Abstract
The new stipitate species of Hypocrea, H. seppoi; is described, based on material freshly collected in Finland. Characterization of this species includes morphology of teleomorph and anamorph, culture studies and molecular phylogeny. H. seppoi is compared to the other European soil-inhabiting, stipitate Hypocrea species. It is characterized by small stromata, thin fibrous stipe, perithecia decurrent on the stipe, and by conidia that are smaller than those of H. leucopus and H. nybergiana. In addition, this work confirms the congenerity of Podostroma and Podocrea with Hypocrea on molecular phylogenetic grounds using sequences of the nuclear genes coding for RNA polymerase II subunit b (rpb2), translation elongation factor 1 alpha (tef1), and endochitinase (chi15-8).
Keywords: chi18-5, endochitinase, Hypocreales, morphology, phylogenetic markers, rpb2, sequence analysis, tef1, Trichoderma
Introduction
Most species of Hypocrea Fr. including its type species H. rufa (Pers. : Fr.) Fr. (Jaklitsch et al. 2006b) form their asci containing disarticulating, 2-celled ascospores in pulvinate stromata only few mm in extent, a lesser number in large stromata several cm long, either in widely effused crustose stromata (Overton et al. 2006a, 2006b), or in upright, stipitate stromata (Chamberlain et al. 2004). The species belonging to the latter group had been classified in Podostroma P. Karst. or Podocrea (Sacc.) Lindau. Chamberlain et al. (2004) synonymized these generic names with Hypocrea, mainly based on anamorph morphology of H. alutacea (Pers.) Tul. & C. Tul. and H. leucopus (P. Karst.) H.L. Chamb., species originally described from Europe. The latter is the type species of Podostroma and was recognized as a species distinct from H. alutacea. Chamberlain et al. (2004) keyed out 17 species and added the new species H. nybergiana T. Ulvinen & H.L. Chamb., the third European species forming upright stromata.
As part of a project established to study the biodiversity of Hypocrea in Europe, a task force was installed in Finland in 2007 to collect stipitate Hypocrea species. In less than two months ten specimens were collected. Two of them turned out to represent an unknown species. This paper therefore serves to describe the new species and to confirm and show congenerity of Podostroma and Podocrea with Hypocrea with molecular phylogeny.
Materials and methods
Isolates and specimens
Single-ascospore isolates were prepared from fresh specimens of Hypocrea stromata and maintained as described by Jaklitsch et al. (2005, 2006a). Isolates investigated in this study, GenBank accession numbers for ITS, chi18-5, rpb2 and tef1 sequences and strain numbers of Hypocrea spp. used in this study are listed in Table 1. Isolates given as C.P.K. are those maintained in the collection of the Institute of Chemical Engineering, Vienna University of Technology. Representative isolates have been deposited at the Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands (CBS). Specimens including the type of Hypocrea seppoi were deposited in the Herbarium of the Institute of Botany, University of Vienna, Austria (WU).
Table 1.
Isolates and accession numbers for rpb2, tef1, and chi18-5 sequences determined in this study. Other sequences used in phylogenetic analyses are retrieved from GenBank (NCBI).
| Name | strain | rpb2 | tef1 | chi18-5 |
|---|---|---|---|---|
|
Hypocrea alutacea (Pers.: Fr.) Tul. & C.Tul. |
CBS 120535 | FJ179600 | FJ179567 | FJ179584 |
| Hypocrea alutacea | CBS 332.69 | FJ179601 | FJ179568 | FJ179585 |
| Hypocrea aureoviridis Plowr. & Cooke | CBS 120536 | FJ179602 | ||
| Hypocrea citrina (Pers.: Fr.) Fr. | C.P.K. 960 | FJ179603 | ||
| Hypocrea gelatinosa (Tode: Fr.) Fr. | C.P.K. 1618 | FJ179604 | FJ179569 | FJ179586 |
| Hypocrea leucopus (P. Karst.) H.L. Chamb. | CBS 122499 | FJ179605 | FJ179571 | FJ179588 |
| Hypocrea leucopus | CBS 122495 | FJ179606 | FJ179570 | FJ179587 |
| Hypocrea leucopus | C.P.K. 3527 | FJ179607 | FJ179572 | FJ179589 |
| Hypocrea lixii Pat. | C.P.K. 1934 | FJ179608 | FJ179573 | FJ179590 |
| Hypocrea lutea (Tode) Petch | CBS 121131 | FJ179609 | ||
