our study (static conditions) washing steps were crucial to remove sedimented bacteria.
When examining bacterial biofilms, several qualitative or quantitative measurements are established. Safranin-staining predominantly detects extracellular substances and is commonly used to quantify biofilm mass [24]–[26]. Viable cell counting identifies cells from biofilms which are able to multiply when transferred on fresh solid medium. Thus, both dead cells and viable but non-cultureable (VBNC) cells [27], [28] are not detected by this method although these cells contribute to total biofilm mass. By SEM, all cells can be visualized irrespective of their viability. Yet, due to the drying process, extracellular matrix is difficult to detect and visualize by this method. Fluorescence microscopy in combination with Live/Dead stain detects all cells, but not the extracellular substance. Multiplying and VBNC cells are simultaneously visualized as live cells. Finally, confocal laser scanning microscopy combined with Live/Dead stain principally detects the same objects as fluorescence microscopy, although the sterical assignment of cells allows to deduce the presence of extracellular matrix. Due to the different targets detected by the diverse methods, results obtained from a given biofilm could vary. The variation in turn allows conclusions about the association of cell numbers and amount of extracellular matrix, which could be produced by multiplying and VBNC cells. The present study demonstrates the necessity to examine bacterial biofilms with at least three different methods, i.e. safranin-staining, viable counts and microscopic inspection to obtain a complete picture.
Only by employing these three complementary methods, it became evident that A. actinomycetemcomitans and S. mitis behaved contrary in their planktonic growth and biofim behavior. Similar observations were previously reported by Fine and colleagues [29]. A. actinomycetemcomitans did not form multi-layered biofilms but covered the plastic surface by monolayers of viable but not multiplying cells. Several A. actinomycetemcomitans strains have been tested for biofilm formation with varying results [30]–[32]. Parameters like surface conditioning, growth medium and environmental conditions have been described to influence A. actinomycetemcomitans biofilm formation [33]. Obviously, different A. actinomycetemcomitans strains vary in their biofilm growth capabilities, with smooth colony formers growing to less biofilm mass and different biofilm structures [32], [34]. The present A. actinomycetemcomitans strain formed smooth colonies. The tendency of such strains to develop monolayers of viable cells for extended incubation periods has not been described so far. In general A. actinomycetemcomitans is known for its dependence on K+ ion concentration [35], slow growth rate, and limited carbon catabolic capabilities [36], which could possibly explain our observations with this species.
For S. mitis no biofilm formation could be observed under all tested conditions. Based on electron microscopy observations, Cowan et al. [37] demonstrated that S. mitis produced few, extremely long fibrils. These fibrils obviously enable the bacteria to adhere to the underlying substrate [38]. However, the cell surface of S. mitis differs from other oral streptococci in its content of nitrogen and oxygen rich polysaccharides [37]. This could be an explanation for the failure of S. mitis to form biofilm structures under the chosen conditions. The plastic surfaces of the used 96- and 24-well plates obviously did not support S. mitis adhesion. Previous studies demonstrated a dependence of S. mitis biofilm formation on the presence of acquired pellicle and lectins [39]. Similarly, the S. oralis strain C104 formed only small biofilm mass, leading to the conclusion that this species lacks effective colonization factors for binding to abiotic surfaces but can participate in complex biofilms by binding to more successful initial colonizers [40]. The latter statement is confirmed by the present observation on mixed S. mitis/S. mutans biofilms.
A notable result of this study is the obvious change in biofilm mass and viable counts in the two-species combinations compared to single-species settings. S. mitis has been described as a bacterium with an ecological control function in the oral cavity. Precisely, S. mitis could inhibit A. actinomycetemcomitans colonization [41]–[45]. Our results support this observation and associate the inhibitory effects of S. mitis to both the initial step as well as the ensuing multiplication ability of A. actinomycetemcomitans. In literature nutrient depletion and/or pH shift are discussed mechanisms for bacterial inhibitory effects [46]–[48]. However, if these mechanisms apply to the effects of S. mitis on A. actinomycetemcomitans is currently unknown.
