Abstract
Specific delivery to tumors and efficient cellular uptake of nucleic acids remain major challenges for gene-targeted cancer therapies. Here we report the use of a Designed Ankyrin Repeat Protein (DARPin) specific for the Epithelial Cell Adhesion Molecule (EpCAM) as carrier for siRNA complementary to the bcl-2 mRNA. For charge complexation of the siRNA, the DARPin was fused to a truncated human protamine-1 sequence. To increase the cell binding affinity and the amount of siRNA delivered into cells, DARPin dimers were generated and used as fusion proteins with protamine. All proteins expressed well in E. coli and, to remove tightly bound bacterial nucleic acids, they were purified under denaturing conditions by immobilized metal ion affinity chromatography, followed by refolding. The fusion proteins were capable of complexing 4-5 siRNA molecules per protamine, and fully retained the binding specificity for EpCAM as demonstrated on MCF-7 breast carcinoma cells. In contrast to unspecific lipofectamine transfection, down-regulation of anti-apoptotic bcl-2 using fusion protein complexed siRNA was strictly dependent on EpCAM binding and internalization. Inhibition of bcl-2 expression facilitated tumor cell apoptosis as demonstrated by increased sensitivity to the anti-cancer agent doxorubicin.
Keywords: tumor targeting, ankyrin repeat proteins, siRNA, nanocomplexes, chemosensitization
Introduction
Specific post-transcriptional gene silencing using antisense oligonucleotides or small interfering RNA (siRNA) sequences is a promising concept in biomedical research, and is also investigated for therapeutic purposes (1). Although the intracellular action of siRNA is highly efficient, activating an intrinsic enzymatic RNA degradation machinery, the therapeutic use is limited by fundamental inherent pharmacological problems. Major obstacles are the failure of efficient delivery of the compounds to diseased tissues and the limited uptake into cells where the target gene of interest is expressed. Since the polyanionic and hydrophilic nature impedes cell penetration of siRNA, and structural modifications are poorly tolerated by the RNA-induced silencing complex (RISC) (2, 3), a main focus of current research is the development of stable and effective delivery systems for therapeutic applications (4-6).
The first report of successful tissue-targeting with siRNA in vivo was the use of a cholesterol-conjugated siRNA, which, corresponding to its site of metabolism, accumulated in the liver (7). In addition, several more versatile nanocarriers have been developed for in vivo delivery of nucleic acids, including liposomal formulations, polymeric nanoparticles, cationic polymer complexes and lipoplexes (4).
Recently, Lieberman and coworkers accomplished targeted delivery of siRNA with fusion proteins composed of a single-chain antibody fragment (scFv) and a truncated protamine peptide to achieve receptor binding on the surface of tumor target cells followed by receptor-mediated endocytosis of the complexed siRNA (8, 9). Protamines are natural small proteins that function as a substitute for histones in sperm chromatin during spermatogenesis. Protamine-1 is a non-antigenic, virtually nontoxic DNA-binding peptide and a well established pharmaceutical excipient for sustained release of insulin. Furthermore, it proved useful for condensing antisense oligonucleotides (10) and plasmids (11) into nanoparticle-like structures to enhance stability and improve cellular uptake and subsequent release of the bioactive compound into the cytoplasm at least to some proportion. Moreover, these nanocomplexes are biocompatible and well tolerated in vivo (12). Complexation is accomplished by strong electrostatic interaction of the guanidinium cations with the phosphate anions of the oligonucleotide, and one protamine fragment was shown to bind about six siRNA molecules (10).
In cancer therapy, protein-based drug delivery to intracellular targets largely exploits the specific binding to a tumor-associated antigen abundantly expressed on the cell surface, followed by quantitative internalization through receptor-mediated endocytosis. Its overall efficiency depends on the degree of tumor localization, the binding to the tumor associated antigen, its endocytosis, and finally the transfer of the payload from the endosome to the cytoplasm. The epithelial cell adhesion molecule (EpCAM) is a 40 kDa transmembrane glycoprotein frequently expressed in solid tumors (13, 14), but with limited expression on normal epithelia. Recent evidence suggests its overexpression particularly on cancer initiating cells isolated from colon, breast, pancreas and prostate carcinomas (15-17). Originally, EpCAM was proposed to be a cell-cell-adhesion molecule, but later insights revealed a more versatile biological role, including cell differentiation, proliferation, migration, and signaling (18). Recently, the mechanism of EpCAM signaling has been elucidated in more depth and intramembrane proteolysis was identified as a key event to activate its mitogenic activity (19). Upon binding to antibodies, EpCAM is rapidly internalized and thus ideally suited for delivery of anti-cancer agents to intracellular targets (20-22). These properties suggest EpCAM as a promising target for highly selective ligand-guided cancer therapy.
Designed Ankyrin Repeat Proteins (DARPins) are a novel class of non-immunoglobulin binding proteins relying on the modularity of ankyrins (23-26). The ankyrin repeat motif consists of 33 amino acids forming a loop, a β-turn and two antiparallel α-helices connected by a tight turn. Their small size, high stability and high-yield expression in E. coli allow engineering procedures usually not well tolerated by antibodies. They have no cysteine, and unique cysteines can thus easily be introduced for site-specific modifications. High affinity binders against soluble or membrane-bound antigens can be readily derived from designed protein libraries consisting of four to six repeats using ribosome or phage display. In summary, DARPins represent a new class of binding molecules with excellent biochemical properties, which can be ideally employed for tumor targeting.
Here we describe for the first time the use of a novel EpCAM-specific DARPin (C9) to generate fusion proteins with protamine-1 for complexation of siRNA complementary to anti-apoptotic bcl-2, a potent inhibitor of apoptosis implicated in cancer drug resistance. Monomeric and dimeric DARPin-protamine fusion protein constructs were tested and their ability to down-regulate bcl-2 expression and facilitate tumor cell apoptosis in an EpCAM-dependent manner is demonstrated.
Materials and Methods
Generation of DARPin and DARPin-protamine fusion proteins
The EpCAM-specific DARPin C9 was selected by ribosome display from a Designed Ankyrin Repeat Protein library essentially as described for Her-2 binders (26), using the biotinylated extracellular domain of EpCAM as target. The detailed procedure and characterization of the binders will be described elsewhere (Martin-Killias et al, manuscript in preparation).
For the generation of C9-protamine fusion proteins, truncated protamine (final sequence RSQSRSRYYRQRQRSRRRRRRSRS) was amplified from the plasmid pACgp67B-Her2mP (10) obtained from Addgene (Cambridge, MA, USA), using primers engineered to include HindIII and NheI restriction sites (forward, 5′-ACTGCATAAGCTTGGTGGTTCTTCTCGCAGCCAGAGCCGG-3′ and reverse, 5′-CATCTCGGCTAGCTCATTAATGGTGGTGGTGATG-3′. After restriction the amplicon was inserted into the vector pQE30ss, to create a C-terminal fusion to the DARPin C9. For expression in E. coli BL21 CodonPlus, the expression insert was religated after EcoRI/NheI digestion into the vector pMPAG6, which encodes the laclq gene. The monomeric protein has the composition MRGSH6-C9-GGSS-Protamine-H6, where the first sequence constitutes an N-terminal his tag, followed by the DARPin C9 sequence, a short linker, the protamine fragment and a C-terminal his tag.
