Abstract
RASSF2 is a recently identified member of a class of novel tumour suppressor genes, all containing a ras association domain. We previously demonstrated that the A isoform of RASSF2, is frequently inactivated by promoter region hypermethylation in colorectal tumours and adenomas, methylation was tumour specific and that expression in methylated tumour lines could be reactivated by treatment with 5-aza-2dc. RASSF2 resides at 20p13, this region has been demonstrated to be frequently lost in human cancers. In this report we investigated methylation status of the RASSF2A promoter CpG island in a series of breast, ovarian and non-small cell lung cancers (NSCLC). RASSF2A was frequently methylated in breast tumour cell lines 65% (13/20) and in primary breast tumours 38% (15/40). RASSF2A gene expression could be switched back on in methylated breast tumour cell lines after treatment with 5-aza-2dC, whilst unmethylated lines showed no difference in level of expression before and after 5-aza-2dC treatment. RASSF2A was also frequently methylated in NSCLC tumours 44% (22/50). Methylation in breast tumours and NSCLC was tumour specific. We did not detect RASSF2A methylation in ovarian tumours (0/17). Furthermore no mutations were found in the coding region of RASSF2A in these ovarian tumours.
RASSF2A suppressed breast tumour cell growth in vitro (through colony formation and soft agar assays) and in vivo. We identified a highly conserved putative bipartite nuclear localisation signal (NLS) between amino acids 151 and 167 in the RASSF2A sequence and demonstrated that endogenous RASSF2A localised to the nucleus. Mutation of the putative nuclear localisation signal abolished the nuclear localisation so RASSF2A became predominantly cytoplasmic. Our data indicates that RASSF2A is frequently methylated in colorectal, breast and NSCLC tumours, furthermore, the methylation is tumour specific. Hence we have identified RASSF2A as a novel methylation marker for multiple malignancies and it has the potential to be developed into a valuable marker for screening several cancers in parallel using promoter hypermethylation profiles.
We also demonstrate that RASSF2 has a functional NLS signal. Furthermore this is the first report demonstrating that RASSF2 suppresses growth of cancer cells in vivo. Hence providing further evidence for its role as a tumour suppressor gene located at 20p13.
INTRODUCTION
Epigenetic silencing of tumour suppressor genes plays an important role in the development of many cancer types. Identification of genes that are inactivated by promoter region CpG island hypermethylation will lead not only to insights into novel disease mechanisms but also to development of new diagnostic molecular markers and potential reactivation therapies. We have identified members of a novel family of genes that contain a ras association domain (reviewed in (Agathanggelou et al., 2005)). So far the Ras association domain family (RASSF) of genes contains 6 members, namely, RASSF1 located at 3p21.3, RASSF2 located at 20p13, RASSF3 located at 12q14.1, AD037 (also known as RASSF4) located at 10q11.21, NORE1 located at 1q32.1, RASSF6 at 4q21.21. RASSF1 was identified from a region of overlapping homozygous deletions at 3p21.3 in lung and breast tumour cell lines (Agathanggelou et al., 2001a; Burbee et al., 2001; Dammann et al., 2000; Lerman and Minna, 2000). Expression of isoform A of RASSF1 is frequently lost following methylation of its promoter in various adult and childhood cancers (reviewed in (Agathanggelou et al., 2005)). So far RASSF1A is thought to play an important role in microtubule stability, it has also shown to be involved in cell cycle regulation, apoptosis and cell migration (see (Agathanggelou et al., 2005; Dallol et al., 2005)). Rassf1a knockout mice showed increased susceptibility to spontaneous and carcinogen induced tumour formation (Tommasi et al., 2005; van der Weyden et al., 2005).
As well as RASSF1A; RASSF2A (Hesson et al., 2005), AD037/RASSF4 (Eckfeld et al., 2004) and NORE1A (Hesson et al., 2003; Irimia et al., 2004), are also epigenetically inactivated in tumours, whilst RASSF3 (Hesson et al., 2004; Tommasi et al., 2002) and RASSF6 (Allen et al., 2007) are not methylated. We have recently demonstrated that RASSF2A is frequently methylated in colorectal tumours and adenomas and showed that methylation was cancer specific (Hesson et al., 2005). Hence, RASSF2A methylation is a potential molecular marker for early detection of colorectal cancer. RASSF2 protein expression is frequently downregulated in human lung tumour cell lines including non-small cell lung cancer (NSCLC) (Vos et al., 2003). Using array-based comparative genomic hybridisation in ovarian tumour cell lines Lambros et al., (Lambros et al., 2005) identified regions of deletions including homozygous deletions on several chromosomes. Candidate tumour suppressor genes localized to these regions included CDKN2C and MADHIP (1p32.3), WW domain protein WWOX (16q23.1) and RASSF2 (20p13).