Timing and spacing are two critical parameters in the development of mixed-species biofilms. Combinations of S. mitis with S. mutans always resulted in biofilm formation, although the final mass was determined by the timing of the bacterial adherence. Previous studies of van Hoogmoed et al. [42] uncovered the inhibition of S. mutans NS adhesion by biosurfactant-releasing S. mitis strains (S. mitis BA and S. mitis BM). These authors found a release of maximal amounts of biosurfactants, identified to be glycolipids, when the S. mitis strains were grown in the presence of sucrose. However, preliminary results from our laboratory indicate that this biosurfactant production could be a strain specific feature (data not shown). In order to introduce spacing as parameter in the line of investigation, transwell experiments were performed. These studies uncovered discrepancies between cfu values and counts of live/dead-stained cells, suggesting the adoption of a VBNC-status of A. actinomycetemcomitans in indirect contact to S. mitis. Furthermore, the experiments with S. mutans as well as A. actinomycetemcomitans in the remote presence of S. mitis suggested a control function for S. mitis under both conditions. At least for the remote effects, production of secreted substances is the most obvious explanation. The chemical nature of these substances needs to be determined. However, it is known from literature that production of detergents, toxic substances as hydrogen peroxide and bacteriocins or bacteriocin-like inhibitory substances are likely candidates for this effect [49], [50].
Analysis of S. mutans mono-species biofilms in the presence of protein-, DNA- or carbohydrate-degrading substances showed an unexpected effect, i.e. an incubation time-dependent increase of biofilm mass induced by DNase and pronase. Others have shown that presence of DNase during biofilm development leads to a significant disturbance of biofilm formation [51], [52]. These discordant observations are most likely due to different experimental setups. In summary, our results indicate that removal of eDNA after complete biofilm maturation has beneficial effects on a further biofilm mass increase. This observation could also explain the biofilm mass-inducing effects of S. mitis in transwell experiments, i.e. a secretion of enzymes with proteolytic or DNase activity.
In summary, the combined analysis of biofilm formation via safranin stain, determination of cfu and fluorescence microscopy after live/dead-stain allowed fast, unambiguous and reproducible results. Scanning electron- and confocal laser scanning microscopy complemented these results. The whole setup for mono-species cultures could be applied to two- and three-species combinations. This allowed first insights in interactions of chosen bacteria, precisely a mutual influence on biofilm formation and structure, as well as on different levels of viability.
Materials and Methods
Bacterial strains and culture conditions
The bacterial strains Streptococcus mitis ATCC 11843, Streptococcus mutans DSM 20523, Streptococcus sanguinis DSM 20567, Fusobacterium nucleatum DSMZ 25586, Porphyromonas gingivalis W83 ATCC BAA-308, Parvimonas micra ATCC 33270 and Aggregatibacter actinomycetemcomitans (A. actinom.) DSMZ 11123 were purchased from commercial providers (DSMZ, Braunschweig, Germany and ATCC, Manassas, USA). Streptococcus intermedius AC 3105 was obtained from the strain collection of the university Aachen, Germany. S. salivarius K12 was kindly provided by Dr. J. Tagg, New Zealand. Unless otherwise specified, all streptococci and A. actinomycetemcomitans were cultured in brain heart infusion medium (BHI; Oxoid) at 37°C under a 5% CO2 – 20% O2 atmosphere. P. gingivalis was cultivated in BHI supplemented with 5 µg/ml hemin and 50 mM galactose, and F. nucleatum and P. micra were cultured in BHI supplemented 0.25% glutamate. The latter three species were grown at 37°C under an anaerobic atmosphere (10% CO2 - 10% H2 – 80% N2).
Culture media for biofilm studies
For the optimization of biofilm formation, brain heart infusion (BHI, Oxoid, Wesel, Germany) was supplemented with human serum (Sigma, Hamburg/Seelze, Germany) (4∶1) or a saliva-like medium (SLM, 0.1% Lab Lemco Powder, 0.2% yeast extract, 0.5% peptone, 0.25% mucine from porcine stomach, type III (Sigma), 6 mM NaCl, 2.7 mM KCl, 3.5 mM KH2PO4, 1.5 mM K2HPO4, 0.05% urea, pH 6.7) (1∶3). Alternatively, a chemically defined medium (CDM, [53]) was used without any glucose or supplemented with either human serum (4∶1), 50 mM glucose or 50 mM sucrose.
