SUMMARY
Eukaryotic protein synthesis begins with assembly of 48S initiation complexes at the initiation codon of mRNA, which requires at least 7 initiation factors (eIFs). First, 43S preinitiation complexes comprising 40S ribosomal subunits, eIFs 3, 2, 1, 1A and tRNAMeti attach to the 5′-proximal region of mRNA, and then scan along the 5′-untranslated region (5′-UTR) to the initiation codon. Attachment of 43S complexes is mediated by three other eIFs: 4F, 4A and 4B, which cooperatively unwind the cap-proximal region of mRNA, and later also assist 43S complexes during scanning. We now report that these 7 eIFs are not sufficient for efficient 48S complex formation on mRNAs with highly structured 5′-UTRs, and that this process requires the DExH-box protein DHX29. DHX29 binds 40S subunits and hydrolyzes ATP, GTP, UTP and CTP. NTP hydrolysis by DHX29 is strongly stimulated by 43S complexes, and is required for DHX29’s activity in promoting 48S complex formation.
Keywords: ribosome, translation initiation, DHX29, structured 5′-UTR, scanning
INTRODUCTION
Eukaryotic protein synthesis begins with assembly of 48S initiation complexes, in which initiator tRNA (Met-tRNAMeti) is base-paired with the initiation codon of mRNA in the P site of the 40S subunit. 48S complex formation on most cellular mRNAs occurs by the scanning mechanism and requires at least seven eIFs (Pestova et al., 2007). First, 43S complexes comprising 40S subunits, eIF2/GTP/Met-tRNAMeti ternary complexes (TCs), eIF3, eIF1 and eIF1A attach to the 5′-proximal region of mRNA and then scan along the 5′-untranslated region (5′-UTR) to the initiation codon where they stop, forming 48S complexes.
Attachment of 43S complexes to mRNA is mediated by eIFs 4F, 4A and 4B. eIF4F comprises eIF4E (cap-binding protein), eIF4A (a DEAD-box RNA helicase, whose activity is enhanced by eIF4G and eIF4B) and eIF4G (which binds eIF4E, eIF4A and also eIF3). eIF4F/4A/4B cooperatively unwind the cap-proximal region of mRNA allowing 43S complexes to bind, and likely promote binding via the eIF4G-eIF3 interaction. The molecular mechanism by which mRNA enters the mRNA-binding cleft of the 40S subunit (e.g. by threading through this entire channel starting from its entrance, or by direct placement of the cap-proximal mRNA segment into the mRNA-binding cleft) is unknown.
Ribosomal scanning consists of two linked processes: unwinding of secondary structure in the 5′-UTR and ribosomal movement along it. During scanning, 43S complexes must be able to reject potential mis-matches between the Met-tRNAMeti and non- and near-cognate codons, but also to recognize the correct initiation triplet. The key role in ensuring accurate initiation codon selection belongs to eIF1, which enables 43S complexes to discriminate against 48S complex formation on non-AUG triplets and on AUG triplets in suboptimal context (Pestova and Kolupaeva, 2002; Pisarev et al., 2006). eIF1 binds to the interface surface of the 40S subunit between the platform and Met-tRNAMeti (Lomakin et al., 2003), and it has been suggested that it performs its monitoring function indirectly, by influencing the conformation of ribosomal complexes. Consistently, binding of eIF1 and eIF1A to yeast 40S subunits induces conformational changes that consist of opening of the entry channel ‘latch’ formed between helix (h) 18 in the body and