|
Hypocrea minutispora B.S. Lu, Fallah & Samuels |
CBS 121276 | FJ179610 | FJ179574 | FJ179591 |
|
Hypocrea nybergiana T. Ulvinen & H.L. Chamb. |
CBS 122500 | FJ179611 | FJ179575 | |
| Hypocrea nybergiana | CBS 122496 | FJ179612 | FJ179576 | FJ179592 |
| Hypocrea pachybasioides Yoshim. Doi | C.P.K. 1989 | FJ179577 | FJ179593 | |
| Hypocrea pachybasioides | C.P.K. 2461 | FJ179613 | ||
|
Hypocrea parapilulifera B.S. Lu, Druzhin. & Samuels |
CBS 120921 | FJ179614 | FJ179578 | FJ179594 |
| Hypocrea pilulifera J. Webster & Rifai | CBS 120927 | FJ179615 | ||
| Hypocrea pilulifera | C.P.K. 3143 | FJ179579 | FJ179595 | |
| Hypocrea placentula Grove | CBS 120924 | FJ179616 | FJ179580 | FJ179596 |
| Hypocrea rufa (Pers.: Fr.) Fr. | CBS 119325 | EU711362 | ||
| Hypocrea seppoi sp. nov. | CBS 122498 | FJ179617 | FJ179581 | FJ179597 |
| Hypocrea seppoi sp. nov. | CBS 122497 | FJ179618 | FJ179582 | FJ179598 |
| Hypocrea sinuosa P. Chaverri & Samuels | C.P.K. 1595 | FJ179619 | ||
|
Hypocrea strictipilosa P. Chaverri & Samuels |
C.P.K. 3135 | FJ179583 | FJ179599 | |
| Hypocrea sulphurea (Schwein.) Sacc. | CBS 119929 | FJ179620 | ||
| Hypocrea viridescens Jaklitsch & Samuels | C.P.K. 2046 | FJ179621 | ||
| Hypocrea voglmayrii Jaklitsch | CBS 117711 | FJ179622 |
Growth characterization
Culture morphology and growth rates were determined in a single experiment under the conditions defined in Jaklitsch et al. (2005, 2006a).
Morphological observations
The methods described by Jaklitsch et al. (2005, 2006a) were used. Quantitative measurements are either reported as maxima and minima in parentheses and the mean plus and minus the standard deviation of a number of measurements given in parentheses, or as minimum–maximum ranges. Colours were determined and cited according to Kornerup and Wanscher (1981).
DNA extraction, PCR amplifications and sequencing
All methods used are defined in Jaklitsch et al. (2005, 2006a), except the following modifications and additions: PCR products were either purified using the QIAquick Kit (Qiagen) according to the manufacturer's instructions or using an enzymatic PCR cleanup (Werle et al. 1994). For the latter, 20 μL PCR reactions were digested with 10 units Exonuclease I (Fermentas, St. Leon-Rot, BRD) and 2 units Calf Intestine Alkaline Phosphatase (Fermentas) for 45 min at 37 C, followed by an enzyme deactivation step at 85°C for 15 min. DNA was cycle-sequenced using the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit v. 3.1 (Applied Biosystems, Warrington) and an automated DNA sequencer (ABI Genetic Analyzers, Applied Biosystems) with the same primers as in PCR or with the internal primers 5′-CCGTGA(T/C)TTCATCAAGAACATG-3′ and 5′-TTGGCAGTGTCCATCTTGTTG-3′ for tef1, 5′-GATCAA(C/T)(A/C)GAGG(C/T)ATGGA (G/A)GT-3′ and 5′-GA(G/A)AAGA TTTT(G/A)AA(C/T)TC(C/T)TG(G/A)T-3′ for rpb2, or 5′-CT(C/T)GACTA(T/C)(G/A)T(C/T)AACCTNATGGC-3′ and 5′-(C/A)(G/A)GGAAC (A/G)CCTCCNTTGAT(A/G)TA-3′ for chi18-5.
Molecular phylogenetic analyses
Sequences of rpb2 and tef1 were subjected to TrichoBLAST (Kopchinskiy et al. 2005), a Hypocrea/Trichoderma similarity search tool implemented on www.ISTH.info. Corresponding sequences of related species were selected for two phylogenetic approaches. To show the phylogenetic placement of the stipitate species within Hypocrea, an extensive rpb2 matrix was produced, including sequences from representative members of major Hypocrea lineages and from the two outgroup species Protocrea farinosa (Berk. & Broome) Petch and Hypomyces stephanomatis Rogerson & Samuels. For a detailed phylogenetic analysis, a combined dataset of rpb2, tef1 and chi15-8 sequences from the closest relatives of stipitate Hypocrea species was prepared. All alignments were produced with Muscle version 3.6 (Edgar 2004). After the exclusion of leading and trailing gap regions, 1033 characters of the rpb2 alignment, 1374 characters of the tef1 alignment and 823 characters of the chi15-8 alignment were included in the subsequent phylogenetic analyses. The resulting alignments were checked and refined using BioEdit version 7.0.4.1 (Hall 1999).