In this report we determined the methylation status of RASSF2A in breast, NSCLC and ovarian tumours. We also undertook experiments to determine tumour suppression properties of RASSF2 and characterised a putative NLS in the RASSF2 sequence.
MATERIAL AND METHODS
Patients and Samples
Forty invasive ductal breast carcinoma, 10 corresponding normal tissue and 20 breast tumour cell lines were analysed and have been described previously (Dallol et al., 2002). 17 primary ovarian tumours were also analysed, these have been described previously (Fullwood et al., 1999). The 50 NSCLC analysed in this study have been described previously (Agathanggelou et al., 2001b).
PCR and bisulphite sequencing
Sodium bisulphite modification was performed as described previously (Agathanggelou et al., 2001b). Breast tumour cell lines were assayed for RASSF2A CpG island hypermethylation using the CoBRA assay. The primers and conditions for the PCR have been described previously (Hesson et al., 2005). PCR products were then assayed by digestion with TaqI or BstUI for 2 hours at 65 or 60 °C respectively. To confirm methylation status and ascertain the extent of methylation the PCR products were purified using QIAquick PCR purification columns (Qiagen) then sequenced using the ABI BigDye Cycle Sequencing Kit version 3.1 (Perkin-Elmer). For primary tumours, PCR products were first cloned in to pGEM-T Easy (Promega) and individual clones were sequenced.
The promoter methylation status of RASSF2A and RASSF1A in breast tumours and corresponding normal tissues was determined using MSP. The primers and conditions have been described previously (Hesson et al., 2004; Hesson et al., 2005).
Cell lines and 5-aza-2dC treatment
Breast tumour cell lines were routinely maintained in DMEM (Gibco) supplemented with 10 % FCS at 37 °C, 5 % CO2. Cells were seeded and grown for 24 hours before addition of 2.5 μM 5′-aza-2′deoxycytidine (Sigma). This medium was replaced every day for 5 days. RNA was prepared using the RNeasy kit (Qiagen) according to manufacturers’ instructions.
Reverse transcriptase polymerase chain reaction (RT-PCR)
cDNA was synthesised from 1 μg of RNA using the SuperScript III cDNA Synthesis Kit (Invitrogen) and 50 ng of this cDNA was amplified by PCR using the RASSF2A isoform and the GAPDH control in breast tumour cell lines by using primers and conditions described previously (Hesson et al., 2005).
Mutation analysis
The coding region of RASSF2A was analysed for mutations by direct sequencing (primer sequences available upon request).
Colony formation assay
The breast tumour cell line MCF7 was used in a colony formation assay. Twenty-four hours before transfection MCF7 cells (not expressing RASSF2A due to promoter methylation) were seeded at 1 × 105 cells per well of a 6 well plate. Each well was transfected with 2μg of pFLAG or pFLAG-RASSF2A using 6 μl of FuGENE 6 (Roche). Forty eight hours after transfection cells were selected with 600 μg/ml Geneticin (Gibco) in DMEM. Twenty one days after transfection surviving colonies were stained with 0.4 % crystal violet.
Anchorage independent growth assay
Anchorage independent growth studies were carried out in soft agar in 6 well plates. Clones 3 and 4 of MCF7 cells stably expressing either pFLAG-RASSF2A or pFLAG-empty were used for the study. A base layer of 2 ml growth medium containing 0.7 % Noble agar was covered with 1 ml of growth medium containing 0.35 % Noble agar and 5 × 104 cells. Finally 2 ml growth medium containing 0.7 % Noble agar was layered on top of the cells. After 35 days colonies comprised of at least 32 cells were counted on an inverted microscope.
In vivo tumourigenicity assay
The tumorigenicity of the parental MCF7 cell line transfected with empty vector or clones 3 or 4 stably expressing FLAG tagged RASSF2A were assayed by s.c. injection as described by Kashuba et al. (Kashuba et al., 2004). In brief, each transfectant clone was grown to obtain about 5×106 cells for one inoculation. Cells were treated with trypsin-EDTA and concentrated by centrifugation at 800 rpm for 2 min and 5 × 106 cells in 0.1 ml serum-free IMDM medium were injected into 4- to 8-week-old SCID mice. A total of eight sites (two inoculations per mice) were tested for each cell line and tumour growth in animals was checked twice a week. If tumour formation was observed, tumours were measured using calipers.