Plasmids for dimers C9D (MRGSH6-C9-(GGGGS)4-C9) and C9D-P (MRGSH6-C9-(GGGGS)4-C9-GGSS-Protamine-H6) were generated by three-fragment ligation of the respective expression vector digested with EcoRI/BamHI, an insert encoding C9, digested with EcoRI/HindIII, and synthetic oligonucleotides for inserting a (GGGGS)4 linker (forward: 5′-AGCTAAATGGTGGCGGAGGTTCAGGTGGAGGCGGTTCAGGTGGCGGAGGTTCAGGTGGAGGCGGTTCAG-3′, reverse: 5′-GATCCTGAACCGCCTCCACCTGAACCTCCGCCACCTGAACCGCCTCCACCTGAACCTCCGCCACCATTT-3′).
Plasmids for proteins with leucine zippers (sequence RMKQLEDKVEELLSKNYHLENEVARLKKLVGERKLN), C9LZ (MRGSH6-C9-GGGG-Leucin Zipper) and C9LZ-P (MRGSH6-C9-GGGG-Leucin Zipper-GGSS-Protamin-H6), were generated by ligation of oligonucleotides (forward: 5′-AGCTAAATGGAGGCGGTGGTCGTATGAAACAGCTGGAGGATAAAGTCGAAGAGTTGCTCTCGAAGAACTATCACCTTGAGAATGAAGTCGCTCGTCTGAAGAAACTTGTCGGTGAACGTA-3′, reverse: 5′-AGCTTACGTTCACCGACAAGTTTCTTCAGACGAGCGACTTCATTCTCAAGGTGATAGTTCTTCGAGAGCAACTCTTCGACTTTATCCTCCAGCTGTTTCATACGACCACCGCCTCCATTT-3′) and the respective expression vector digested by HindIII.
Protein expression and refolding
All proteins were expressed in E. coli BL21 CodonPlus (Stratagene, CA, USA). Bacteria were grown in 250 ml LB broth at 30°C to an OD600 of 0.6, when expression was induced by addition of 0.5 mM IPTG (Sigma, Buchs, Switzerland). After 5 h, bacteria were centrifuged and stored overnight at −20°C.
For native protein purification (for proteins not containing protamine tails), cells were thawed on ice and resuspended in 12 mL 100 mM NaH2PO4, 10 mM Tris, 300 mM NaCl, 10 mM imidazol, 20% glycerol at pH 8.0. After sonication with a MSE 150 watt Ultrasonic Disintegrator (MSE Scientific Instruments, Crawles, Sussex, England) and centrifugation, supernatants were added to 400 μL Ni-NTA (Qiagen, Hombrechtikon, Switzerland) and shaken at 4°C for 60 min. The lysate/resin mixture was loaded on a polypropylene column (Qiagen) and the resin was washed thoroughly with lysis buffer. His-tagged proteins were eluted with 100 mM NaH2PO4, 10 mM Tris, 300 mM NaCl, 250 mM imidazol, 20% glycerol at pH 8.0. For buffer exchange, protein solutions were dialyzed against PBS with 20% glycerol using SpectraPor FloatALyzers (Spectrum Labs, Breda, Netherlands) with a molecular weight cut-off of 8 kDa.
Fusion proteins containing a protamine fragment were purified under denaturing conditions. Cells were thawed and resuspended in 12 ml 100 mM NaH2PO4, 10 mM Tris, 8 M urea at pH 8.0. After sonication and centrifugation, supernatants were added to 400 μL Ni-NTA and shaken for 30 min at RT. The lysate/resin mixture was then loaded on a polypropylene column. The resin was washed thoroughly with lysis buffer, and subsequently with a urea gradient (8 to 0 M urea) for on-column refolding. His-tagged proteins were eluted with 100 mM NaH2PO4, 10 mM Tris, 300 mM NaCl, 250 mM imidazol, 20% glycerol, pH 8.0. For buffer exchange, protein solutions were dialyzed against PBS with 20% glycerol.
Cell lines
The breast carcinoma cell line MCF-7, the colon carcinoma cell line HT-29 and the human embryonic kidney cells HEK293T were obtained from the American Type Culture Collection (Manassas, VA). Cells were grown in DMEM including GlutaMax (Invitrogen, Carlsbad, CA). Culture media were supplemented with 10% FCS, 50 IU/mL penicillin and 50 μg/mL streptomycin (all Invitrogen), and cell cultures were maintained at 37°C in a humidified atmosphere containing 5% CO2.
siRNA
The sequence of the siRNA complementary to the bcl-2 mRNA was sense, 5′-UCAGGUACUCAGUCAUCCACA-3′, antisense, 5′-UGUGGAUGACUGAGUACCUGA-3′ (underlined nucleotides are 2′-O-methylated, others are unmodified ribonucleotides). Scrambled siRNA (sense, 5′-UGUGGAUGACUGAGUACCUGA-3′, antisense, 5′-UCAGGUACUCAGUCAUCCACA-3′) was used as specificity control. For internalization studies, 5′-terminal FITC-labeled siRNA and control oligonucleotides were used. All sequences were synthesized on a PolyGen 10 column DNA Synthesizer (PolyGen, Langen, Germany), desalted by Sep-Pak cartridges (Waters, Milford, MA, USA) and analyzed for purity by gel electrophoresis.
Analysis of EpCAM binding
Binding of DARPin and fusion proteins to purified EpCAM was analyzed by ELISA using the extracellular domain of EpCAM. The wells of a 96-well ELISA plate (Greiner Bio-One GmbH, Frickenhausen, Germany) were coated with 66 nM neutravidin (Pierce Biotechnology Rockford, IL, USA) at 4°C and washed with Tris-buffered saline containing 0.1% Tween 20 (TBST). Following saturation with 0.2% bovine serum albumin (Sigma-Aldrich, St. Louis, MI, USA), a 50 nM solution of EpCAM was added to the wells for 1 h at RT. After thorough washing with TBST, a 50 nM solution of DARPin or the fusion proteins C9-P, C9D-P or C9-LZ-P was added. Binding was detected using an anti-RGS-His antibody (Qiagen; 1:1000), an anti-mouse-IgG-alkaline phosphatase conjugate (Pierce; 1:10000) and p-nitrophenylphosphate (Fluka). The absorbance of the NPP product was measured after 60 min in a plate reader (DigiScan, Asys Hitech GmbH, Austria).
To measure binding of the ligands to EpCAM on the cell surface, cells were trypsinized, washed and resuspended in PBS containing 1% FCS (PBS/FCS). Thereafter, cells were put on ice and incubated with DARPin or the fusion proteins C9-P, C9D-P or C9-LZ-P at the indicated concentrations for 1 h. After washing twice, cells were further incubated with a secondary anti-Penta-His Alexa Fluor 488-conjugate antibody (1:200, Qiagen) for 1 h on ice in the dark. Following repeated washing, cells were resuspended in 500 μl PBS/FCS and analyzed using a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ, USA). Secondary antibody alone was used as a negative control.