General setup of biofilm cultures
Bacteria were grown in BHI to stationary phase, washed with phosphate buffered saline (PBS, pH 7.4), and adjusted to a strain specific OD600 to obtain 1×10∧8 cells ml∧-1. Subsequently, each bacterial suspension was diluted 10-fold in culture medium and inoculated in polystyrene 24-well-plates (Greiner Bio-One, Frickenhausen, Germany). The bacteria were cultivated alone to establish mono-species biofilms. Alternatively, S. mutans or A. actinomycetemcomitans were cultivated in combination with S. mitis resulting in two-species biofilms.
Biofilm-cultures were grown in an anaerobic incubator under an appropriate atmosphere (80% N2, 10% CO2, 10% H2) at 37°C for periods up to 5 days under static conditions (unless otherwise indicated). The atmosphere of the incubator was saturated with water vapor to prevent exsiccation of the cultures and was constantly exposed to a platinum catalyst to decrease the content of short-chained fatty acids in the atmosphere.
For comparison, planktonic growth of the bacteria in each medium was monitored by batch culture under anaerobic conditions and measuring absorbance at 600 nm.
Biofilm mass and viable counts
For this type of assay, 96-well polystyrene microtiter plates (Greiner Bio-One, Frickenhausen, Germany) were employed. The plastic surfaces of the 96-well plates were either used uncoated or were coated with human fibronectin (Roche) at a concentration of 50 µg/ml∧-1 overnight at 4°C. Prior to the inoculation of the bacteria, fibronectin was removed and wells were washed and air-dried. After incubation of the bacterial cultures, liquid medium was removed and wells were washed gently with PBS in order to remove non-adherent sedimented cells.
For determination of biofilm mass, wells were stained with 0.1% safranin for 15 min, washed with PBS and air-dried. Biofilm mass was quantified in the air-dried wells by measuring the absorbance at 492 nm with a microplate reader (Tecan reader).
Viable cell numbers from biofilm bacteria were obtained by thorough scraping and washing of the wells with PBS. The resulting suspensions were serially diluted in PBS and plated in 100 µl aliquots on BHI-agar. Colony forming units (cfu) were counted after two days of incubation. The distinct colony morphology allowed for differentiation between the species.
Biofilm structure documentation
Mono- or two-species biofilms were cultured in uncoated 24-well polystyrene cell culture plates (Greiner Bio-One, Frickenhausen, Germany), each well containing a sterile, uncoated 13-mm-diameter plastic microscope coverslip (Nunc, Wiesbaden, Germany). After one to five days of incubation under anaerobic conditions, biofilms were gently washed with PBS, stained with BacLight Live/Dead (Molecular Probes, Eugene, Oregon) and inspected by fluorescence microscopy (BX60 microscope, Olympus, Hamburg, Germany). Visible biofilms were documented with an attached digital camera (Leica, Solms, Germany).
In parallel experiments, samples were prepared for scanning electron microscopy (SEM) studies as follows: biofilms on the coverslips were fixed for 24 h in a solution containing 2.5% glutardialdehyde. The coverslips were washed with 0.1 M Na-acetate buffer (pH 7.3) and dehydrated in a graded series of ethanol. Subsequently, coverslips were subjected to critical point drying with CO2, sputter-coated with gold (thickness approx. 10 nm), and examined with a Zeiss DSM 960A electron microscope.
For confocal laser scanning microscopy (CLSM) studies, biofilms were grown in glass-bottom chamber slides (Nunc) and cultured for up to three days under anaerobic conditions. Following incubation, biofilms were gently washed with PBS and stained with BacLight Live/Dead (Molecular Probes, Eugene, Oregon). Preparations were inspected with a Zeiss inverted microscope attached to a Leica TCS SP2 AOBS laser scanning confocal imaging system with an Argon laser at 488-nm excitation wavelength and an Helium/Neon laser at 546-nm excitation wavelength. 3D images were obtained using the IMARIS x64 software.