Maximum parsimony (MP) analyses of the rpb2 and the combined DNA matrices were performed with PAUP* version 4.0 b10 (Swofford 2002), using 1000 replicates of heuristic search with random addition of sequences and subsequent TBR branch swapping (MULTREES option in effect, COLLAPSE=MAXBRLEN, steepest descent option not in effect). All molecular characters were unordered and given equal weight; analyses were performed with gaps treated as missing data. MP bootstrap analysis with 1000 replicates was performed in the same way, but using 10 rounds of random sequence addition and subsequent TBR branch swapping during each bootstrap replicate.
The Bayesian approach to phylogenetic reconstructions (Rannala & Yang 1996, Yang & Rannala 1997) was implemented with MrBayes 3.1.2 (Huelsenbeck & Ronquist 2001). Modeltest 3.6 (Posada & Crandall 1998) was used to compare the likelihood of different nested models of DNA substitution and to select the best-fit model for the investigated data set. Three parallel runs of four incrementally heated simultaneous Markov chains were performed over 1 million generations from which every 100th tree was sampled in each run. The trees before apparent stationary probability distribution of the cold chain were discarded. A 90 % majority rule consensus of the remaining trees was computed to obtain estimates for the probabilities that groups are monophyletic given the sequence data (posterior probabilities). According to the protocol of Leache and Reeder (2002), posterior probabilities (PP) lower then 95 % were not considered significant, while values below 90 % are not shown on phylograms.
Results
Phylogeny
Sequences of four genes used for identification (ITS) or phylogenetic reconstruction (rpb2, tef1, chi18-5) were determined for all stipitate species of Hypocrea occurring in Europe. The species differ in all four genes, whereas different isolates of the same species had highly similar or identical sequences. Identifications using TrichOKey (Druzhinina et al. 2005) on the ISTH webpage failed, because the respective ITS sequences are not available in the respective database yet.
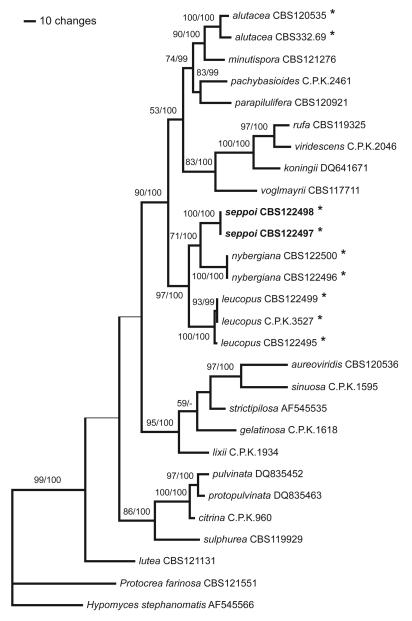
Of the 1033 characters of the extended rpb2 alignment included in the analyses, 325 were parsimony informative. MP analyses revealed three trees of length 1262, one of which was selected as Fig. 1. For Bayesian analyses, the general time reversible model, additionally assuming a proportion of invariant sites with gamma-distributed substitution rates of the remaining sites (GTR+I+G), was selected by Modeltest under the Akaike Information Criterion. The three Bayesian runs revealed almost identical posterior probabilities (PP). The Bayesian trees had topologies identical to the MP strict consensus tree. MP bootstrap values above 50 % and Bayesian posterior probabilities above 90 %, respectively, are indicated in Fig. 1 above/below the branches.
Fig. 1.
One of 3 phylograms of length 1262 revealed by a maximum parsimony (MP) analysis of 1033 characters of the rpb2 alignment. The thin horizontal branches denote the nodes collapsing in the MP strict consensus tree. MP bootstrap values above 50 % and Bayesian posterior probabilities above 90 %, respectively, are indicated above/below the branches. The stipitate Hypocrea species are denoted by an asterisk (*). The numbers after the taxon names are strain numbers, or accession numbers for sequences retrieved from GenBank.
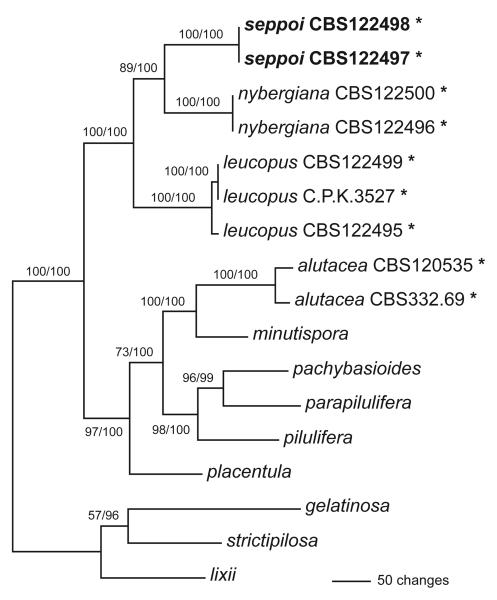
Of the 3230 characters of the combined rpb2 – tef1 – chi15–8 alignment included in the analyses, 734 were parsimony informative. MP analyses revealed one tree of length 1946 (Fig. 2). Using Modeltest, the TIM+I+G model was selected under the Akaike Information Criterion; however, as this model cannot be implemented in Mr-Bayes 3.1.2, the GTR+I+G model was used for the Bayesian analyses. The three Bayesian runs revealed almost identical posterior probabilities (PP). The Bayesian and MP trees had identical topologies. MP bootstrap values above 50 % and Bayesian probabilities above 90 %, respectively, are indicated in Fig. 2 above/below the branches.