h34 and ribosomal protein (rp) S3 in the neck, and establishment of a new head-body connection likely mediated by h16 and rpS3 (Passmore et al., 2007). But what is the role of different factors in ribosomal movement per se? 43S complexes containing TCs, eIF3, eIF1 and eIF1A can bind to the 5′ end of an unstructured 5′-UTR and scan to the initiation codon without ATP or factors associated with ATP hydrolysis and RNA unwinding, revealing the intrinsic ability of 43S complexes to move along mRNA (Pestova and Kolupaeva, 2002). Importantly, omission of eIF1A greatly reduces the ability of 43S complexes to form 48S complexes in the absence of eIF4A/4G/4B, and omission of eIF1 almost abrogates it. Although eIF3 is indispensable for 48S complex formation, it is difficult to separate its role in scanning from functions in recruitment of TCs to 40S subunits and initial attachment of 43S complexes. Scanning on 5′-UTRs containing even weak internal secondary structure, on the other hand, requires ATP and eIF4A/4G/4B, and the requirement for ATP and eIF4A is proportional to the degree of secondary structure in the 5′-UTR (Pestova and Kolupaeva, 2002; Jackson, 1991; Svitkin et al., 2001). Continued association of eIF4G with ribosomal complexes (Pöyry et al. 2004) ensures eIF4A’s processivity and couples mRNA unwinding with ribosomal movement. It is unknown whether eIF4F/4A/4B bind at the 40S subunit’s leading edge and unwind mRNA before it enters the mRNA-binding cleft, or at the trailing edge near the E-site and assist scanning by helicase-mediated “pulling” of mRNA through the mRNA-binding channel and/or preventing backward movement. eIF4A/4G/4B also stimulate 48S complex formation on mRNAs with unstructured 5′-UTRs and make this process less dependent on eIF1/1A (Pestova and Kolupaeva, 2002). Thus, it is clear that scanning requires ATP-dependent unwinding of RNA secondary structure by eIF4A/4G/4B and induction by eIF1/1A of the scanning-competent conformation of 43S complexes.
The efficiency of translation of different mRNAs in vivo and in vitro depends on the degree of secondary structure in their 5′-UTRs. Thus, stems of ΔG=−30 kcal/mol located distally in the 5′-UTR inhibit but do not abolish initiation, whereas stems of ΔG=−60 kcal/mol abrogate translation by obstructing ribosomal movement to the initiation codon (Kozak, 1991).
In our in vitro reconstituted initiation system containing eIF2/3/1/1A/4A/4B/4F, 48S complexes did not form efficiently on mRNAs containing GC-rich stems of even moderate stability in their 5′-UTRs although they are translated well in cell-free extracts (Pestova and Kolupaeva, 2002). Moreover, during 48S complex formation on β-globin mRNA, additional toe-prints appear +8-9 nt from the AUG codon, equal to as much as 30-40% of the +15-17 nt toe-print of properly assembled 48S complexes, that most likely represent an initiation complex, in which the 3′-portion of mRNA was not properly fixed in the 40S subunit’s mRNA-binding cleft (Battiste et al., 2000). Such aberrant toe-prints do not appear when 48S complexes are assembled in cell-free translation extracts. Here, we report that we have purified and identified the DExH-box protein DHX29 as a factor that is required for efficient 48S complex formation on mRNAs with highly structured 5′-UTRs and that also suppresses the aberrant +8-9 nt toe-print.