Transwell biofilm assay
For transwell studies, uncoated 24-transwell polystyrene cell culture plates (Corning) with one coverslip per well were inoculated with 600 µl (10∧7 cfu/ml) of the first bacterial species in the lower compartment and 200 µl (10∧7 cfu/ml) of the second bacterial species in the upper compartment (transwell inserts). Furthermore, uncoated 96 transwell polystyrene microtiter plates were used, containing 200 µl of the first/50 µl of the second bacterial species in the lower/upper compartment, respectively. After one to three days of incubation, transwell inserts and liquid medium were removed. The wells were gently washed with PBS, and biofilms were analyzed for biofilm mass, cell number, as well as biofilm structure using microscopic techniques (see above).
Successive seeding assay
Medium suspensions with 10∧7 S. mitis were inoculated as first bacterial species in an uncoated 96- or 24-well polystyrene plate, the latter with one coverslip per well, and incubated for 24 h under anaerobic conditions at 37°C. Following this incubation time, the liquid medium (containing remaining planktonic bacteria) was removed and 10∧7 S. mutans or A. actinomycetemcomitans suspended in growth medium were inoculated as the second bacterial species into the wells. Subsequently, well plates were again incubated anaerobically at 37°C for up to two days. Biofilm formation was analyzed on a daily basis for two consecutive days by determination of biofilm mass via safranin stain, cell number by counting of cfu/ml and fluorescence microscopy after staining with BacLight Live/Dead (Molecular Probes, Eugene, Oregon). This assay was also performed in reverse order with S. mitis as the second bacterial species inoculated.
Disorganization of biofilms
The disorganization of biofilm was performed as described by Inoue et al. [29] with minor modifications. Mono-species biofilms were cultured in uncoated 96-well polystyrene microtiter plates (Greiner Bio-One, Frickenhausen, Germany) for 1 to 3 days under anaerobic conditions. Following incubation time, liquid medium was removed and wells were washed gently with PBS. Subsequently, 200 µl of pronase (500 µg/ml), DNase (90 units), or sodiummetaperiodate (10 mM) diluted in PBS were added into the wells and the microtiter plates were incubated for further two hours at 37°C under anaerobic conditions. Finally, the liquid was removed and biofilm mass was quantified by safranin-stain (see above).
Reproducibility and statistics
Each assay was performed in at least 3 wells at a given time (technical replicates) and was repeated on at least 3 independent occasions (biological replicates). Where appropriate, statistical parameters (mean, standard deviation of mean, p-Values) were determined employing the Windows Excel program and the Mann-Whitney U Test. P-values less than 0.05 were considered as significant.
Supporting Information
Regular growth curves of A. actinomycetemcomitans, S. mitis, and S. mutans in CDM/sucrose. Growth was monitored by OD600 nm measurements in hourly intervals. One representative experiment of at least three replicates is shown.
(0.20 MB TIF)
Results of safranin-staining assay for the mono-species biofilms on a fibronectin-coated surface. The graph shows the result of safranin-staining assay for the tested mono-species on uncoated and fibronectin-coated surfaces. Fn - fibronectin.
(0.15 MB TIF)
Results of safranin-staining assay for the mono-species biofilm disorganization with pronase, DNase and sodiummetaperiodate. A), B) and C) Results for 500 µg/ml pronase, 90 units DNase, 10 mM sodiummetaperiodate, respectively. SMP - sodiummetaperiodate, * means significance of p<0.05 and ** means significance with p<0.01. PBS was used as control.
(0.38 MB TIF)
Results of Safranin-staining assay, number of colony froming units and microscopic analysis of the S. mitis/S. mutans/A. actinomycetemcomitans three-species combination. A) Safranin-staining assay of the mono- and three species-biofilms of S. mitis, S. mutans and A. actinomycetemcomitans. B) Number of colony forming units for the mono- and three-species cultures. Bacteria in brackets were the corresponding combination partner in the three-species culture. C–D) SEM and fluorescence microcopy of the S. mitis/S. mutans/A. actinomycetemcomitans combination. White arrow: S. mitis; Black arrow: S. mutans.