Generation of RASSF2A-GFP constructs
Full length RASSF2A was subcloned into the BglII and SalI sites of the pEGFP-C1 vector (Clontech).
The QuikChange® site directed mutagenesis kit (Stratagene) was used to mutate the nuclear localisation signal of RASSF2A. The mutagenic primers were forward AGTGATGTTGGGGTGGCTGCCGCTGGCAATGTGAGGACG and reverse CGTCCTCACATTGCCAGCGGCAGCCACCCCAACATCACT
Antibodies and western blotting
Cell lines were fractionated by standard CLB buffer protocol. The antibody against RASSF2A is described in (Vos et al., 2003). The anti FLAG monoclonal antibody was clone M5 from Sigma. The antibody against alpha tubulin was from Santa-Cruz.
Preparation of cells for microscopy
Twenty four hours prior to transfection Cos7 cells were seeded at 2×105 cells per well on to coverslips in a 6 well plate. Cells were transfected with 1μg DNA using 3μl FuGENE6. After 24 or 48 h as indicated in the figure legends the cells were washed twice with PBS and fixed with 4 % paraformaldehyde for 10 minutes at room temperature. Cells were mounted with Vectashield (containing 4′, 6-diamidino-2-phenyllindole) after fixing.
RESULTS
RASSF2A methylation analysis in breast tumour cell lines
Using combined bisulphite restriction analysis (CoBRA) (Hesson et al., 2005) and bisulphite sequencing we determined the methylation status of RASSF2A promoter region CpG island in a series of breast tumour cell lines. RASSF2A was methylated in 13 of 20 (65%) of these (Fig 1a). Direct sequencing confirmed these CoBRA results and demonstrated dense methylation across the region (Fig 1b). Treatment with 5′-aza-2′deoxycytidine (5-aza-2dC) of cell lines where RASSF2A was methylated and expression silenced reactivated RASSF2A expression, whilst no change in RASSF2A expression was seen in unmethylated tumour lines (Fig 1c).
FIGURE 1.

RASSF2A expression is lost following promoter methylation in breast tumour cell lines.
(a) Frequent methylation of the RASSF2A CpG island is observed in breast tumour cell lines as determined by CoBRA digestion of PCR products. Using the restriction enzymes TaqI or BstUI to digest PCR products methylation was observed in 65 % (13/20) breast tumour cell lines. The breast tumour cell lines HCC38 and BT174 were unmethylated while HTB19, MCF7 and HCC712 were heavily methylated.
(b) Direct bisulphite sequencing of the RASSF2A CpG island in breast tumour cell lines. CG dinucleotides within the amplification region are numbered 1-23 the underlined CpGs are contained within BstUI or TaqI recognition sequences. White and black squares represent unmethylated and methylated CpGs respectively.
(c) Expression of RASSF2A in breast tumour cell lines. RASSF2A CpG island hypermethylation corresponds with loss of RASSF2A expression in the cell lines MDA-MB-435, MCF7 and HTB19. Expression was restored after treatment with the demethylating agent 5-aza-2dC. GAPDH was used as a control to ensure RNA integrity and equal loading. Breast tumour line HCC38 is not methylated for RASSF2A, hence expression is unchanged after 5-aza-2dC treatment.
RASSF2A methylation analysis in breast, NSCLC and ovarian tumours
We had previously developed a methylation specific PCR (MSP) assay to analyse RASSF2A methylation in colorectal tumours and adenomas (Hesson et al., 2005). The same assay was utilized for analysing a series of breast, NSCLC and ovarian tumours for RASSF2A methylation. RASSF2A was found to be methylated in 38% (15/40) of breast tumours, and the corresponding normal DNA was unmethylated, again demonstrating tumour specific methylation (Fig 2). To determine the extent of methylation, CoBRA PCR was performed on three methylated and one unmethylated breast tumour and the PCR products were cloned and sequenced. Sequencing confirmed the extent of methylation of the promoter region in the methylated primary breast tumours, and revealed no methylation in samples deemed unmethylated by digestion. There was no correlation between RASSF2A methylation and breast tumour grade. We also analysed methylation status of RASSF1A in the same series of breast tumours. RASSF1A was methylated in 18 of 32 (56%) of these. There was no correlation between RASSF1A and RASSF2A methylation status.
FIGURE 2.