Fig. 2.
Phylogram showing the single most parsimonious tree of length 1946 revealed by a MP analysis of 3230 characters of the combined rpb2 – tef1 – chi15–8 alignment. MP bootstrap values above 50 % and Bayesian posterior probabilities above 90 % respectively, are indicated above/below the branches. The tree was rooted according to the topology of Fig. 1. The stipitate Hypocrea species are denoted by an asterisk (*).The numbers after the names of the stipitate Hypocrea species are strain numbers; for information on other species see Table 1.
The analyses of the extended rpb2 and the combined datasets revealed fully compatible trees. The trees clearly show a close relationship of H. alutacea with H. minutispora B.S. Lu, Fallah & Samuels, while H. seppoi, H. leucopus and H. nybergiana form a distinct, highly supported clade not closely related to H. alutacea. Sister group relationship between H. seppoi and H. nybergiana was moderately and highly supported by MP bootstrap and Bayesian analyses, respectively (Figs. 1 and 2). As can be also seen on the phylograms, the status of H. seppoi, H. nybergiana and H. leucopus as genetically distinct species is clearly shown by the molecular data.
Taxonomy
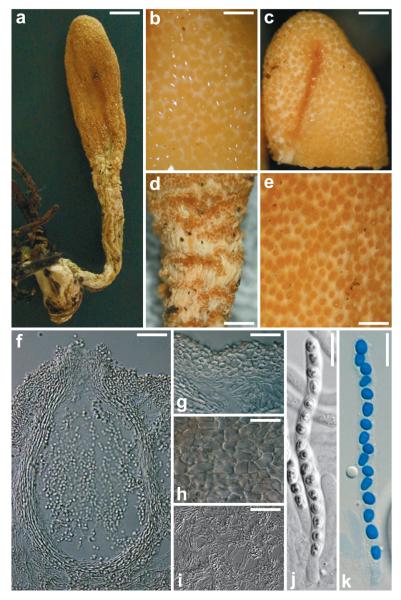
Hypocrea seppoi W. M. Jaklitsch, sp. nov. Fig. 3
Fig. 3.
Teleomorph of Hypocrea seppoi. a) Dry stroma. b) Stroma surface in 3% KOH. c) Fertile part of stroma after reconstitution with water. d) Groups of perithecia on stipe of stroma. e) Stroma surface after reconstitution with water. f) Perithecium in section. g) Cortical and subcortical tissue in section. h) Stroma surface in face view. i) Subperithecial tissue in section. j, k) Asci with ascospores (j in 3% KOH, k in cotton blue/lactic acid). a, d, e, h from WU 28698. b, c, f, g, i, j, k from WU 28699. Scale bars: a = 2.5 mm, b, e = 0.3 mm, c = 0.6 mm, d = 0.8 mm, f, g, i = 30 μm, h, j, k = 10 μm.
Anamorph
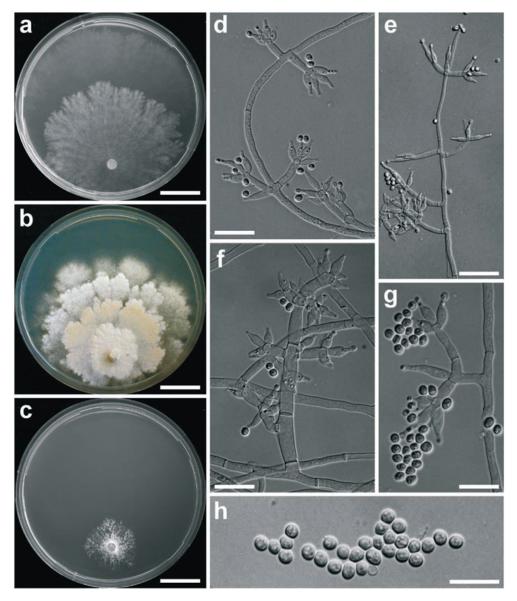
Trichoderma seppoi W. M. Jaklitsch, sp. nov. Fig. 4
Fig. 4.