(4.10 MB TIF)
Summarized results for planktonic and biofilm growth of the different species in the tested media.
(0.09 MB DOC)
Number of colony forming units obtained for the mono- and two-species cultures in CDM/sucrose.
(0.03 MB DOC)
Number of colony forming units for the successive seeding experiment.
(0.02 MB DOC)
Number of colony forming units obtained in transwell experiments.
(0.02 MB DOC)
Acknowledgments
The authors would like to thank Prof. Dr. Dieter Weiss and Dr. Sergei Kuznetsov (Institute of Cell Biology, University of Rostock) for granting access to the IMARIS X64 software (Bitplane).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the Federal Ministry of Education and Research (BMBF) in the framework of the PathoGenoMic Plus program (FKZ 0313801M) and ERANet PathoGenoMics I program (FKZ 0313936B). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol. 2003;30:644–654. doi: 10.1034/j.1600-051x.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- 2.Haffajee AD, Teles RP, Socransky SS. The effect of periodontal therapy on the composition of the subgingival microbiota. Periodontol 2000. 2006;42:219–258. doi: 10.1111/j.1600-0757.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- 3.Diaz PI, Chalmers NI, Rickard AH, Kong C, Milburn CL, et al. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol. 2006;72:2837–2848. doi: 10.1128/AEM.72.4.2837-2848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawes C. Salivary flow patterns and the health of hard and soft oral tissues. J Am Dent Assoc. 2008;139(Suppl):18S–24S. doi: 10.14219/jada.archive.2008.0351. [DOI] [PubMed] [Google Scholar]
- 5.Socransky SS, Haffajee AD; Cugini MA, Smith C, Kent RL. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 6.Slots J, Ting M. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease: occurence and treatment. Periodontol 2000. 1999;20:82–121. doi: 10.1111/j.1600-0757.1999.tb00159.x. [DOI] [PubMed] [Google Scholar]
- 7.Van Winkelhoff AJ, Loos BG, van der Reijden WA, van der Velden U. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. J Clin Periodontol. 2002;29:1023–1028. doi: 10.1034/j.1600-051x.2002.291107.x. [DOI] [PubMed] [Google Scholar]
- 8.Riep B, Edesi-Neuβ L, Claessen F, Skarabis H, Ehmke B, et al. Are putative periodontal pathogens reliable diagnostic markers? J Clin Microbiol. 2009;47:1705–1711. doi: 10.1128/JCM.01387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlafer S, Riep B, Griffen AL, Petrich A, Hübner J, et al. Filifactor alocis – involvement in periodontal biofilms. BMC Microbiol. 2010;10:66. doi: 10.1186/1471-2180-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stingu C-S, Eschrich K, Rodloff AC, Schaumann R, Jentsch H. Periodontitis is associated with a loss of colonization by Streptococcus sanguinis. J Med Microbiol. 2008;57:495–499. doi: 10.1099/jmm.0.47649-0. [DOI] [PubMed] [Google Scholar]
- 12.Sjollema J, van der Mei HC, Uyen HM, Busscher HJ. Direct observations of cooperative effects in oral streptococcal adhesion to glass by analysis of the spatial arrangement of adhering bacteria. FEMS Microbiol Lett. 1990;57:263–269. doi: 10.1016/0378-1097(90)90078-5. [DOI] [PubMed] [Google Scholar]
- 13.Larsen T, Fiehn NE. Development of a flow method for susceptibility testing of oral biofilms in vitro. APMIS. 1995;103:339–344. doi: 10.1111/j.1699-0463.1995.tb01117.x. [DOI] [PubMed] [Google Scholar]
- 14.Bradshaw DJ, Marsh PD, Schilling KM, Cummins D. A modified chemostat system to study the ecology of oral biofilms. J Appl Bacteriol. 1996;80:124–130. doi: 10.1111/j.1365-2672.1996.tb03199.x. [DOI] [PubMed] [Google Scholar]
- 15.Bowden GH. Controlled environment model for accumulation of biofilms of oral bacteria. Methods Enzymol. 1999;310:216–224. doi: 10.1016/s0076-6879(99)10019-3. [DOI] [PubMed] [Google Scholar]