To measure the association of siRNA with target cells, the fusion proteins C9-P, C9D-P and C9-LZ-P were complexed with FITC-labeled siRNA at a molar ratio of 1:4 for 30 min on ice, and suspended cells were added to the solution to give a final protein concentration of 50 nM. After 1 h on ice, cells were washed at least 3 times with 1 ml PBS/FCS, resuspended in 500 μl PBS/FCS and subjected to FACS analysis.
Internalization of siRNA and confocal microscopy
Cells were seeded on coverslips at densities of 5×104 cells/well. FITC-labeled siRNA was mixed with the fusion protein C9-P at a molar ratio of 4:1 in 100 μl PBS, and incubated at RT for 30 min before adding to the cells in 400 μl medium containing 10% FCS. Lipofectamine 2000 (Invitrogen) transfection of cells was done according to the manufacturer′s instructions. After 4 h, cells were washed with PBS and fixed with 2 % formaldehyde in PBS (10 min, RT). Coverslips were mounted on glass slides and cells were analyzed with a LSM 510 confocal microscope selecting a confocal plane to ensure that intracellular fluorescence was measured (Carl Zeiss MicroImaging, Feldbach, Switzerland).
Quantitative RT-PCR
MCF-7 cells were cultured in 24-well plates at a density of 1×105 cells per well. The siRNA was mixed with the fusion proteins C9-P, C9D-P or C9-LZ-P at a molar ratio of 4:1 in 100 μl PBS and incubated at RT for 30 min before being added to cells in 400 μl medium containing 10% FCS. Lipofectamine 2000 transfection of cells was done according to the manufacturer's instructions. Addition of siRNA was repeated after 24 h. Cells were harvested for mRNA quantification after a total of 48 h and total RNA was isolated from cells using Trizol reagent (Sigma). For cDNA synthesis, 0.5 μg of extracted RNA was transcribed by RevertAid™ M-MuLV reverse transcriptase (Fermentas, Vilnius, Lithuania) using a random hexamer primer. Real-time monitoring of PCR amplification of the cDNA was done in an iCycler (Bio-Rad) using SybrGreen MasterMix (Bio-Rad). Primers were: forward, 5′-ATGTGTGTGGAGAGCGTCAACC-3′, reverse, TGAGCAGAGTCTTCAGAGACAGCC. Relative quantification of gene expression was calculated by the ΔCt method using 18S-rRNA (primer forward, CGATTCCGTGGGTGGTGGTG, reverse, 5′-CATGCCAGAGTCTCGTTCGTTATC-3′) as internal standard. Statistical evaluation was done by one-way ANOVA.
Western blotting
Treatment of cells with siRNA complex to fusion proteins and lipofectamine mediated transfection was done as described above. Cells were lysed after 48 h (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10% Triton X, 10% glycerol, 2 mM EDTA, 2 mM EGTA, 40 mM β-gylcerolphosphate, 50 mM sodium fluoride, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 μM pepstatin, 1 mM PMSF). 30 μg of soluble total protein, quantified by a Bradford assay, were separated on a 12.5% polyacrylamide SDS gel and transferred to a nitrocellulose membrane in a tank blotter (Bio-Rad). The membranes were blocked in TBST containing 5% nonfat milk and incubated overnight at 4°C with rabbit anti-bcl-2 monoclonal antibody (Epitomics, CA, USA). β-Actin staining with goat anti-actin antibodies (Santa Cruz Biotechnolgy, CA, USA) was used as loading control. After incubation with secondary anti-rabbit or anti-goat polyclonal antibodies conjugated to horseradish peroxidase (Santa Cruz), visualization was achieved using the ECL kit (GE Healthcare, Chalfont St. Giles, UK) and Hyperfilm ECS (GE Healthcare). Protein bands were quantified using QuantityOne Software (Bio-Rad) and normalized to the respective β-actin bands.
Determination of chemosensitization
Cells were seeded at a density of 2×104 cells/well in 96-well plates and cultured overnight. The bcl-2-targeted siRNA (200 nM) was complexed with the fusion proteins C9-P, C9D-P or C9-LZ-P at a molar ratio of 4:1 in 20 μl PBS, and incubated at RT for 30 min before adding to cells in 80 μl medium containing 10% FCS. Treatment with siRNA was repeated after 24 h. After 48 h, the medium was replaced with 100 μl medium containing the indicated concentrations of doxorubicin and cells were incubated for another 24 h before viability was determined in CellTiter 96® AQueous One Solution Cell Proliferation Assays (Promega, Madison, WI, USA) according to the manufacturer's instructions. Curves were fitted using the Hill-Slope model to calculate the IC50 values. Statistical analysis was done by one-way ANOVA.
Results
Expression and purification of DARPin and fusion proteins
DARPins binding to EpCAM were selected from a Designed Ankyrin Repeat Protein library using ribosome display (27, 28) and the extracellular domain of EpCAM as target. As a first generation binder, DARPin C9 was obtained which expressed well (48 mg/L purified yield) in an E. coli strain co-expressing rare arginine tRNA and showed half-maximal binding activity at about 25 nM, as determined by ELISA using the extracellular domain of EpCAM. DARPin C9 was used for the generation of a fusion protein with truncated protamine for complexation of siRNA complementary to bcl-2 (C9-P; Fig. 1) at the C-terminus of the DARPin.
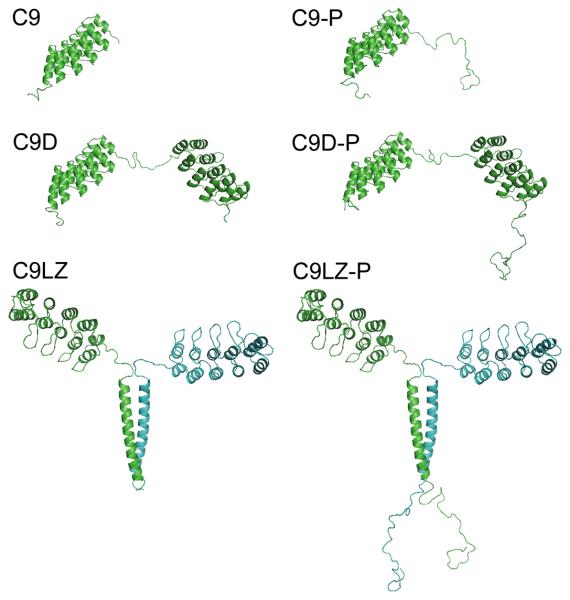
Figure 1.
Formats of DARPin C9 and its derivatives used in the study in ribbon representation. C9 is an EpCAM-recognizing designed ankyrin repeat protein (DARPin). C9D is a dimer that consists of two C9 sequences joined by a flexible linker, resulting in a head-to-tail arrangement. C9LZ results from self-dimerization of the leucine zipper motif engineered to the C-terminus of C9. P indicates the respective fusion proteins containing the human protamine-1 peptide. Unstructured parts of homology models were modelled in a molecular dynamics simulation using the Insight II software package (Accelrys Software 2000).