Cultures and anamorph of Hypocrea seppoi. a) Culture on CMD (21 d, 25°C). b) Culture on PDA (21 d, 25°C). c) Culture on SNA (21 d, 15°C). d–g) Conidiophores with phialides on SNA (18 d, 15°C). h) Conidia (SNA, 18 d, 15°C). a, b, c from WU 28698. d–h from WU 28699. Scale bars: a, b, c = 19 mm, d, e = 20 μm, f = 15 μm, g, h = 10 μm.
Differt a Hypocrea leucopus et a H. nybergiana stromatibus minoribus, conidiis minoribus et conidiophoribus irregulariter verticillatis, a H. nybergiana etiam ascosporis minoribus et a H. leucopus peritheciis stipite decurrentibus.
Anamorphosis Trichoderma seppoi. Conidiophora effusa, irregulariter verticillata, phialides (5−)6−11(−18) × (2.5−)2.8−3.6(−4.0) μm, lageniformes. Conidia ellipsoidea vel subglobosa, hyalina, glabra, (2.5−)3.0−3.5(−4.0) × (2.2−)2.5−2.8(−3.2) μm in agaro ‘SNA’.
HOLOTYPE
Finland, Pohjois-Savo, Heinävesi, close to Hernelampi, grid 27°E 6925:586, on soil in a grass-rich spruce forest slope, 19 Sep 2007 Stig Jakobsson 4741 (WU 28698; culture CBS 122498 = C.P.K. 3161).
Stromata not seen in fresh condition, reported by the collectors to have a pale yellow colour like H. leucopus. Stromata when dry 8–24 mm long; fertile part 3–12 mm long, 1.5–4.5 mm wide and 0.5–3 mm thick; stipe 5–13 mm long, 1–3 mm wide and 0.3–2 mm thick, base slightly thickened, 1.2–3 mm wide (n=4). Fertile part clavate to spathulate, distinctly laterally compressed or longitudinally furrowed or folded, gradually tapered downwards. Stroma surface white, smooth, largely concealed by numerous, densely disposed, convex to papillate, ochre ostiolar/perithecial dots (30−)50−110(−165) μm diam (n=60), giving the surface a finely roughened appearance. Colour yellow-ochre 5AB4, 5B5, 6B4. Stipe thin, cylindrical, fibrous to delicately longitudinally striate, slightly compressed, off-white to creamochre, straight or strongly curved, also twisted around its axis. Stipe surface dotted by scattered or aggregated perithecia decurrent nearly its whole length. Base not or slightly thickened, typically bearing needles of Picea colonized by brown rhizomorphs. Spore deposits on stroma surface fine, white. After reconstitution in water stromata somewhat larger than dry, lighter, light yellow-ochre, stroma white, perithecia yellow. No change or slightly lighter after addition of 3% KOH. Entostroma white.
Stroma anatomy
Ostioles (43−)53−70(−75) μm long, plane or projecting to 30(−47) μm, (30−)45−65(−85) μm wide at apex (n=30), cylindrical, periphysate, apex widened; apical cells palisadic, cylindrical to clavate, to ca 5 μm wide. Perithecia (125−)180−250(−280) × (70−)100−175(−220) μm (n=30), crowded, mostly laterally compressed, flask-shaped, ellipsoidal or subglobose. Peridium (11−)15−19(−20) μm wide at base, (9−)12−16(−18) μm (n=30) at sides, hyaline to pale yellowish, of compressed, elongated, thick-walled, refractive cells, isodiametric around ostioles. Cortical layer (16−)20−30(−36) μm wide (n=30), a subhyaline to pale yellow t. angularis of cells (4−)6−14(−18) × (4−)5−9(−12) μm in face view and (2.5−)4−10(−16) × (2−)3−6(−8) μm (n=30) in vertical section; cells isodiametric to oblong, similar to peridial cells, but smaller and thin-walled. Subcortical tissue absent or a loose hyaline t. intricata of thin-walled hyphae (2−)3−5(−6) μm (n=35) wide. Subperithecial tissue a loose hyaline t. intricata of thin-walled hyphae (2−)3−5(−7) μm (n=30) wide, often collapsed, with variable orientation, therefore in part appearing as irregular t. epidermoidea upon strong magnification. Asci (62−)68−84(−87) × 4.0−4.5(−5.0) μm, including a stipe (6−)10−25(−30) μm (n=20) long, base thickened, apex thickened to 1–1.5 μm and with a minute, inconspicuous ring. Ascospores hyaline, finely verruculose, with 1 or several guttules per cell, cells dimorphic, distal cell (2.7−)3.0−3.5(−4.0) × (2.5−)2.7−3.2(−3.5) (n=70), l/w (0.9−)1−1.2(−1.5) (n=70), (sub)globose to nearly wedge-shaped, proximal cell (3.0−)3.5−4.5(−5.5) × (2.0−)2.3−2.7(−3.0) (n=70), l/w (1.2−)1.4−2.0(−2.4) (n=70) oblong to wedge-shaped; sometimes inverted inside asci.