RESULTS
Efficient 48S complex formation on mRNAs with structured 5′-UTRs requires DExH-box protein DHX29
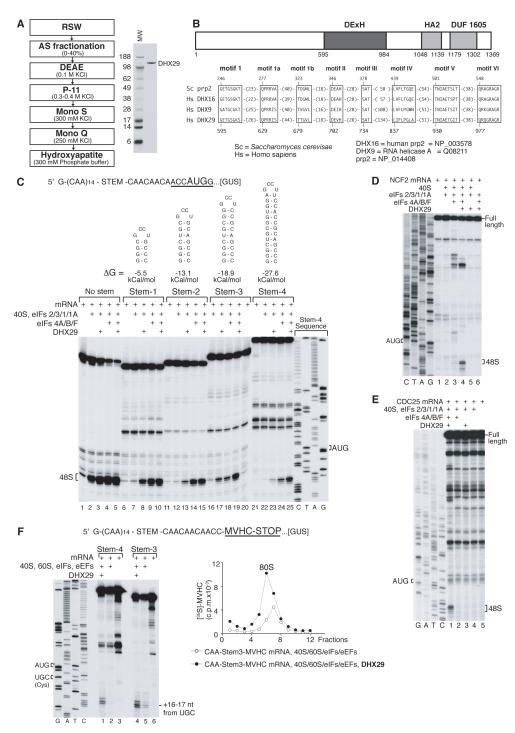
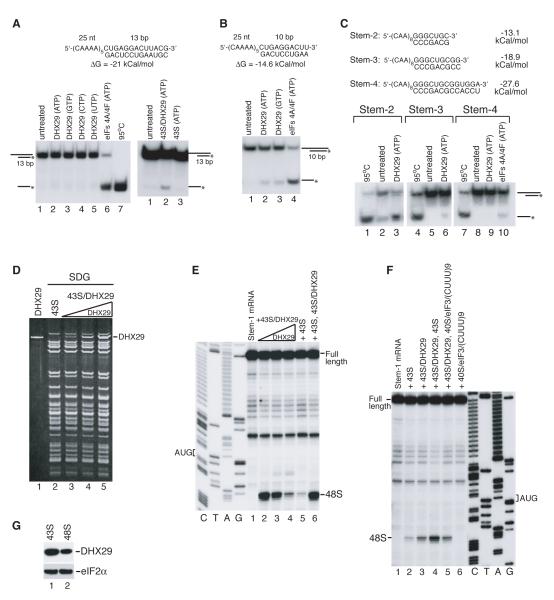
Although in an in vitro reconstituted system, eIF2/3/1/1A/4A/4B/4F promoted efficient 48S complex formation on model synthetic mRNAs comprising the β-glucuronidase (GUS) coding region and an unstructured 5′-UTR consisting of 19 CAA repeats (CAA-GUS mRNA; Pestova and Kolupaeva, 2002) or 5′-UTRs containing GC-rich stems of relatively low stability flanked by CAA repeats (CAA-GUS Stem-1 and Stem-2 mRNAs), yielding intense toe-prints +15-17 nt from the AUG codon (Figure 1C, lanes 4, 9, 14), they did not support high-level 48S complex formation on CAA-GUS Stem-3 and Stem-4 mRNAs containing more stable stems with ΔG=−18.9 and −27.6 kcal/mol, respectively (Figure 1C, lanes 18, 24), even though they translated efficiently in rabbit reticulocyte lysate (RRL) (data not shown). These eIFs also supported only very weak 48S complex assembly on neutrophil cytosolic factor 2 (NCF2) mRNA containing a 168 nt-long 5′UTR (55% GC; ΔG~-54 kCal/mol) (Figure 1D, lane 3), and did not promote 48S complex formation at all on CDC25 mRNA containing a 271 nt-long 5′-UTR (44% GC; ΔG~-120 kCal/mol) (Figure 1E, lane 2), even though both mRNAs were relatively efficiently translated in RRL (data not shown). We therefore undertook extensive purification from RRL of missing factor(s) required for efficient 48S complex formation on mRNAs with structured 5′-UTRs. Purification yielded an apparently homogenous ~150 kDa protein (Figure 1A) that was identified as DHX29 (Supplementary Table 1A), a putative DExH-box helicase (Figure 1B). DHX29 has a central helicase domain with consensus sequence motifs that are characteristic of DEAH helicases such as DHX9 and the splicing factor Prp2/DHX16 (de la Cruz et al., 1999; Figure 1B), and C-terminally located helicase-associated HA2 and DUF1605 domains of unknown function.
Figure 1. DHX29 is essential for efficient initiation on mRNAs with structured 5′-UTRs.