Native IMAC purification with Ni-NTA resulted in high yields with low contamination by other proteins, but a high amount of nucleic acid was found to be co-purified. Attempts to degrade the bacterial nucleic acid binding tightly to the protamine part of the fusion protein by benzonase treatment failed to remove the contaminants. Nucleic acid contaminations, however, were readily removed when the proteins were adsorbed to Ni-NTA under denaturing conditions (Fig. 2a). Subsequent washing of the Ni-NTA-bound protein, still under denaturing conditions, followed by on-column renaturation using an urea gradient, provided a highly pure C9-P fusion protein at a yield of 21 mg/L.
Figure 2.
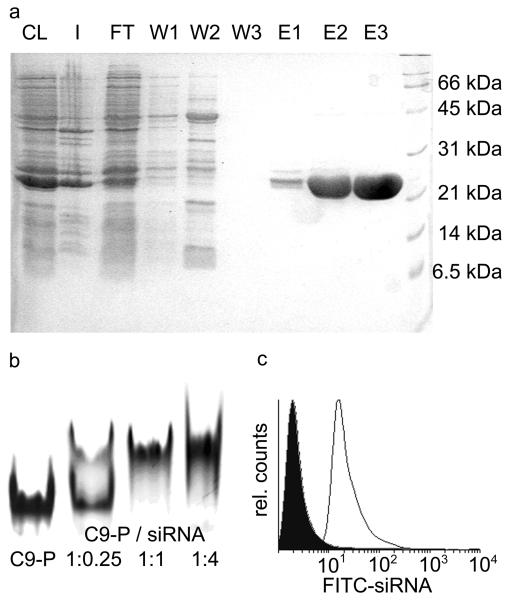
(a) SDS-PAGE of purification fractions of fusion protein C9-P (22.6 kDa). CL; cleared lysate, I; insoluble fraction, FT; IMAC flow through, W1-3; three wash fractions, E1-3; three elution fractions; M; molecular weight marker (6.5, 14.4, 21.5, 31.0, 45.0, 66.2, 97.4, 200.0 kDa). After protein expression and cell lysis in denaturing buffer (8 M urea), the protein solution was purified by Ni-NTA in a batch purification, and subsequently loaded on a polypropylene column. Non-his-tagged impurities and nucleic acids were washed from the column using denaturing buffer. Subsequently, the his-tagged DARPin was refolded on the column by gradually lowering of urea concentrations followed by elution in buffer containing 250 mM imidazol.
(b) Binding of C9-P to the 21mer bcl-2 siRNA. From left to right: C9-P, C9-P/siRNA 1:0.25, C9-P/siRNA 1:1, C9-P/siRNA 1:4. Samples were run on a native 15% polyacrylamide gel and stained with Coomassie blue.
(c) Binding of FITC-labeled siRNA to MCF-7 cells in the presence of C9 (black filled peak) or as a complex with C9-P (open grey peak). Cells were incubated on ice with a 4:1 mixture of FITC-labeled siRNA and C9 or C9-P for 1 h. After repeated washing, cell binding was determined by FACS analysis.
Cell binding activity of DARPin and fusion proteins
Efficient and specific binding to a surface antigen on tumor cells is mandatory for tumor-targeted therapy. Cell binding was determined by FACS analysis on EpCAM-positive and negative control cells. To improve the functional affinity of DARPin C9 on cells, two different types of C9 dimers were constructed (Fig. 1): (i) by joining two C9 sequences with a 20 aa flexible linker, which results in a head-to-tail arrangement (C9D, C9D-P) and (ii) by adding a leucine zipper between C9 and the protamine peptide to produce a tail-to-tail dimer (C9LZ, C9LZ-P) caused by parallel arrangement of the two zipper helices (29). The latter approach also doubles the possible payload of siRNA which can be complexed per protein molecule as two protamine motifs are present.
The dimers expressed equally well in E. coli as the monomers. As demonstrated by FACS analysis, both C9 and C9-P bound specifically to EpCAM-positive MCF-7 and HT-29 cells, whereas binding to EpCAM-negative HEK293T cells was negligible (Fig. 3a). This shows that cell binding is mediated by the DARPin interacting with EpCAM, and not by protamine interacting with negative charges on the cell surface. Cell binding of C9-P was independent of its loading state (Fig. 2c and 3b). The dimer C9LZ as well as its fusion protein C9LZ-P indeed showed higher apparent affinity both in a sandwich ELISA assay using the purified extracellular domain of EpCAM (data not shown) and in a FACS-based cell binding assay using EpCAM positive MCF-7 cells. In contrast, dimerization in a head-to-tail arrangement as in C9D, while also resulting in higher apparent affinity to purified EpCAM in ELISA, did not enhance the apparent binding activity for EpCAM expressed on target cells. This argues for an arrangement of the epitopes on cellular EpCAM such that the C9-LZ dimer, possessing a two-fold axis of symmetry, can simultaneously bind to EpCAM monomers. Such an arrangement is not possible by the head-to-tail arrangement of the DARPins. As expected, no binding was measured to EpCAM-negative HEK293T control cells (Fig. 3b).
Figure 3.
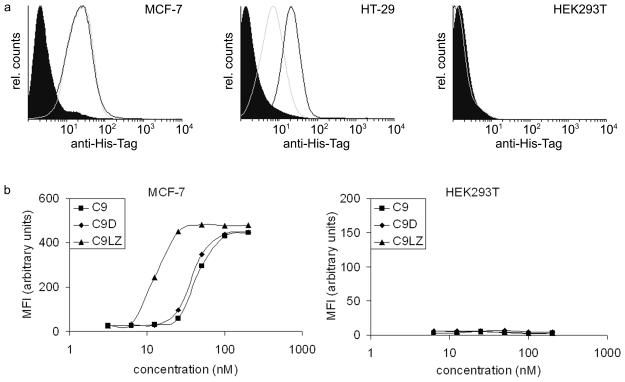
(a) Binding of EpCAM-targeted DARPin C9 and the fusion protein C9-P to EpCAM-positive MCF-7 and HT-29, and to EpCAM-negative HEK293T cells. Black filled peak, secondary antibody alone; open grey peak, C9 (without protamine fusion), open black peak, C9-P. Cells were trypsinized, resuspended in ice-cold PBS containing 1% FCS, and stained with the indicated proteins (1 μM). FACS analysis was performed following visualization with Alexa 488-conjugated anti-His-Tag antibodies.
(b) Cell binding activity of monomeric and dimeric C9 to EpCAM-positive MCF-7 and negative HEK293T control cells. Varying concentrations of DARPin C9 or the dimers C9D or C9LZ were added to the cells for 1 h before adding Alexa 488-conjugated anti-His-Tag antibodies to determine the maximum binding concentrations by flow cytometry analysis.