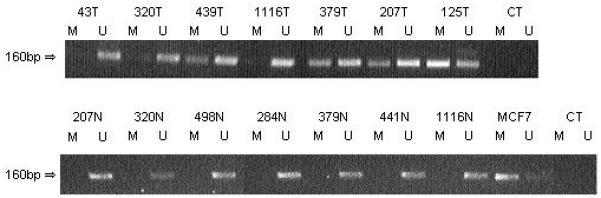
The RASSF2A promoter is frequently methylated in breast tumours.
Methylation status of the RASSF2A CpG island in primary breast tumours (T) was determined by MSP where M represents methylated-specific PCR and U represents unmethylated-specfic PCR. The MCF7 cell line was used as a positive control for the M PCR, CT represents water control. The RASSF2A CpG island promoter region was hypermethylated in 38 % (15/40) primary breast tumours. The corresponding normal DNA (N) was unmethylated suggesting that the methylation was tumour specific.
RASSF2A was methylated in 44% (22/50) of NSCLC tumours (Fig 3a), where corresponding normal DNA was available it was not methylated, suggesting that the methylation was a tumour specific event (Fig 3b). Methylation was found at an equal frequency in all grades of tumour (I/II = 7/16 (44%); IIIA = 4/9 (44%); IV = 10/23 (44%) and 1of 2 NSCLC of unknown grade was also methylated) suggesting that it is an early event in lung tumourigenesis. The methylation status of RASSF1A in this series of tumours had previously been determined (Agathanggelou et al., 2001b). There was no association between RASSF1A and RASSF2A methylation status (p=0.26).
FIGURE 3.

The RASSF2A promoter is frequently methylated in NSCLC primary tumours but not in ovarian tumours.
(a) Methylation status of the RASSF2A promoter was determined in NSCLC primary tumours using MSP, where M represents methylated-specific PCR and U represents unmethylated-specfic PCR. Methylation was found in 22/50 (44%) primary tumours. Methylation was found in all grades of tumor (I/II = 7/16 (44%); IIIA = 4/9 (44%); IV = 10/23 (44%)).
(b) RASSF2A CpG island methylation was tumour specific. (T represents tumour; N represents corresponding normal lung)
(c) The RASSF2A CpG island promoter region was unmethylated in primary ovarian tumours. OVCA28, an ovarian tumour cell line partially methylated for RASSF2A, acts as a positive control for both M and U sets of primers. RASSF2A CoBRA was also carried out on these samples to confirm the result (data not shown).
We also analysed ovarian tumours for RASSF2A methylation using both MSP and CoBRA assays. None of the 17 ovarian tumours analysed demonstrated methylation of the RASSF2A promoter CpG island (Fig 3c). Since no methylation was found in ovarian tumours, we looked for mutations in the coding region of RASSF2A in these samples. No somatic inactivating mutations were found but one silent substitution was detected in exon 8 in two tumours. We also analysed methylation status of RASSF1A in the same series of ovarian tumours. RASSF1A was methylated in 10 of 17 (59%) of ovarian tumours respectively. This frequency of RASSF1A methylation in ovarian tumors is similar to that which has been reported previously (Yoon et al., 2001).
RASSF2A expression inhibits growth of breast cancer cells in vitro and in vivo
Flag-tagged wild-type RASSF2A or empty vector were transfected into the MCF7 breast tumour cell line (where endogenous RASSF2A expression is completely suppressed by promoter methylation). Using a colony-formation assay we demonstrated that re-expression of RASSF2A reduces colony formation ability by 66% (P = 0.0005, Fig 4a). This effect was consistent through 3 independent experiments. Two independent MCF7 clones stably expressing RASSF2A were generated (Cl.3 and Cl.4) these clones inhibited anchorage-independent growth in a soft agar assay (Cl.3 and Cl.4 vs vector P < 0.05) (Fig 4b-c). Cells from these RASSF2A expressing clones were also inoculated in to SCID mice and over the course of 140 days no tumours were observed, whereas tumours were formed within 120 days in mice inoculated with vector control cells Fig 4d.
FIGURE 4.

RASSF2A suppresses growth of breast tumour cells.
(a) Colony formation. The number of Geneticin resistant colonies in cells transfected with empty vector was set at 100 %. Values are the mean ± SD of three separate experiments each calculated from triplicate plates.
(b) Immunoblot stained with anti-FLAG (Sigma clone M5) to detect RASSF2A in protein extracts from MCF7 cells stably transfected with pFLAG-RASSF2A (stable clones, Cl.3 and Cl.4).
(c) Anchorage independent growth studies. Growth of MCF7-RASSF2A stable clones (Cl.3 and Cl.4) and MCF7 cells stably transfected with an empty vector were compared using a soft agar assay. The number of Geneticin resistant colonies in cells transfected with empty vector was set at 100 %. Values are the mean ± SD calculated from triplicate plates.