(A) Left panel - purification scheme for DHX29, right panel - purified DHX29 resolved by SDS-PAGE. (B) Model of the domain organization of DHX29 (upper panel) and alignment of conserved motifs in the helicase core domains of human DHX29 and representative DExH-box proteins (lower panel). (C-E) Toe-printing analysis of 48S complex assembly on (C) CAA-GUS mRNAs containing stems of various stabilities, (D) NCF2 and (E) CDC25 mRNAs. (F) Formation of elongation complexes on CAA-Stem3,4-MVHC-STOP mRNAs assayed by toe-printing (left panel) and by SDG centrifugation with subsequent monitoring of 35S-MVHC tetrapeptide (right panel). P-site mRNA codons and positions of assembled ribosomal complexes are indicated. Lanes C/T/A/G depict corresponding DNA sequences.
Inclusion of DHX29 in an in vitro reconstituted system strongly (~5-20-fold) increased 48S complex formation on CAA-GUS Stem-3 and Stem-4 mRNAs (Figure 1C, lanes 19, 25) and on NCF2 mRNA (Figure 1D, lane 4), and allowed 48S complex formation on CDC25 mRNA (Figure 1E, lane 1). DHX29 also slightly (~20-30%) stimulated the already efficient 48S complex formation on CAA-GUS Stem-1 and Stem-2 mRNAs (Figure 1C, lanes 10, 15). Toe-prints that appeared at intermediate positions on NCF2 and CDC25 5′-UTRs in reaction mixtures containing 43S complexes and eIF4A/4B/4F with/without DHX29 (Figure 1D, lanes 3, 4; Figure 1E, lane 1) likely corresponded to scanning ribosomal complexes arrested upstream of the initiation codon by stable secondary structures. Moderate stimulation of 48S complex formation on Stem-containing CAA-GUS mRNAs by DHX29 occurred even in the absence of eIF4A/4B/4F (Figure 1C, lanes 3, 8, 13, 17, 23), but it was lower than by eIF4A/4B/4F (Figure 1C, lanes 4, 9, 14, 18, 24). In contrast to CAA-GUS mRNAs, DHX29 did not promote 48S complex formation on NCF2 or CDC25 mRNAs in the absence of eIF4A/4B/4F (Figure 1D, lane 5; Fig. 1E, lane 3), and mediated only marginal 48S complex assembly on β-globin mRNA (Figure 2B, lane 6). We speculate that this difference was due to the presence of 43 unstructured 5′-terminal nts in CAA-GUS mRNAs that can promote eIF4A/4B/4F-independent attachment of 43S complexes (Pestova and Kolupaeva, 2002). If this assumption is correct, then DHX29 likely assists scanning but does not function during attachment of 43S complexes. To verify that 48S complexes assembled with DHX29 were elongation-competent, formation of ribosomal complexes was assayed on derivatives of CAA-GUS Stem-3 and Stem-4 mRNAs encoding a MVHC tetrapeptide followed by a UAA stop codon. Addition of 60S subunits, eIF5/5B, elongation factors and aminoacylated tRNAs to 48S complexes assembled on both mRNAs with DHX29 yielded prominent toe-prints +16-17 nt from the UGC Cys codon that occupies the P-site of elongating ribosomes arrested at the stop codon (Figure 1F, left panel). As with 48S complexes, substantially more elongation complexes formed on both mRNAs in the presence of DHX29, assayed by toe-printing and sucrose density gradient centrifugation (Figure 1F).
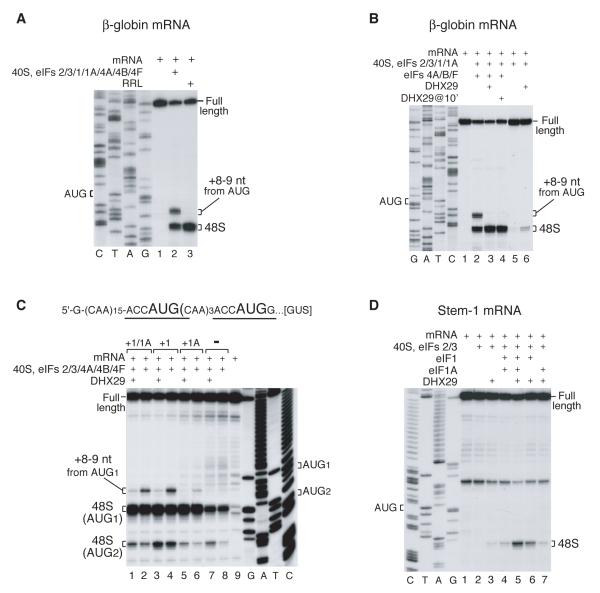
Figure 2. DHX29 suppresses the aberrant toe-print +8-9 nt from the AUG codon.