EpCAM-specific cell binding of siRNA complexed to C9-P
To be useful as a delivery vehicle, the DARPin-protamine fusion protein must complex siRNA on one site and bind to EpCAM on tumor cells with the other site. The electrophoretic mobility of C9-P in a native polyacrylamide gel shifted after addition of the siRNA to a position identical to that of C9 alone, indicating that the basic protamine was fully neutralized by complexation with the nucleic acid (Fig. 2b). A gel filtration assay showed that about 80% of a 5-fold molar excess was complexed to the fusion protein (data not shown), which is in line with findings from other studies reporting a capacity of about 5 siRNA molecules per protamine fragment (10). The protamine fragment in the C9-P fusion protein was therefore capable of complexing about four siRNA molecules under these conditions. We also tested the specificity of siRNA interaction with EpCAM positive cells. FITC-labeled siRNA was found associated with EpCAM positive cells only when complexed to C9-P, but not when applied as a mixture with C9 alone (Fig. 2c).
Internalization of C9-P-siRNA complexes in tumor cells
To investigate whether the fusion proteins can deliver their siRNA payload into cells upon binding to EpCAM on the cell surface, the uptake of FITC-labeled siRNA in MCF-7 cells was measured. Binding to and accumulation of fluorescence inside cells occurred only when cells were treated with C9-P complexed FITC-siRNA, but not upon treatment with C9 plus uncomplexed FITC-siRNA. Binding was quantified by flow cytometry, while internalization of the siRNA in MCF-7 cells was verified by confocal microscopy (Fig. 4). When complexed to C9-P a punctate pattern indicative for endo/lysosomal localization of the siRNA was found, however, fluorescence was also found diffusely distributed in the cytoplasm. Similarly, lipofectamine transfected siRNA also showed preferentially vesicular localization, although the signal intensity was slightly higher compared to C9-P.
Figure 4.
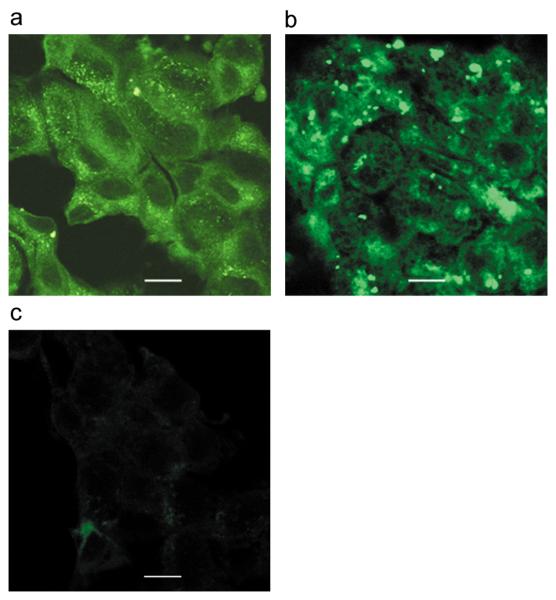
Internalization of FITC-conjugated bcl-2 siRNA in MCF-7 cells. (a) The siRNA was complexed to C9-P in a molar ratio of 4:1, (b) to lipofectamine, or (c) mixed with C9 lacking an oligonucleotide binding domain as negative control and added to cells grown on coverslips. After incubation at 37°C for 4 h, cells were fixed and visualized by confocal microscopy. Z-stacks were examined to ensure the signal is inside the cells. Scale bar = 10 μm.
Bcl-2 down-regulation with C9-P-siRNA complexes in tumor cells
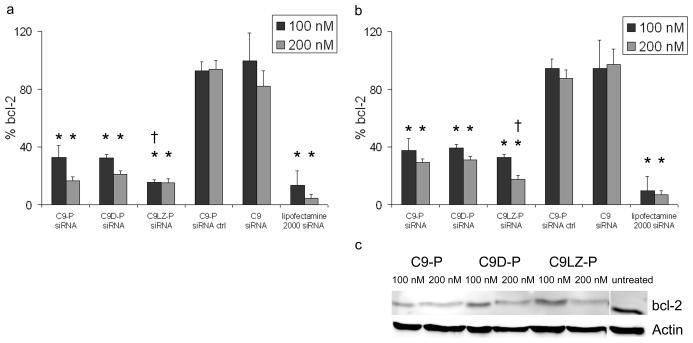
To confirm that the delivered siRNA was at least partly capable of escaping into the cytoplasm and activating the RNAi pathway, bcl-2 mRNA and protein levels in MCF-7 cells were determined 48 h after addition of C9-P complexed siRNA. As measured by qPCR, the bcl-2 mRNA was significantly reduced in a dose-dependent manner upon treatment with siRNA complexed to C9-P, C9D-P, and C9LZ-P (p<0.001 for all samples compared to the untreated control), but not after the addition of C9 plus uncomplexed siRNA or a scrambled control siRNA sequence, not complementary to any human mRNA, complexed to C9-P (Fig. 5). As a positive control, lipofectamine transfected siRNA was used, resulting in the highest target down-regulation. Compared to the C9-P monomer, the increase in bcl-2 down-regulation by the head-to-tail C9D-P dimer was only moderate. The bcl-2 mRNA was reduced to 33 ± 7% (100 nM) and 16 ± 2% (200 nM) after treatment with siRNA complexed to C9-P, and 33 ± 2% (100 nM) and 21 ± 2% (200 nM) after delivery of the siRNA complexed with C9D-P. This is consistent with the observation that this orientation of the dimer does not lead to an avidity increase, presumably because bivalent binding is geometrically impossible. In contrast, the effect after delivery with the tail-to-tail dimer C9LZ-P was more pronounced, and it reduced mRNA expression to 16 ± 1% (100 nM, p<0.01 compared to C9-P) and 15 ± 2% (200 nM). Unspecific lipofectamine transfection of siRNA decreased bcl-2 expression to 7 ± 2% and 3 ± 1%, underlining the influence of membrane permeabilization by the detergent.
Figure 5.
Down-regulation of (a) bcl-2 mRNA and (b) bcl-2 protein in MCF-7 cells upon treatment with bcl-2-targeted siRNA in the presence of C9 or complexed to the various C9 fusion proteins in a ratio of 4:1 or with lipofectamine. Cells were treated for a total of 48 h, then lysed and subjected to mRNA or protein analysis by qPCR or Western blotting, respectively. Values were standardized to ribosomal RNA or actin. Following lysis, bcl-2 protein levels were determined by densitometric quantification of bcl-2 bands and normalization to actin. As negative controls, an irrelevant non-functional siRNA sequence delivered with C9-P and a mixture of anti-bcl-2 siRNA with C9 (without protamine) were used. Error bars are mean + SD, n=3. *: p<0.01, compared to untreated control, †: p<0.05 compared to C9-P + siRNA.
(c) Representative Western blot of bcl-2 protein expression in MCF-7 cells upon treatment with bcl-2-targeted siRNA delivered with the indicated fusion proteins.