(d) Tumour formation in SCID mice. Over 140 days tumours develop in mice inoculated with MCF7 cells, however they are not observed in mice inoculated with MCF7 cells expressing RASSF2A.
RASSF2A contains a functional nuclear localisation signal
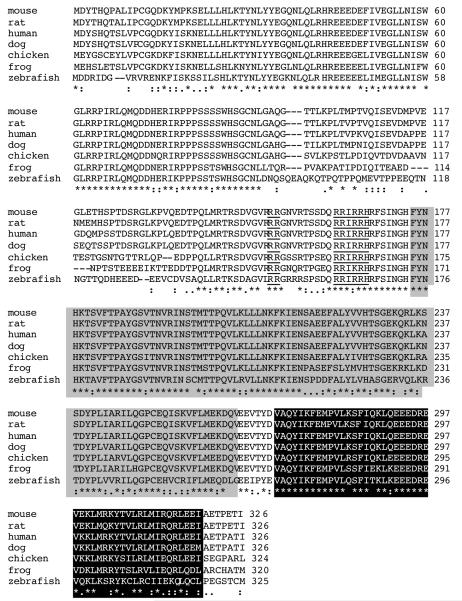
Bioinformatic analysis of RASSF2A was performed using PSORT (www.psort.org) and two putative nuclear localisation signals (NLS) were predicted; a monopartite NLS of the pat 4 subcategory (RRHR between amino acids 165 and 168) and a bipartite NLS (Robbins et al., 1991) between amino acids 151 and 167. The two motifs of basic amino acids RR(N)10RIRRH are perfectly conserved amongst vertebrates Fig 5, and the only alteration between vertebrates and in Xenopus converts an R to K thereby retaining the positive charge.
FIGURE 5. RASSF2A has a putative nuclear localisation signal.
RASSF2A protein sequences were aligned using the ClustalW program (www.ebi.ac.uk). The sequences of human (NM_014737), rat (NM_001037096), chicken (NM_001030884), frog (NM_001016903), mouse (NM_175445), dog (XM_542904) and zebra fish (NM_001004676) RASSF2A are shown. The * illustrates the location of amino acids identical in all proteins; : denotes the location of amino acids that undergo conservative substitution; . indicates the location of amino acids that undergo semi-conservative substitution. The area highlighted in grey represents the Ras-association (RA) domain and the region highlighted in black illustrates the coiled-coil SARAH domains. The putative bipartite nuclear localisation domain is underlined
Therefore we generated a GFP-tagged RASSF2A and a version with basic amino acids within the bipartite NLS mutated (R150A/R151A/R152A) transfection of these constructs into Cos7 cells demonstrated predominantly nuclear localisation of wild-type (Fig 6a) and predominantly cytoplasmic localisation of the mutant NLS (Fig 6b). More than 65 cells from triplicate experiments were analysed for localisation of the GFP signal, and the results are presented in Fig 6c. Similar results were obtained in HeLa and 293 cells, and the breast tumour cell lines MCF7 and HTB19 and also with FLAG tagged RASSF2A constructs.
FIGURE 6.
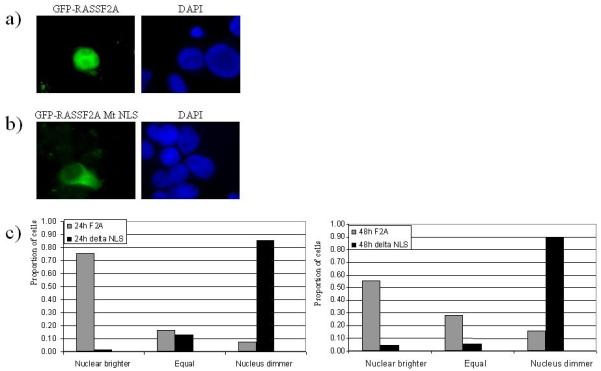
RASSF2A is localised to the nucleus, mutation of a putative nuclear localization signal can translocate RASSF2A to the cytoplasm.
a) Cos7 cells transfected with GFP-RASSF2A
b) Cos7 cells transfected with GFP-RASSF2A MtNLS
c) Cos7 cells were transfected with GFP-RASSF2A or GFP-RASSF2A MtNLS and the localization scored as more intense in the nucleus, more intense in the cytoplasm or equal intensity. For each vector at each timepoint at least 65 cells were analysed
In order to demonstrate the localisation of endogenously expressed RASSF2A; lung cancer cell lines H417 and H720 were fractionated into nuclear and cytoplasmic fractions, and western blotting confirmed that endogenous RASSF2A partitions to the nucleus (Fig 7).