Toe-printing analysis of 48S complex assembly on (A, B) β-globin mRNA, (C) mRNA containing two AUG triplets, and (D) CAA-GUS Stem-1 mRNA in RRL (A, lane 3) and in an in vitro reconstituted initiation system (A-D) with eIFs as indicated. Initiation codons and positions of assembled ribosomal complexes are indicated. Lanes C/T/A/G depict corresponding DNA sequences.
Taken together, these data indicate that eIF4A/4B/4F and DHX29 synergistically promote efficient 48S complex formation on mRNAs with structured 5′-UTRs.
DHX29 suppresses the aberrant toe-print +8-9 nt downstream from the AUG codon
Although eIF2/3/1/1A/4A/4B/4F ensured efficient 48S complex formation on native capped β-globin mRNA, we have noted additional toe-prints +8-9 nt from the AUG codon at up to 30-40% of the level of the +15-17 nt toe-prints that correspond to properly assembled 48S complexes (Fig. 2A, lane 2; Battiste et al., 2000). The +8-9 toe-prints were apparent on some other mRNAs, for example, on the first AUG codon of mRNA containing two AUG triplets flanked by CAA repeats (Fig. 2C, lanes 2, 4). In contrast, 48S complexes assembled on β-globin or other mRNAs in RRL yielded toe-prints exclusively at +15-17 positions (e.g. Figure 2A, lane 3). Appearance of the +8-9 nt toe-print required 40S subunits, Met-tRNAMeti, eIFs and an AUG codon suggesting that it corresponds to a 48S complex, in which the 3′-portion of mRNA is not fixed in the 40S subunit’s mRNA-binding cleft, thus allowing reverse transcriptase to penetrate further. Formation of the +8-9 nt toe-print was eIF1-dependent: almost no such toe-print was observed on the first AUG codon of mRNA with two AUG triplets in reaction mixtures lacking eIF1 (Figure 2C, compare lanes 2, 4 and 6, 8). The +8-9 nt toe-print was also exacerbated by some eIF1A mutants (Battiste et al., 2000). Although DHX29 did not influence the overall yield of 48S complex formation on β-globin mRNA, it suppressed this aberrant toe-print (Figure 2B, lane 3). Importantly, DHX29 had the same effect on the +8-9 toe-print upon its delayed addition to preformed 48S complexes (Figure 2B, lane 4). DHX29 also suppressed the aberrant +8-9 nt toe-print on other mRNAs, including the mRNA with two AUG triplets (Figure 2C, lanes 1, 3). These data suggest that binding of DHX29 to ribosomal complexes induces conformational changes near the mRNA-binding cleft that influence accommodation of the 3′-portion of mRNA.
DHX29 also increased leaky scanning, enhancing 48S complex formation on the second AUG codon of mRNA with two AUG triplets, irrespective of the presence of eIF1 or eIF1A (Figure 2C, lanes 1, 3, 5, 7). This increase in leaky scanning is consistent with enhanced processivity of ribosomal complexes.
In reaction mixtures lacking eIF4F/4A/4B, DHX29 promoted low-level 48S complex formation on CAA-GUS Stem-1 even without eIF1/1A (Figure 2D, lane 3). However eIF1, particularly with eIF1A, substantially increased initiation (Figure 2D, lanes 5, 6).