Cultures and anamorph (growth rate determined in a single experiment): optimum growth at 25°C on PDA and SNA, on CMD at 30°C; no growth at 35°C.
On CMD 3 mm at 15°C, 6–7 mm at 25°C, 8–9 mm at 30°C after 72 h; mycelium covering plate after ca 3 wk at 25°C. Colony hyaline, thin, in-distinctly zonate, margin of central zone irregularly lobed, broad marginal zone distinctly separated, growing faster; surface becoming slightly farinose by white conidiation; mycelium dense, hyphae narrow. Aerial hyphae none to inconspicuous. No autolytic excretions, no coilings seen, no pigment, no odour, no chlamydospores noted. Conidiation noted after 3 d at 25°C, effuse, starting around plug, spreading across plate; conidiophores short, ca 30–60 μm long, with 1–2 branching levels; phialides solitary or in whorls of 2–6, straight or curved to sinuous, strongly inclined upwards. Conidia formed in small numbers in variable wet heads, ellipsoidal(-subglobose-oblong), (3−)3.3−4(−4.6) × (2−)2.5−3.0(−3.7), l/w (1.1−)1.2−1.4(−1.5) (n=30), hyaline, smooth, with some fine guttules, scar indistinct.
On PDA 1 mm at 15°C, 7–8 mm at 25°C, 1–1.5 mm at 30°C after 72 h; mycelium covering plate after ca 4 wk at 25°C. Colony dense, of several irregularly lobed zones, surface flat, farinose, mottled, white to cream, reverse in central areas becoming yellowish to light brown 5CD5–6. Aerial hyphae inconspicuous, short, becoming fertile. No autolytic excretions, no coilings noted. Odour none to slightly mushroomy. Conidiation noted after 3 d at 25°C, effuse, spreading from plug, dense, short, white, irregularly Verticillium-like. At 30°C little growth, no conidiation seen.
On SNA 1 mm at 15°C, 2 mm at 25 and 30°C after 72 h. Colony irregularly lobed, radial, white farinose streaks formed, hyphae narrow, forming pegs. No autolytic excretions, no coilings, no pigment, no odour, no chlamydospores noted. Conidiation noted after 9 d at 25°C, effuse, on short conidiophores, irregularly Verticillium-like, particularly in streaks, numerous drops appearing. At 30°C dense white mycelium, little growth, conidiation effuse. At 15°C colony circular, hyaline, dense, narrow, white, farinose ring formed around plug. Conidiation effuse, better developed than at 25°C, noted after 9 d, examined after 18 d. Conidiophores in dense lawns and erect on surface hyphae and in right angles (paired or unpaired) on aerial hyphae; simple, short, 20−60(−150) μm long, 2−5 (−7) μm wide, with some thickenings to 9.5 μm wide, 1–3 celled, with 0–4 branchings, branches 1(−2) celled, right-angled or inclined slightly upwards, mostly paired, often thickened in the middle. Phialides formed on cells 3–5 μm wide, solitary or in whorls of 2–6, often inclined upwards in steep angles, sometimes nearly cruciform. Conidia formed mostly in dry heads <10 μm diam and in fewer wet heads <40 μm diam. Phialides (5−)6−11(−18) × (2.5−)2.8−3.6(−4) μm, l/w (1.4−)1.8−3.5(−7.3), (1.3−)1.7−2.5(−3.0) μm (n=63) wide at base; lageniform, mostly symmetrical and with long, abruptly attenuated narrow tip, also base often thin; straight, some strongly curved, generally strongly thickened in or below middle; solitary phialides often much longer (>11 μm) and nearly subulate. Conidia (2.5−)3.0−3.5(−4) × (2.2−)2.5−2.8(−3.2) μm, l/w 1.1−1.3(−1.5) (n=63), hyaline, smooth, subglobose to ellipsoidal, with 1 to few guttules, scar indistinct.
Habitat: on the ground in Picea dominated forests.
Distribution: Finland, only known from type locality.
Etymology: named after the Finnish mycologist Seppo Huhtinen, who established a ‘Podostroma task force’ in 2007 to collect stipitate species of Hypocrea in Finland.
Other specimen examined: ca. 100 m from type location, grid 27°E 6925:587, on soil in mossy and old spruce-dominated forest, 19 Sep 2007, Mauri Lahti (WU 28699; culture CBS 122497 = C.P.K. 3162).
Notes: This species forms the smallest stromata (maximum length 2.5 cm) of the stipitate Hypocrea species found in Europe. The colour of the fertile part of dry stromata is between the lighter yellow H. leucopus and the ‘reddish-brown’ H. nybergiana, being closer to the latter. Also the decurrent perithecia on the stipe of stromata are shared with H. nybergiana. However, the latter species has slightly larger ascospores, while H. leucopus cannot be differentiated from H. seppoi by ascospore characters.