FIGURE 7.

Endogenous RASSF2 is localized to the cytoplasm.
Protein from two human lung cancer cell lines was fractionated into nuclear (N) and cytoplasmic (C) fractions by standard CLB buffer protocol, and the blot immunoprobed with anti-RASSF2, then reprobed with anti-alpha tubulin to confirm that the nuclear fraction was free of contaminating cytoplasmic proteins.
DISCUSSION
Previously we had identified RASSF2 (Hesson et al., 2005; Vos et al., 2003) as a member of the recently discovered RASSF family of Ras effectors/tumour suppressors (Agathanggelou et al., 2005) and demonstrated that it binds directly to K-Ras via the Ras effector domain. K-Ras is a member of a family of oncoproteins that play an important role in many biological processes (Malumbres and Barbacid, 2003; Rommel and Hafen, 1998; Schubbert et al., 2007). These abilities are mediated by the ability of RAS proteins to interact with a wide range of effector proteins (Cox and Der, 2003; Shields et al., 2000); for example via signalling through phosphatidylinositol 3-kinase and AKT, RAS proteins can induce cell division and oncogenesis, whereas interactions with RASSF2 can mediate apoptosis and cell cycle arrest (Vos et al., 2003). We had previously demonstrated that RASSF2A was frequently methylated in colorectal tumours and adenomas (Hesson et al., 2005) and that methylation was tumour specific. Furthermore, using our sets of methylation primers (Hesson et al., 2005), two recent reports (Park et al., 2007; Zhang et al., 2007) have demonstrated that methylation of RASSF2A corresponds to loss of expression in primary tumors as well as tumour cell lines.
In this report we demonstrated for the first time that the RASSF2A promoter was frequently methylated in breast (38%) and NSCLC (44%) tumours but was not methylated in ovarian tumors (0/17). We showed methylation correlated with loss of expression in cancer cell lines and that treatment with 5-aza-2dC could reintroduce expression of RASSF2A in the cancer cell lines where previously methylation was observed. Neither corresponding normal lung nor normal breast samples demonstrated methylation, this in conjunction with other published data from colon (Hesson et al., 2005; Park et al., 2007) and nasopharyngeal epithelia (Zhang et al., 2007) suggests that RASSF2A promoter methylation is uncommon in normal tissues implicating it as a potential biomarker. The frequency of methylation was similar for all grades of NSCLC tumour analysed, and it was not correlated with breast cancer grade suggesting that RASSF2A inactivation by methylation is an early event in lung and breast tumourogenesis as also appears to be the case in colorectal cancer (Akino et al., 2005; Hesson et al., 2005; Park et al., 2007). We did not detect a correlation between RASSF1A and RASSF2A promoter methylation suggesting that methylation of these loci are independent events and not part of a generalised methylator phenotype.
Other studies have suggested that RASSF2A may function as a tumour suppressor gene in vitro in cell migration, cell cycle progression and colony formation assays (Akino et al., 2005; Vos et al., 2003; Zhang et al., 2007), we have confirmed and extended this work demonstrating that RASSF2A can function as a tumour suppressor gene in vitro inhibiting the growth of breast cancer cell lines in colony formation and soft agar growth assays, and in vivo inhibiting tumour formation when cells expressing RASSF2A were subcutaneously injected into SCID mice. This is the first report demonstrating that RASSF2 can suppress tumour cell growth in vivo hence providing further evidence for its role as a tumour suppressor gene.
Analysis of the RASSF2A amino acid sequence identified a putative bipartite nuclear localisation signal, which was highly conserved in vertebrates. In order to determine if it is a functional NLS motif we performed transient transfection assays using GFP tagged RASSF2A and demonstrated that wild type RASSF2A is a predominantly nuclear protein. Mutation of conserved arginines within the putative NLS relocated RASSF2A to a predominantly cytoplasmic localisation, thus the RR(N)10RIRRH sequence within RASSF2A can function as a nuclear localisation signal. While this work was being completed, a report by Kumari et al (Kumari et al., 2007) appeared, also identifying the same NLS in RASSF2A, however this report is the first to show that endogenous RASSF2 partitions to the nucleus. The nuclear localization of RASSF2A is expected to be important for its function as a tumour suppressor.