DHX29 specifically binds to 40S subunits
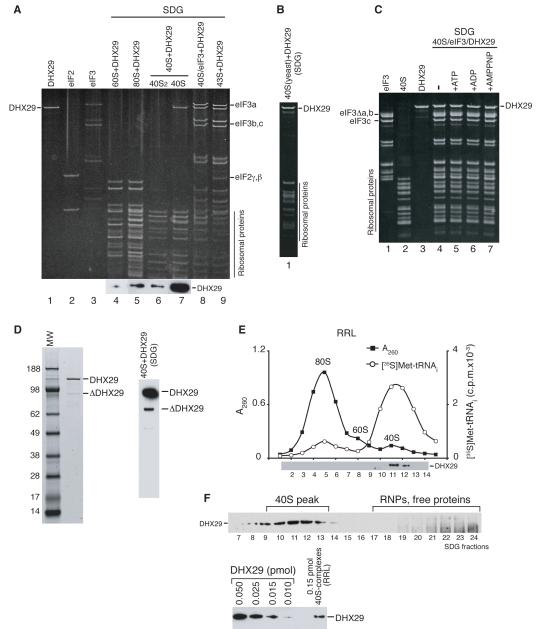
Experiments done to identify interactions between DHX29 and translational components revealed that it bound stably to 40S subunits, but not 60S subunits or 80S ribosomes, and remained associated with them during sucrose density gradient (SDG) centrifugation (Figure 3A, lanes 4, 5, 7). Importantly, DHX29 associated only with 40S monomers, but not the dimers (Figure 3A, lanes 6, 7) that always occur in mammalian 40S subunit preparations (Unbehaun et al., 2004). DHX29 also bound stably and stoichiometrically to 40S/eIF3 complexes formed with (CUUU)9 RNA (Kolupaeva et al., 2005), to 43S complexes (Figure 3A, lanes 8, 9) and to yeast 40S subunits, indicating that it associated with a conserved region of 40S subunits (Figure 3B). DHX29’s ribosomal binding was nucleotide-independent (Figure 3C). Some DHX29 preparations contained a ~90-95 kDa band (Figure 3D, left panel) that we identified as truncated DHX29 (Supplementary Table 1B). One of its tryptic peptides corresponded to amino acids 98-106, indicating that ΔDHX29 cannot lack more than 96 N-terminal amino acids, and must thus be significantly truncated at its C-terminus. If the lower band on the western blot of 40S/DHX29 ribosomal complexes obtained with such preparations of DHX29 (Figure 3D, right panel) corresponds to C-terminally truncated ΔDHX29, the region of DHX29 responsible for ribosomal binding is likely located in the N-terminal two thirds of the protein. Consistently, DHX29 in RRL was bound to 40S-containing ribosomal complexes, but not to 60S subunits or 80S ribosomes (Figure 3E). About 10% of 40S-ribosomal complexes were associated with DHX29, and all of the DHX29 was involved in this interaction (Figure 3F).
Figure 3. Interaction of DHX29 with 40S subunits.
Association of DHX29 with (A) individual 40S and 60S subunits, 80S ribosomes, 40S/eIF3/(CUUU)9 complexes and 43S complexes containing 40S subunits and eIFs 2/3/1/1A, (B) yeast 40S subunits, and (C) 40S/eIF3/(CUUU)9 complexes in the presence/absence of nucleotides as indicated (lanes 4-7). (D) DHX29 preparation containing a C-terminally truncated fragment resolved by SDS-PAGE (left panel) and its association with 40S subunits (right panel). Ribosomal peak fractions obtained by SDG centrifugation were analyzed by SDS-PAGE and fluorescent SYPRO staining (A-C) and/or western blotting using DHX29 antibodies (A, D). (E) Association of DHX29 with ribosomal complexes in RRL in the presence of GMPPNP assayed by SDG centrifugation. In addition to optical density, the ribosomal profile of RRL was analyzed by scintillation counting to monitor [35S]Met-tRNAMeti incorporation. Gradient fractions were analyzed by western blotting using DHX29 antibodies. (F) Estimation of the proportion of 40S-bound DHX29 relative to free protein (upper panel) and of the ratio of DHX29-bound vs. unbound 40S-ribosomal complexes assayed by western blotting using DHX29 antibodies (lower panel).