The results with bcl-2 mRNA were confirmed on the protein level by Western blotting (p<0.005 for all samples compared to the untreated control). Bcl-2 protein was reduced to 37 ± 4% (100 nM) and 29 ± 7% (200 nM) by siRNA complexed to C9-P, to 39 ± 4% and 31 ± 6% complexed to C9D-P, and to 33 ±7% and 18 ± 2% (p=0.04 compared to C9-P) using complexation with C9LZ-P (Fig. 5b). Again, unspecific transfection of the siRNA with lipofectamine 2000 resulted in the strongest reduction of bcl-2 protein among the fusion proteins tested (10 ± 4% and 7 ± 4%). Both the results from measuring mRNA and protein levels suggest that the gain in cell binding activity by leucine zipper dimerization of C9 indeed improved the delivery of siRNA into the target cells. All experiments were performed with the indicated siRNA concentrations using a complexation ratio of siRNA/protamine of 4:1, and all protein concentrations given of C9 derivatives refer to the single DARPin unit. Delivery of siRNA with the DARPin nanocomplexes also led to bcl-2 mRNA and protein down-regulation in EpCAM positive HT-29 colon carcinoma cells (Supplementary Materials).
Chemosensitization of tumor cells using C9-P-siRNA complexes
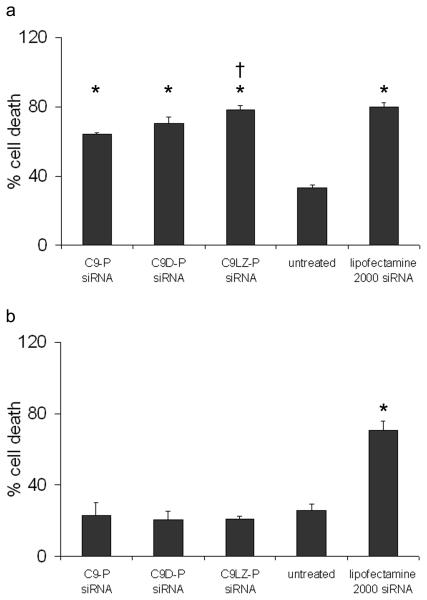
Down-regulation of bcl-2 in tumor cells was previously shown to facilitate apoptosis or sensitize cells to cytotoxic stimuli (30). To investigate the biological consequences of bcl-2 down-regulation by siRNA delivered by the fusion proteins, chemosensitization of MCF-7 cells to the anti-cancer agent doxorubicin was determined in cell viability assays (Fig.6). When the bcl-2 siRNA was delivered as complexes with the fusion proteins C9-P, C9D-P and C9LZ-P, MCF-7 cells, but not EpCAM-negative HEK293T cells became more susceptible to the cytotoxic effect of doxorubicin (p<0.001 for all samples compared to the untreated control for 0.1 μM doxorubicin concentrations). siRNA complexation with C9LZ-P, but not with C9D-P, led to significantly higher chemosensitization than with monomer C9-P (p=0.003). In contrast, and as expected from its ability to deliver siRNA independent of EpCAM expression on the cell surface, lipofectamine-transfected siRNA also sensitized HEK293T cells to doxorubicin. As demonstrated in Table 1, the decrease in the IC50 for doxorubicin was between 2- and 5-fold, compared to cells treated with doxorubicin alone or with an inactive mixture of C9 plus siRNA, which did not result in chemosensitization. On EpCAM-positive MCF-7 cells, sensitization by lipofectamine was similar to the effect of C9LZ-P. Thus, C9LZ-P facilitated apoptosis of MCF-7 cells by targeted bcl-2 down-regulation similar to lipofectamine, but most importantly, in contrast to lipofectamine, its effect was strictly dependent on EpCAM expression on the cell surface.
Figure 6.
Chemosensitization of (a) EpCAM-positive MCF-7 and (b) EpCAM-negative HEK293T cells to doxorubicin upon treatment with bcl-2-targeted siRNA. The siRNA was added to cells as indicated in the presence of C9 or as complexes with the various C9 fusion proteins (in a ration of 4:1) or lipofectamine 2000. 48 h after transfection, all cell samples were treated with 0.1 μM doxorubicin and another 24 h later the effect on cell viability was determined in MTT assays. Error bars are mean + SD, n=3. *: p<0.001 compared to untreated control, †: p<0.01 compared to C9P +siRNA.
Table 1.
Chemosensitization of MCF-7 breast cancer cells to doxorubicin. Treatment was performed as described in the legend to Fig. 6. Doxorubicin concentrations were used in the range of 1 nM to 100 μM, and cell viability was determined in cell proliferation assays. IC50: concentration at which cell viability/proliferation was inhibited by 50%. Data are reported as mean ± SD, n=3. Fold sensitization was determined as the ratio of IC50 values of doxorubicin after control treatment and treatment with the various complexes.
| Treatment (+doxorubicin)a | IC50 (nM)b | Fold sensitizationc |
|---|---|---|
| - | 144 ± 36 | |
| siRNA+C9-P | 55 ± 4 | 2.6 |
| siRNA+C9D-P | 53 ± 1 | 2.7 |
| siRNA+C9LZ-P | 35 ± 20 | 4.2 |
| siRNA+lipofectamine 2000 | 27 ± 6 | 5.3 |
| siRNA+C9 | 133 ± 5 | 1.1 |
Discussion
Targeted inhibition of anti-apoptotic proteins, either by modulating their expression or activity, has been extensively studied as a strategy to improve the efficacy and specificity of cancer therapy, especially in combination with cytotoxic anti-cancer agents (31, 32). A great number of nucleic-acid-based cancer therapeutics has been investigated in preclinical models, but only few have entered clinical trials (1, 6). Major obstacles are the lack of tumor-specific delivery and the inability of the highly charged molecules to penetrate the plasma membrane. Here, we describe the generation of a novel non-vesicular delivery system for therapeutic siRNA, which is based on the use of an EpCAM-specific Designed Ankyrin Repeat Protein (DARPin), fused to an arginine-rich protamine sequence for complexation and delivery of nucleic acids into cells (8, 9). The DARPin-protamine fusion proteins were loaded with siRNA complementary to bcl-2 to specifically facilitate apoptosis in EpCAM-positive tumor cells.
The natural human protamine sequence consists of abundant rare arginine codons limiting efficient production in commonly used E. coli strains XL1-Blue and BL21, since the expression of heterologous proteins can deplete the pool of rare arginine tRNAs and stall translation. To overcome this limitation, we used the E. coli strain BL21 CodonPlus, which contains extra copies of genes encoding tRNAs recognizing the arginine codons AGA and AGG. After removing contaminating bacterial DNA tightly bound to the protamine fragment by a denaturing purification protocol, we succeeded to obtain the fusion proteins C9-P, C9D-P, and C9LZ-P at a yield of approximately 20 mg/L from shake flask cultures. This high production rate and purification yield can be attributed to the favorable intrinsic properties of the DARPins (33), including efficient refolding after denaturation.
EpCAM is overexpressed in many solid human tumors, and cancer initiating cells are sites of abundant expression. EpCAM is also efficiently internalized by receptor-mediated endocytosis and thus ideally suited for ligand-mediated drug delivery into cells. Flow cytometric analyses with MCF-7 breast and HT29 colon carcinoma cells demonstrated that even after complexation with four siRNA molecules per protamine the complexes fully retained their binding activity and specificity for EpCAM. This indicates that siRNA binding and EpCAM binding are spatially localized to the DARPin and the protamine unit, respectively, as expected from the design. The marginal increase of C9-P binding to HT29 cells compared to C9 suggests some background binding of the uncomplexed cationic protamine, which was more pronounced on this cell line.