The conidiophores of Trichoderma seppoi are not as regularly verticillate as in the anamorph of H. leucopus; the phialides are wider and shorter, and the conidia tend to be subglobose, smaller-sized than in both H. leucopus and H. nybergiana.
Key to the stipitate species of Hypocrea in Europe.
1. Stromata on wood and bark, clavate to irregular, fertile part yellowish, anamorph green-conidial ...............................................................................................H. alutacea
1. Stromata on the ground, typically in leaf litter of Picea, anamorphs white-conidial .................... 2
2. Stromata large, fertile part reddish-brown to brownish orange, pigment inhomogeneously distributed, distal ascospore cell (3.0−)3.5−4.0(−5.5) × 3.0−3.5(−4.2) μm, proximal ascospore cell (3.0−)4.0−5.0(−5.5) × (2.2−)2.7−3.0(−4.0) μm .................................. H. nybergiana
2. Stromata small or medium-sized, fertile part pale yellowish when fresh, distal ascospore cell (2.7−)3.0−3.5(−4.0) × (2.5−)2.7−3.2(−3.5) μm, proximal ascospore cell (3.0−)3.5−4.5(−5.5) × (2.0−)2.3−2.7(−3.0) μm ....................................................... 3
3. Stromata medium-sized, colour not changing upon drying, perithecia not decurrent on stipe, anamorph Verticillium-like, conidia 3.0−5.5(−8.0) μm long on CMD, phialide length on SNA (8−)11−17(−23) μm ................................................................ H. leucopus
3. Stromata small, to 2.5 cm long, colour changing to ochre upon drying, solitary or grouped perithecia decurrent on stipe, anamorph irregularly Verticillium-like, conidia (3.0−)3.3−4.0(−4.5) μm long on CMD, phialide length on SNA (5−)6−11(−18) μm ............................................................................................... H. seppoi
Discussion
Molecular phylogeny based on rpb2 sequences (Fig. 1) clearly shows that the type species of Podostroma, P. leucopus P. Karst., and the type species of Podocrea, P. alutacea (Pers.) Lindau, are unequivocal members of the genus Hypocrea. Consequently the synonymization of Podocrea and Podostroma with Hypocrea by Chamberlain et al. (2004) is correct. Thus the the application of the stipitate condition of stromata as a generic character is artificial and no reason to recognize and segregate different genera.
Surprisingly, the wood-inhabiting H. alutacea is closely related to H. minutispora (Lu et al. 2004), a common species forming pulvinate stromata. Not only the morphology of stromata of these species is quite different, but also the Pachybasium structure (Bissett 1991) of T. minutisporum Bissett differs from the more slender conidiophores and narrower/longer phialides in H. alutacea. Only the similarly pale green colour of conidia and the tendency of not forming compact pustules on CMD is comparable (data not shown).
On the other hand, the three stipitate European species that occur on soil and forest debris form a clade of their own in a sister relationship to a large clade comprising the Pachybasioides Clade (cf. Jaklitsch et al. 2006a) and section Trichoderma (Rufa Clade plus section Pachybasium A according to Jaklitsch et al. 2006a), including H. voglmayrii.
To gain further information on the distributional stability of the stipitate species two additional loci were analyzed after addition of data from other species (H. pilulifera J. Webster & Rifai, H. placentula Grove) of the Pachybasioides Clade. One of these loci is tef1 (a long fragment including introns 4 and 5 and a part of exon 6), of which intron 4 is currently used for high resolution in the phylogeny of Hypocrea/Trichoderma on the specific level. Individual trees computed from tef1 and chi18-5 sequences (not shown) resulted in identical topologies, that are also compatible with the rpb2 tree. Accordingly, the requirements of the GCPSR concept (Taylor et al. 2000) are met for all stipitate species included in the phylogeny. Subsequently a combined tree was calculated from the data of all three genes (Fig. 2). This tree confirms the conclusions provided by the rpb2 tree, i.e. that H. alutacea is closely related to H. minutispora and that H. leucopus, H. nybergiana and H. seppoi are well-supported species in a distinct clade.
Also morphology corroborates the phylogeny-driven view that the stipitate species including H. seppoi are unequivocally well-defined and recognizable. Apart from the occurrence on wood and bark, H. alutacea differs from the soil-dwelling species by forming an anamorph with green conidia, while the latter all produce white conidia in culture. H. nybergiana forms large conidia on solitary phialides (unpublished results), while H. leucopus produces effuse Verticillium-like conidiophores. The anamorph of H. seppoi resembles that of H. leucopus, but the structure deviates somewhat from the Verticillium-like type by more irregular branching, an increased number of unpaired branches, a more irregular arrangement of phialides; the phialides are wider and shorter, and the conidia are smaller and rather subglobose than ellipsoidal.