The data presented in this study and our previously published work (Hesson et al., 2005) suggest that methylation of the RASSF2A promoter is an early, frequent and tumour specific event in a range of epithelial derived tumours (including NSCLC, breast and colorectal). Thus RASSF2A promoter methylation shows potential as a biologically relevant biomarker for the early detection of these cancers and for developing novel therapeutic intervention regimes. We also demonstrate that RASSF2A can function as a TSG both in vitro and in vivo and has a functional NLS that may be important for its tumour suppressor gene function(s).
Acknowledgments
Research in FL laboratory funded in part by Breast Cancer Campaign, Cancer Research UK, Association for International Cancer Research and Birmingham Children’s Hospital Research Foundation. ERZ was supported by research grants from the Swedish Cancer Society, the Swedish Research Council, the Swedish Foundation for International Cooperation in Research and Higher Education (STINT), the Swedish Institute, the Royal Swedish Academy of Sciences, INTAS and Karolinska Institute.
REFERENCES
- Agathanggelou A, Astuti D, Honorio S, Dallol A, Martinsson T, Cummins C, et al. RASSF1A promoter region CpG island hypermethylation in pheochromocytoma and neuroblastoma tumors. Am J Hum Genet. 2001a;69:485. doi: 10.1038/sj.onc.1204968. [DOI] [PubMed] [Google Scholar]
- Agathanggelou A, Cooper WN, Latif F. Role of the ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 2005;65:3497–3508. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- Agathanggelou A, Honorio S, Macartney DP, Martinez A, Dallol A, Radar J, et al. Methylation associated inactivation of RASSF1A from region 3p21.3 in lung, breast and ovarian tumours. Oncogene. 2001b;20:1509–1518. doi: 10.1038/sj.onc.1204175. [DOI] [PubMed] [Google Scholar]
- Akino K, Toyota M, Suzuki H, Mita H, Sasaki Y, Ohe-Toyota M, et al. The Ras effector RASSF2 is a novel tumor-suppressor gene in human colorectal cancer. Gastroenterology. 2005;129:156–169. doi: 10.1053/j.gastro.2005.03.051. [DOI] [PubMed] [Google Scholar]
- Allen NPC, Donninger H, Vos MD, Eckfeld K, Hesson L, Gordon L, et al. RASSF6 is a novel member of the RASSF family of tumor suppressors. Oncogene. 2007 doi: 10.1038/sj.onc.1210440. in press. [DOI] [PubMed] [Google Scholar]
- Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao BN, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Der CJ. The dark side of Ras: regulation of apoptosis. Oncogene. 2003;22:8999–9006. doi: 10.1038/sj.onc.1207111. [DOI] [PubMed] [Google Scholar]
- Dallol A, Agathanggelou A, Tommasi S, Pfeifer GP, Maher ER, Latif F. Involvement of the RASSF1A tumor suppressor gene in controlling cell migration. Cancer Res. 2005;65:7653–7659. doi: 10.1158/0008-5472.CAN-05-0247. [DOI] [PubMed] [Google Scholar]
- Dallol A, Da Silva NF, Viacava P, Minna JD, Bieche I, Maher ER, et al. SLIT2, a human homologue of the Drosophila Slit2 gene, has tumor suppressor activity and is frequently inactivated in lung and breast cancers. Cancer Res. 2002;62:5874–5880. [PubMed] [Google Scholar]
- Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- Eckfeld K, Hesson L, Vos MD, Bieche I, Latif F, Clark GJ. RASSF4/AD037 is a potential ras effector/tumor suppressor of the RASSF family. Cancer Res. 2004;64:8688–8693. doi: 10.1158/0008-5472.CAN-04-2065. [DOI] [PubMed] [Google Scholar]
- Fullwood P, Marchini S, Rader JS, Martinez A, Macartney D, Broggini M, et al. Detailed genetic and physical mapping of tumor suppressor loci on chromosome 3p in ovarian cancer. Cancer Res. 1999;59:4662–4667. [PubMed] [Google Scholar]
- Hesson L, Bieche I, Krex D, Criniere E, Hoang-Xuan K, Maher ER, et al. Frequent epigenetic inactivation of RASSF1A and BLU genes located within the critical 3p21.3 region in gliomas. Oncogene. 2004;23:2408–2419. doi: 10.1038/sj.onc.1207407. [DOI] [PubMed] [Google Scholar]
- Hesson L, Dallol A, Minna JD, Maher ER, Latif F. NORE1A, a homologue of RASSF1A tumour suppressor gene is inactivated in human cancers. Oncogene. 2003;22:947–954. doi: 10.1038/sj.onc.1206191. [DOI] [PubMed] [Google Scholar]