Comparing the intravesicular signal, internalization of FITC-labeled siRNA mediated by C9-protamine derivatives was lower than with lipofectamine complexes. Lipofectamine 2000 and other formulations of cationic lipids are optimized for transfection of large amounts of nucleic acids in cell culture experiments. Lipoplexes (complexes of cationic lipids with DNA) exploit the hydrophobic character leading to partial membrane permeability of such complexes. However, delivery by cationic detergents is cell-type unspecific and unable to discriminate between malignant and normal tissues. Consequently, their in vivo use is associated with high plasma protein binding, immune reactivity, unspecific cytotoxicity, lack of stability and poor biodistribution (34). PEGylation can reduce the toxicity and stabilize the particles, but in turn is associated with decreased cellular uptake due to weaker interactions with cell membranes.
Confocal microscopy demonstrated that transfection of siRNA into cells with either C9-P or lipofectamine resulted in predominantly intravesicular localization. Nevertheless, a certain amount was delivered into the cytoplasm, which was sufficient for RISC-activation to diminish bcl-2 expression at the mRNA and protein level, and consequently facilitated tumor cell apoptosis as demonstrated in chemosensitization experiments. Similarly, in our previous study EpCAM-targeted nanovesicles containing antisense oligonucleotides also resulted in significant target down-regulation, despite a major fraction remaining entrapped in endocytotic vesicles (21). These findings are in line with data reported from other approaches using protamine or similar arginine-rich peptides for targeted siRNA delivery (5, 8). An interaction of the cationic protamine chain or a cationic lipid with anionic membrane components has been discussed to destabilize vesicles (34). It remains unclear, however, whether there is a specific role of the C9-protamine fusion, after it has reached its endosomal location, for the ultimate transfer of a fraction of the siRNA molecules to the cytosol. It is nevertheless encouraging that the limited amount of payload transferred to the cytoplasm was sufficient to achieve biological effects.
Two different types of DARPin dimers were easily produced with essentially the same yield as monomers. In one, the two domains are joined by a flexible linker, in the other by a dimerization motif, here a leucine zipper, as previously demonstrated with antibody fragments (35, 36). The relative orientation of the two domains in space is very different (Fig. 1). While in ELISA both strategies lead to an avidity increase, suggesting that epitopes properly oriented for bivalent binding can be found on the plate surface containing immobilized EpCAM, this is not the case on cells. Here, only the zipper-mediated (tail-to-tail) arrangement lead to an avidity gain, suggesting that only in this case, two epitopes can be reached by one molecule.
With all fusion protein (C9-P, C9D-P, C9LZ-P), efficient bcl-2 down-regulation was achieved in EpCAM positive cancer cell lines MCF-7 (Fig. 5) and HT-29 (Supplementary Materials). The siRNA was designed as a 2′-O-methyl gapmer sequence (see Materials and Methods), with the flanking region consisting of 2′-O-methyl ribonucleotides and the central part of unmodified ribonucleotides, which provides higher stability against degrading enzymes and reduces off-target effects, especially immune cell activation by interferon-release (37-40).
Consistent with their effect on apparent functional affinity on cells, the different dimerization strategies behaved very different in activity assays. The directly linked bivalent fusion protein C9D-P, not showing an avidity gain with respect to C9-P, also showed no gain in down-regulation of bcl-2 expression in MCF-7 cells. In contrast, C9LZ-P more effectively down-regulated bcl-2 expression, and it had shown an avidity gain. In control experiments, neither a random control siRNA sequence nor the bcl-2-specific siRNA applied in a mixture with C9 (lacking the oligonucleotide binding motif) affected bcl-2 expression. Decreased bcl-2 expression resulted in a significant sensitization of cells to doxorubicin, almost to the same level as upon transfection of siRNA complexed to lipofectamine. However, unlike lipofectamine, the fusion proteins did not sensitize EpCAM-negative HEK293T cells, confirming that siRNA transfection at high efficiency and specificity is feasible.
Altogether, the strict EpCAM-dependency of the demonstrated biological effects suggests particularly the leucine zipper construct C9LZ-P as a promising nanocarrier for tumor-targeted gene therapy as an alternative to liposomal approaches. Optimization of tumor-targeted siRNA delivery is possible by DARPin affinity maturation and/or multimerization, by modification of the siRNA itself (6, 41) and by new approaches that help improving the translocation of siRNA from endosomes into the cytoplasm.
Supplementary Material
Acknowledgements
We thank Dr. Annemarie Honegger for support in preparation of the protein models.
This work was supported by an Erwin Schrödinger Fellowship of the Austrian Science Fund FWF for J.W. [J2593], the Krebsliga of the Kanton Zürich and the Sassella Foundation of the Zürcher Kantonalbank.
Abbreviations
- DARPin
Designed Ankyrin Repeat Protein
- EpCAM
Epithelial Cell Adhesion Molecule
- RISC
RNA induced silencing complex
- VEGF
vascular endothelial growth factor
- Bcl-2
B-cell lymphoma 2
References
- 1.Wilkinson A. Oligonucleotide Therapeutics: The Next Big Thing. Scrip Executive Briefing. 2008;1:1–12. [Google Scholar]
- 2.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–30. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 3.Corey DR. Chemical modification: the key to clinical application of RNA interference? J Clin Invest. 2007;117:3615–22. doi: 10.1172/JCI33483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Fougerolles AR. Delivery vehicles for small interfering RNA in vivo. Hum Gene Ther. 2008;19:125–32. doi: 10.1089/hum.2008.928. [DOI] [PubMed] [Google Scholar]
- 5.Juliano R, Alam MR, Dixit V, Kang H. Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides. Nucl Acids Res. 2008;36:4158–71. doi: 10.1093/nar/gkn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen T, Menocal EM, Harborth J, Fruehauf JH. RNAi therapeutics: An update on delivery. Curr Opin Mol Ther. 2008;10:158–67. [PubMed] [Google Scholar]