- 16.Guggenheim B, Giertsen E, Schüpbach P, Shapiro S. Validation of an in vitro Biofilm Model of Supragingival Plaque. J Dent Res. 2001;80:363–370. doi: 10.1177/00220345010800011201. [DOI] [PubMed] [Google Scholar]
- 17.Palmer RJ, Jr, Kazmerzak K, Hansen MC, Kolenbrander PE. Mutualism versus Independence: Strategies of Mixed-Species Oral Biofilms In Vitro Using Saliva as the Sole Nutrient Source. Infect Immun. 2001;69:5794–5804. doi: 10.1128/IAI.69.9.5794-5804.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster JS, Palmer RJ, Jr, Kolenbrander PE. Human oral cavity as a model for the study of genome-genome interactions. Biol Bull. 2003;204:200–204. doi: 10.2307/1543559. [DOI] [PubMed] [Google Scholar]
- 19.Hansen SK, Rainey PB, Haagensen JA, Molin S. Evolution of species interactions in a biofilm community. Nature. 2007;445:533–536. doi: 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- 20.Periasamy S, Kolenbrander PE. Aggregatibacter actinomycetemcomitans Builds Mutualistic Biofilm Communities with Fusobacterium nucleatum and Veillonella Species in Saliva. Infect Immun. 2009;77:3542–3551. doi: 10.1128/IAI.00345-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowen WH. Do we need to be concerned about dental caries in the coming millennium? Crit Rev Oral Biol Med. 2002;13:126–131. doi: 10.1177/154411130201300203. [DOI] [PubMed] [Google Scholar]
- 22.Paes Leme AF, Koo H, Bellato CM, Bedi G, Cury JA. The role of sucrose in cariogenic dental biofilm formation – new insight. J Dent Res. 2006;85:878–887. doi: 10.1177/154405910608501002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomez-Suarez C, Busscher HJ, van der Mei HC. Analysis of bacterial detachment from substratum surfaces by the passage of air-liquid interfaces. Appl Environ Microbiol. 2001;67:2531–2537. doi: 10.1128/AEM.67.6.2531-2537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christensen GD, Simpson WA, Bisno AL, Beachey EH. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Immun. 1982;37:318–326. doi: 10.1128/iai.37.1.318-326.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fessia SL, Griffin MJ. A method for assaying biofilm capacity on polyurethane-coated slides. Perit Dial Int. 1991;11:144–146. [PubMed] [Google Scholar]
- 26.Lembke C, Podbielski A, Hidalgo-Grass C, Jonas L, Hanski E, et al. Characterization of biofilm formation by clinically relevant serotypes of group A streptococci. Appl Environ Microbiol. 2006;72:2864–2875. doi: 10.1128/AEM.72.4.2864-2875.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu H-S, Roberts N, Singleton FL, Attwell RW, Grimes DJ, et al. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb Ecol. 1982;8:313–323. doi: 10.1007/BF02010671. [DOI] [PubMed] [Google Scholar]
- 28.Oliver JD. The Viable but Nonculturable State in Bacteria. J Microbiol. 2005;43:93–100. [PubMed] [Google Scholar]
- 29.Fine DH, Furgang D, Kaplan J, Charlesworth J, Figurski DH. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch Oral Biol. 1999;44:1063–1076. doi: 10.1016/s0003-9969(99)00089-8. [DOI] [PubMed] [Google Scholar]