- Hesson LB, Wilson R, Morton D, Adams C, Walker M, Maher ER, et al. CpG island promoter hypermethylation of a novel Ras-effector gene RASSF2A is an early event in colon carcinogenesis and correlates inversely with K-ras mutations. Oncogene. 2005;24:3987–3994. doi: 10.1038/sj.onc.1208566. [DOI] [PubMed] [Google Scholar]
- Irimia M, Fraga MF, Sanchez-Cespedes M, Esteller M. CpG island promoter hypermethylation of the Ras-effector gene NORE1A occurs in the context of a wild-type K-ras in lung cancer. Oncogene. 2004;23:8695–8699. doi: 10.1038/sj.onc.1207914. [DOI] [PubMed] [Google Scholar]
- Kashuba VI, Li JF, Wang FL, Senchenko VN, Protopopov A, Malyukova A, et al. RBSP3 (HYA22) is a tumor suppressor gene implicated in major epithelial malignancies. Proc Natl Acad Sci U S A. 2004;101:4906–4911. doi: 10.1073/pnas.0401238101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari G, Singhal PK, Rao MR, Mahalingam S. Nuclear Transport of Ras-associated Tumor Suppressor Proteins: Different Transport Receptor Binding Specificities for Arginine-rich Nuclear Targeting Signals. J Mol Biol. 2007 doi: 10.1016/j.jmb.2007.01.026. [DOI] [PubMed] [Google Scholar]
- Lambros MB, Fiegler H, Jones A, Gorman P, Roylance RR, Carter NP, et al. Analysis of ovarian cancer cell lines using array-based comparative genomic hybridization. J Pathol. 2005;205:29–40. doi: 10.1002/path.1681. [DOI] [PubMed] [Google Scholar]
- Lerman MI, Minna JD. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: Identification and evaluation of the resident candidate tumor suppressor genes. Cancer Res. 2000;60:6116–6133. [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- Park HW, Kang HC, Kim IJ, Jang SG, Kim K, Yoon HJ, et al. Correlation between hypermethylation of the RASSF2A promoter and K-ras/BRAF mutations in microsatellite-stable colorectal cancers. Int J Cancer. 2007;120:7–12. doi: 10.1002/ijc.22276. [DOI] [PubMed] [Google Scholar]
- Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Rommel C, Hafen E. Ras--a versatile cellular switch. Curr Opin Genet Dev. 1998;8:412–418. doi: 10.1016/s0959-437x(98)80111-1. [DOI] [PubMed] [Google Scholar]
- Schubbert S, Bollag G, Shannon K. Deregulated Ras signaling in developmental disorders: new tricks for an old dog. Curr Opin Genet Dev. 2007;17:15–22. doi: 10.1016/j.gde.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding Ras: ‘it ain’t over ‘til it’s over’. Trends Cell Biol. 2000;10:147–154. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- Tommasi S, Dammann R, Jin SG, Zhang XF, Avruch J, Pfeifer GP. RASSF3 and NORE1: identification and cloning of two human homologues of the putative tumor suppressor gene RASSF1. Oncogene. 2002;21:2713–2720. doi: 10.1038/sj.onc.1205365. [DOI] [PubMed] [Google Scholar]
- Tommasi S, Dammann R, Zhang ZQ, Wang Y, Liu LM, Tsark WM, et al. Tumor susceptibility of Rassf1a knockout mice. Cancer Res. 2005;65:92–98. [PubMed] [Google Scholar]
- van der Weyden L, Tachibana KK, Gonzalez MA, Adams DJ, Ng BL, Petty R, et al. The RASSF1A isoform of RASSF1 promotes microtubule stability and suppresses tumorigenesis. Mol Cell Biol. 2005;25:8356–8367. doi: 10.1128/MCB.25.18.8356-8367.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos MD, Ellis CA, Elam C, Ulku AS, Taylor BJ, Clark GJ. RASSF2 is a novel K-Ras-specific effector and potential tumor suppressor. J Biol Chem. 2003;278:28045–28051. doi: 10.1074/jbc.M300554200. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Damman R, Pfeifer GP. Hypermethylation of the CpG island of the RASSF1A gene in ovarian and renal cell carcinomas. Int J Cancer. 2001;94:212–217. doi: 10.1002/ijc.1466. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Sun D, Van do N, Tang A, Hu L, Huang G. Inactivation of RASSF2A by promoter methylation correlates with lymph node metastasis in nasopharyngeal carcinoma. Int J Cancer. 2007;120:32–38. doi: 10.1002/ijc.22185. [DOI] [PubMed] [Google Scholar]