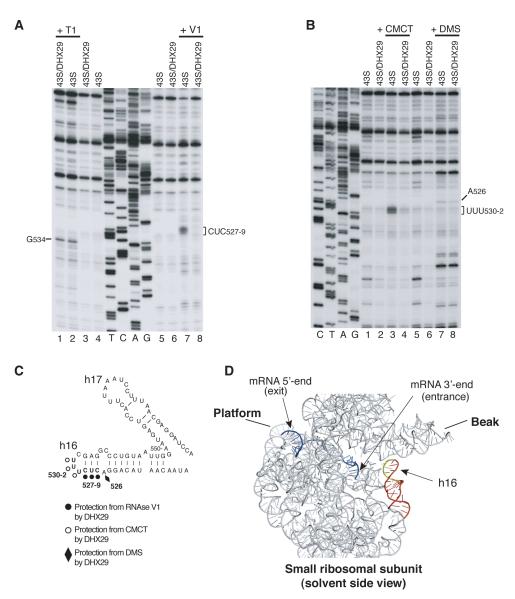
To obtain insights into the ribosomal position of DHX29, we compared chemical/enzymatic foot-printing of 18S rRNA in 43S and 43S/DHX29 complexes. DHX29 strongly protected CUC527-9 and UUU530-2 in h16 from RNase V1 cleavage and CMCT modification, respectively (Figure 4A, lanes 7, 8; Figure 4B, lanes 3, 4; Figure 4C) and weakly protected A526 from DMS modification (Figure 4B, lanes 7, 8), but did not protect G534 on the opposite strand of the stem from RNase T1 cleavage (Figure 4A, lanes 1, 2). If the observed protections resulted from direct interaction between h16 and DHX29, rather than from induced conformational changes, then DHX29 likely binds to the 40S subunit near the mRNA entrance (Figure 4D).
Figure 4. Ribosomal position of DHX29.
Enzymatic (A) and chemical (B) foot-printing analysis of 18S rRNA in 43S and 43S/DHX29 complexes. The positions of residues protected by DHX29 from RNase V1 cleavage and CMCT/DMS modification or cleaved by RNase T1 are indicated. (C, D) 18S rRNA nucleotides protected by DHX29 mapped onto (C) the secondary structure of rabbit 18S rRNA and (D) the crystal structure of the mRNA/T. thermophilus 30S subunit complex (PDB 2HGR). mRNA (blue) and 16S rRNA (grey) are in ribbon representation. Helix 16 is red, protected nucleotides are yellow.
Stimulation of 48S complex formation by DHX29 requires its NTPase activity
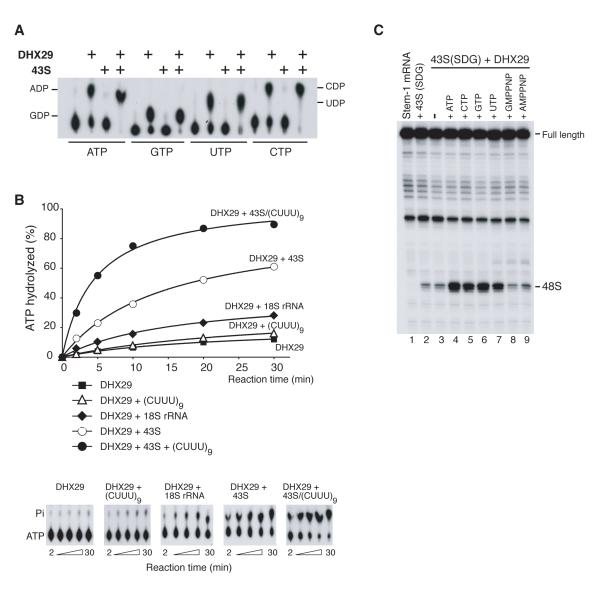
Like other DExH-box proteins (Lee et al., 1997; Tanaka and Schwer, 2005), DHX29 lacked nucleotide specificity and hydrolyzed ATP, GTP, CTP and UTP (Figure 5A). These proteins all lack the Q-motif upstream of the helicase domain that has been implicated in determining the specificity of adenine recognition by the related DEAD box helicases (Tanner et al., 2003). DHX29’s NTPase activity was strongly stimulated by 43S complexes, whereas stimulation by single-stranded (CUUU)9 RNA was low (Figures 5A, B; we note that the concentration of DHX29 in experiments shown in panel A was substantially higher than in panel B). 18S rRNA had higher stimulatory activity than (CUUU)9 RNA, but lower than 43S complexes (Figure 5B). If the ribosome binding site for DHX29 is formed by 18S rRNA, and is to some extent preserved in naked 18S rRNA, specific binding of DHX29 to 18S rRNA could account for its relatively high stimulatory activity. However, stimulation was greatest in the presence of 43S complexes and (CUUU)9 RNA, and it is tempting to speculate that this combination simulates mRNA-attached or scanning 43S complexes. To verify whether NTP hydrolysis by DHX29 is required for stimulation of 48S complex formation, we investigated eIF4A/4B/4F-independent 48S complex assembly on CAA-GUS Stem-1 mRNA in the presence of DHX29 and different NTPs (Figure 5C). 