- 30.Inoue T, Shingaki R, Sogawa N, Sogawa CA, Asaumi J, et al. Biofilm Formation by a Fimbriae-Deficient Mutant of Actinobacillus actinomycetemcomitans. Microbiol Immunol. 2003;47:877–881. doi: 10.1111/j.1348-0421.2003.tb03454.x. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan JB, Meyenhofer MF, Fine DH. Biofilm Growth and Detachment of Actinobacillus actinomycetemcomitans. J Bacteriol. 2003;185:1399–1404. doi: 10.1128/JB.185.4.1399-1404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amarasinghe JJ, Scannapieco FA, Haase EM. Transcriptional and translational analysis of biofilm determinants of Aggregatibacter actinomycetemcomitans in response to environmental perturbation. Infect Immun. 2009;77:2896–2907. doi: 10.1128/IAI.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haase EM, Bonstein T, Palmer RJ, Jr, Scannapieco FA. Environmental influences on Actinobacillus actinomycetemcomitans biofilm formation. Archives of Oral Biology. 2006;51:299–314. doi: 10.1016/j.archoralbio.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Fine DH, Furgang D, Schreiner HC, Goncharoff P, Charlesworth J, Ghazwan G, Fitzgerald-Bocarsly P, Figurski DH. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology. 1999;145:1335–47. doi: 10.1099/13500872-145-6-1335. [DOI] [PubMed] [Google Scholar]
- 35.Ohta H, Onoue T, Fukui K. Energy metabolsim of Actinobacillus actinomycetemcomitans during anaerobic and microaerobic growth in low- and high potassium contiuous culture. Microbiology. 2001;147:2461–2468. doi: 10.1099/00221287-147-9-2461. [DOI] [PubMed] [Google Scholar]
- 36.Brown SA, Whiteley M. A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J Bacteriol. 2007;189:6407–6414. doi: 10.1128/JB.00554-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cowan MM, van der Mei HC, Rouxhet PG, Busscher HJ. Physico-chemical and structural properties of the surfaces of Peptostreptococcus micros and Streptococcus mitis as compared to those of mutans streptococci, Streptococcus sanguis and Streptococcus salivarius. J Gen Microbiol. 1992;138:2707–2714. doi: 10.1099/00221287-138-12-2707. [DOI] [PubMed] [Google Scholar]
- 38.Vadillo-Rodríguez V, Busscher HJ, Norde W, de Vries J, van der Mei HC. Relations between macroscopic and microscopic adhesion of Streptococcus mitis strains to surfaces. Microbiology. 2004;150:1015–1022. doi: 10.1099/mic.0.26828-0. [DOI] [PubMed] [Google Scholar]
- 39.Oliveira MR, Napimoga MH, Cogo K, Goncalves RB, Macedo MLR, Freire MGM, Groppo FC. Inhibition of bacterial adherence to saliva-coated through plant lectins. J Oral Science. 2007;49:141–145. doi: 10.2334/josnusd.49.141. [DOI] [PubMed] [Google Scholar]
- 40.Loo CY, Corliss DA, Ganeshkumar N. Streptococcus gordonii Biofilm Formation: Identification of Genes that Code for Biofilm Phenotypes. J Bacteriol. 2000;182:1374–1382. doi: 10.1128/jb.182.5.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teughels W, Kinder Haake S, Sliepen I, Pauwels M, van Eldere J, et al. Bacteria Interfere with A. actinomycetemcomitans Colonization. J Dent Res. 2007;86:611–617. doi: 10.1177/154405910708600706. [DOI] [PubMed] [Google Scholar]
- 42.Van Hoogmoed CG, van der Kuijl-Booij M, van der Mei HC, Busscher HJ. Inhibition of Streptococcus mutans NS adhesion to glass with and without a salivary conditioning film by biosurfactant-releasing Streptococcus mitis strains. Appl Environ Microbiol. 2000;66:659–663. doi: 10.1128/aem.66.2.659-663.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Hoogmoed CG, Geertsema-Doornbusch GI, Teughels W, Quirynen M, Busscher HJ, et al. Reduction of periodontal pathogens adhesion by antagonistic strains. Oral Microbial Immunol. 2008;23:43–48. doi: 10.1111/j.1399-302X.2007.00388.x. [DOI] [PubMed] [Google Scholar]
- 44.Sliepen I, Hofkens J, Van Essche M, Quirynen M, Teughels W. Aggregatibacter actinomycetemcomitans adhesion inhibited in a flow cell. Oral Microbiol Immunol. 2008;23:520–524. doi: 10.1111/j.1399-302X.2008.00456.x. [DOI] [PubMed] [Google Scholar]
- 45.Sliepen I, Van Essche M, Loozen G, Van Eldere J, Quirynen M, Teughels W. Interference with Aggregatibacter actinomycetemcomitans: colonization of epithelial cells under hydrodynamic conditions. Oral Microbiol Immunol. 2009;24:390–395. doi: 10.1111/j.1399-302X.2009.00531.x. [DOI] [PubMed] [Google Scholar]