- 7.Soutschek J, Akinc A, Bramlage B, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–8. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 8.Song E, Zhu P, Lee SK, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–17. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 9.Peer D, Zhu P, Carman CV, Lieberman J, Shimaoka M. Selective gene silencing in activated leukocytes by targeting siRNAs to the integrin lymphocyte function-associated antigen-1. Proc Natl Acad Sci U S A. 2007;104:4095–100. doi: 10.1073/pnas.0608491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayer G, Vogel V, Weyermann J, et al. Oligonucleotide-protamine-albumin nanoparticles: Protamine sulfate causes drastic size reduction. J Control Release. 2005;106:181–7. doi: 10.1016/j.jconrel.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 11.Medberry P, Dennis S, Van Hecke T, DeLong RK. pDNA bioparticles: comparative heterogeneity, surface, binding, and activity analyses. Biochem Biophys Res Commun. 2004;319:426–32. doi: 10.1016/j.bbrc.2004.04.188. [DOI] [PubMed] [Google Scholar]
- 12.Vogel V, Lochmann D, Weyermann J, et al. Oligonucleotide-protamine-albumin nanoparticles: preparation, physical properties, and intracellular distribution. J Control Release. 2005;103:99–111. doi: 10.1016/j.jconrel.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 13.Went P, Vasei M, Bubendorf L, et al. Frequent high-level expression of the immunotherapeutic target Ep-CAM in colon, stomach, prostate and lung cancers. Br J Cancer. 2006;94:128–35. doi: 10.1038/sj.bjc.6602924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spizzo G, Gastl G, Obrist P, et al. High Ep-CAM Expression is Associated with Poor Prognosis in Node-positive Breast Cancer. Breast Cancer Res Treat. 2004;86:207–13. doi: 10.1023/B:BREA.0000036787.59816.01. [DOI] [PubMed] [Google Scholar]
- 15.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 16.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 18.Trzpis M, McLaughlin PMJ, de Leij LMFH, Harmsen MC. Epithelial cell adhesion molecule: More than a carcinoma marker and adhesion molecule. Am J Pathol. 2007;171:386–95. doi: 10.2353/ajpath.2007.070152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maetzel D, Denzel S, Mack B, et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11:162–71. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- 20.Di Paolo C, Willuda J, Kubetzko S, et al. A recombinant immunotoxin derived from a humanized epithelial cell adhesion molecule-specific single-chain antibody fragment has potent and selective antitumor activity. Clin Cancer Res. 2003;9:2837–48. [PubMed] [Google Scholar]
- 21.Hussain S, Plückthun A, Allen TM, Zangemeister-Wittke U. Chemosensitization of carcinoma cells using epithelial cell adhesion molecule-targeted liposomal antisense against bcl-2/bcl-xL. Mol Cancer Ther. 2006;5:3170–80. doi: 10.1158/1535-7163.MCT-06-0412. [DOI] [PubMed] [Google Scholar]
- 22.Hussain S, Plückthun A, Allen TM, Zangemeister-Wittke U. Antitumor activity of an epithelial cell adhesion molecule targeted nanovesicular drug delivery system. Mol Cancer Ther. 2007;6:3019–27. doi: 10.1158/1535-7163.MCT-07-0615. [DOI] [PubMed] [Google Scholar]
- 23.Binz HK, Amstutz P, Kohl A, et al. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat Biotech. 2004;22:575–82. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- 24.Binz HK, Stumpp MT, Forrer P, Amstutz P, Plückthun A. Designing repeat proteins: Well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J Mol Biol. 2003;332:489–503. doi: 10.1016/s0022-2836(03)00896-9. [DOI] [PubMed] [Google Scholar]
- 25.Zahnd C, Wyler E, Schwenk JM, et al. A designed ankyrin repeat protein evolved to picomolar affinity to Her2. J Mol Biol. 2007;369:1015–28. doi: 10.1016/j.jmb.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 26.Zahnd C, Pecorari F, Straumann N, Wyler E, Plückthun A. Selection and characterization of Her2 binding-designed ankyrin repeat proteins. J Biol Chem. 2006;281:35167–75. doi: 10.1074/jbc.M602547200. [DOI] [PubMed] [Google Scholar]
- 27.Hanes J, Plückthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci U S A. 1997;94:4937–42. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zahnd C, Amstutz P, Plückthun A. Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat Meth. 2007;4:269–79. doi: 10.1038/nmeth1003. [DOI] [PubMed] [Google Scholar]
- 29.Plückthun A, Pack P. New protein engineering approaches to multivalent and bispecific antibody fragments. Immunotechnology. 1997;3:83–105. doi: 10.1016/s1380-2933(97)00067-5. [DOI] [PubMed] [Google Scholar]
- 30.Jansen B, Wacheck V, Heere-Ress E, et al. Chemosensitisation of malignant melanoma by BCL2 antisense therapy. Lancet. 2000;356:1728–33. doi: 10.1016/S0140-6736(00)03207-4. [DOI] [PubMed] [Google Scholar]
- 31.Gleave ME, Monia BP. Antisense therapy for cancer. Nat Rev Cancer. 2005;5:468–79. doi: 10.1038/nrc1631. [DOI] [PubMed] [Google Scholar]
- 32.Lessene G, Czabotar PE, Colman PM. BCL-2 family antagonists for cancer therapy. Nat Rev Drug Discov. 2008;7:989–1000. doi: 10.1038/nrd2658. [DOI] [PubMed] [Google Scholar]
- 33.Wetzel SK, Settanni G, Kenig M, Binz HK, Plückthun A. Folding and unfolding mechanism of highly stable full-consensus ankyrin repeat proteins. J Mol Biol. 2008;376:241–57. doi: 10.1016/j.jmb.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 34.Li W, Szoka F. Lipid-based nanoparticles for nucleic acid delivery. Pharm Res. 2007;24:438–49. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- 35.Willuda J, Kubetzko S, Waibel R, Schubiger PA, Zangemeister-Wittke U, Plückthun A. Tumor targeting of mono-, di-, and tetravalent anti-p185HER-2 miniantibodies multimerized by self-associating peptides. J Biol Chem. 2001;276:14385–92. doi: 10.1074/jbc.M011669200. [DOI] [PubMed] [Google Scholar]
- 36.Kubetzko S, Balic E, Waibel R, Zangemeister-Wittke U, Plückthun A. PEGylation and multimerization of the anti-p185HER-2 single chain Fv fragment 4D5: effects on tumor targeting. J Biol Chem. 2006;281:35186–201. doi: 10.1074/jbc.M604127200. [DOI] [PubMed] [Google Scholar]
- 37.Noe CR, Winkler J, Urban E, Gilbert M, Haberhauer G, Brunar H. Zwitterionic oligonucleotides: a study on binding properties of 2′-O-aminohexyl modifications. Nucleosides Nucleotides Nucleic Acids. 2005;24:1167–85. doi: 10.1081/NCN-200067400. [DOI] [PubMed] [Google Scholar]
- 38.Sioud M. Does the understanding of immune activation by RNA predict the design of safe siRNAs? Front Biosci. 2008;13:4379–92. doi: 10.2741/3011. [DOI] [PubMed] [Google Scholar]
- 39.Winkler J, Gilbert M, Kocourková A, Stessl M, Noe CR. 2′-O-Lysylaminohexyl Oligonucleotides: Modifications for Antisense and siRNA. ChemMedChem. 2008;3:102–10. doi: 10.1002/cmdc.200700169. [DOI] [PubMed] [Google Scholar]
- 40.Stessl M, Marchetti-Deschmann M, Winkler J, Lachmann B, Allmaier G, Noe CR. A proteomic study reveals unspecific apoptosis induction and reduction of glycolytic enzymes by the phosphorothioate antisense oligonucleotide oblimersen in human melanoma cells. J Prot. 2009 doi: 10.1016/j.jprot.2009.06.001. doi:10.1016/j.jprot.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Winkler J, Saadat K, Díaz-Gavilán M, Urban E, Noe CR. Oligonucleotide polyamine conjugates: Influence of length and position of 2′-attached polyamines on duplex stability and antisense effect. Eur J Med Chem. 2009;44:670–7. doi: 10.1016/j.ejmech.2008.05.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.