However, the description of H. seppoi is solely based on two specimens collected on the same day at two sites with only ca. 100 m distance from each other. Therefore it remains to be assessed by additional collections whether some morphological characters emphasized in the notes to the description of H. seppoi, and implemented in the key to species, are stable, e.g. stroma size, the decurrent perithecia on the stipe or the darkening of stromata upon drying. At present the most reliable way to distinguish H. seppoi from H. leucopus is a direct microscopic comparison of the anamorphs of the two species in culture, particularly on SNA.
Acknowledgments
We thank Seppo Huhtinen for collecting and organizing collection of ‘Podostromas’, Stig Jakobsson and Mauri Lahti for collecting the specimens of H. seppoi. The financial support by the Austrian Science Fund (FWF Project P19143-B17) to WMJ is gratefully acknowledged.
References
- Bissett J. A revision of the genus Trichoderma. III. Section Pachybasium. Canad. J. Bot. 1991;69:2373–2417. [Google Scholar]
- Chamberlain HL, Rossman AY, Stewart EL, Ulvinen T, Samuels GJ. The stipitate species of Hypocrea (Hypocreales, Hypocreaceae) including Podostroma. Karstenia. 2004;44:1–24. [Google Scholar]
- Druzhinina IS, Kopchinskiy AG, Komon M, Bissett J, Szakacs G, Kubicek CP. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet. Biol. 2005;42:813–828. doi: 10.1016/j.fgb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jaklitsch WM, Komon M, Kubicek CP, Druzhinina IS. Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea / Trichoderma. Mycologia. 2005;97:1365–1378. doi: 10.3852/mycologia.97.6.1365. [DOI] [PubMed] [Google Scholar]
- Jaklitsch WM, Komon M, Kubicek CP, Druzhinina IS. Hypocrea crystalligena sp. nov., a common European species with a white-spored Trichoderma anamorph. Mycologia. 2006a;98:499–513. doi: 10.3852/mycologia.98.3.499. [DOI] [PubMed] [Google Scholar]
- Jaklitsch WM, Samuels GJ, Dodd SL, Lu B-S, Druzhinina IS. Hypocrea rufa / Trichoderma viride: a reassessment, and description of five closely related species with and without warted conidia. Stud. Mycol. 2006b;56:135–177. doi: 10.3114/sim.2006.56.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopchinskiy AG, Komon M, Kubicek CP, Druzhinina IS. TrichoBLAST: A multilocus database for Trichoderma and Hypocrea identifications. Mycol. Res. 2005;109:657–660. doi: 10.1017/s0953756205233397. [DOI] [PubMed] [Google Scholar]
- Kornerup A, Wanscher JH. Taschenlexikon der Farben. Zürich & Göttingen; Muster-Schmidt Verlag: 1981. [Google Scholar]
- Leache AD, Reeder TW. Molecular systematics of the eastern fence lizard Sceloporus undulatus: A comparison of parsimony, likelihood, and Bayesian approaches. Syst. Biol. 2002;51:44–68. doi: 10.1080/106351502753475871. [DOI] [PubMed] [Google Scholar]
- Lu B-S, Druzhinina IS, Fallah P, Chaverri P, Gradinger C, Kubicek CP, Samuels GJ. Hypocrea /Trichoderma species with pachybasium-like conidiophores: teleomorphs for T. minutisporum and T. polysporum and their newly discovered relatives. Mycologia. 2004;96:310–342. [PubMed] [Google Scholar]
- Overton BE, Stewart EL, Geiser DM, Jaklitsch WM. Systematics of Hypocrea citrina and related taxa. Stud. Mycol. 2006a;56:1–38. doi: 10.3114/sim.2006.56.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overton BE, Stewart EL, Geiser DM. Taxonomy and phylogenetic relationships of nine species of Hypocrea with anamorphs assignable to Trichoderma section Hypocreanum. Stud. Mycol. 2006b;56:39–65. doi: 10.3114/sim.2006.56.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rannala B, Yang Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996;43:304–311. doi: 10.1007/BF02338839. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sinauer Associates; Sunderland, MA: 2002. [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC. Phylogenetic species recognition and species concepts in Fungi. Fungal Genet. Biol. 2000;31:21–31. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- Werle E, Schneider C, Renner M, Völker M, Fiehn W. Convenient single-step, one tube purification of PCR products for direct sequencing. Nucleic Acids Res. 1994;22:4354–4355. doi: 10.1093/nar/22.20.4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Bayesian phylogenetic inference using DNA sequences: a Markov chain Monte Carlo method. Mol. Biol. Evol. 1997;14:717–724. doi: 10.1093/oxfordjournals.molbev.a025811. [DOI] [PubMed] [Google Scholar]