- 46.Talarico TL, Dobrogosz WJ. Chemical characterization of an antimicrobial substance produced by Lactobacillus reuteri. . Antimicrob Agents Chemother. 1989;33:674–679. doi: 10.1128/aac.33.5.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sreenivasan PK, Meyer DH, Fives-Taylor PM. Factors influencing the growth and viability of Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 1993;8:361–369. doi: 10.1111/j.1399-302x.1993.tb00612.x. [DOI] [PubMed] [Google Scholar]
- 48.Leriche V, Carpentier B. Limitation of adhesion and growth of Listeria monocytogenes on stainless steel surfaces by Staphylococcus sciuri biofilms. J Appl Microbiol. 2000;88:594–605. doi: 10.1046/j.1365-2672.2000.01000.x. [DOI] [PubMed] [Google Scholar]
- 49.Law DJ, Dajani AS. Interactions between Neisseria sicca and viridin B, a bacteriocin produced by Streptococcus mitis. Antimicrob Agents Chemother. 1978;13:473–478. doi: 10.1128/aac.13.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.García-Mendoza A, Liébana J, Castillo AM, de la Higuera A, Piédrola G. Evaluation of the capacity of oral streptococci to produce hydrogen peroxide. J Med Microbiol. 1993;39:434–439. doi: 10.1099/00222615-39-6-434. [DOI] [PubMed] [Google Scholar]
- 51.Petersen FC, Tao L, Scheie AA. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J Bacteriol. 2005;187:4392–4400. doi: 10.1128/JB.187.13.4392-4400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perry JA, Cvitkovitch DG, Levesque CM. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol Lett. 2009;299:261–266. doi: 10.1111/j.1574-6968.2009.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van de Rijn I, Kessler RE. Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun. 1980;27:444–448. doi: 10.1128/iai.27.2.444-448.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Regular growth curves of A. actinomycetemcomitans, S. mitis, and S. mutans in CDM/sucrose. Growth was monitored by OD600 nm measurements in hourly intervals. One representative experiment of at least three replicates is shown.
(0.20 MB TIF)
Results of safranin-staining assay for the mono-species biofilms on a fibronectin-coated surface. The graph shows the result of safranin-staining assay for the tested mono-species on uncoated and fibronectin-coated surfaces. Fn - fibronectin.
(0.15 MB TIF)
Results of safranin-staining assay for the mono-species biofilm disorganization with pronase, DNase and sodiummetaperiodate. A), B) and C) Results for 500 µg/ml pronase, 90 units DNase, 10 mM sodiummetaperiodate, respectively. SMP - sodiummetaperiodate, * means significance of p<0.05 and ** means significance with p<0.01. PBS was used as control.
(0.38 MB TIF)
Results of Safranin-staining assay, number of colony froming units and microscopic analysis of the S. mitis/S. mutans/A. actinomycetemcomitans three-species combination. A) Safranin-staining assay of the mono- and three species-biofilms of S. mitis, S. mutans and A. actinomycetemcomitans. B) Number of colony forming units for the mono- and three-species cultures. Bacteria in brackets were the corresponding combination partner in the three-species culture. C–D) SEM and fluorescence microcopy of the S. mitis/S. mutans/A. actinomycetemcomitans combination. White arrow: S. mitis; Black arrow: S. mutans.
(4.10 MB TIF)
Summarized results for planktonic and biofilm growth of the different species in the tested media.
(0.09 MB DOC)
Number of colony forming units obtained for the mono- and two-species cultures in CDM/sucrose.
(0.03 MB DOC)
Number of colony forming units for the successive seeding experiment.
(0.02 MB DOC)
Number of colony forming units obtained in transwell experiments.
(0.02 MB DOC)