43S complexes formed with eIF2/3/1/1A were separated from unincorporated GTP by SDG centrifugation and incubated with DHX29 and mRNA in the presence/absence of GTP, ATP, CTP, UTP, GMPPNP or AMPPNP. DHX29’s stimulatory activity was higher with GTP or ATP than with CTP or UTP (Figure 5C, lanes 4-7). No stimulation occurred without nucleotides or with non-hydrolysable GMPPNP or AMPPNP (Figure 5C, lanes 3, 8, 9). NTP hydrolysis by DHX29 was therefore required for its activity in 48S complex formation.
Figure 5. Stimulation of 48S complex formation by DHX29 requires its NTPase activity.
(A) Thin-layer chromatography analysis of DHX29′s NTPase activity in the presence/absence of SDG-purified 43S complexes comprising 40S subunits and eIF2/3/1/1A. 10 μl reaction mixtures containing 1 pmol DHX29, 1 pmol 43S complexes and 6.7 μM [α-32P]ATP, [α-32P]GTP, [α-32P]UTP or [α-32P]CTP, as indicated, were incubated at 37°C for 40 minutes. The positions of [32P]-NDPs are indicated. (B) Time courses of ATP hydrolysis by DHX29 in the presence/absence of (CUUU)9 RNA, 18S rRNA, 43S complexes, or 43S/(CUUU)9, as indicated. 10 μl reaction mixtures containing 0.3 pmol DHX29, 6.7 μM [γ-32P]ATP and 20 pmol (CUUU)9 RNA, 0.3 pmol 18S rRNA, 0.3 pmol 43S complexes, or 0.3 pmol 43S complexes with 20 pmol (CUUU)9 RNA, as indicated, were incubated at 37°C. Aliquots were removed after 2-30 minutes. (C) Toe-printing analysis of 48S complexes assembled on CAA-GUS Stem-1 mRNA in the presence of SDG-purified 43S complexes, DHX29 and NTPs or non-hydrolyzable NTP analogues, as indicated. The position of 48S complexes is indicated.
DHX29 does not possess a processive helicase activity
To investigate the potential helicase activity of DHX29, we used RNA duplexes comprising overhanging 25 nt-long 5′- or 3′-ends and 13 nt- or 10 nt-long double-stranded regions (ΔG=−21 and −14.6 kCal/mol, respectively), corresponding blunt duplexes, as well as duplexes resembling stems 2, 3 and 4 of CAA-GUS Stem2-4 mRNAs. DHX29 could not unwind 13 nt-long duplexes with overhanging 5′- or 3′-ends in the presence of any NTP, whereas unwinding by eIF4A/4F was efficient (Figure 6A, left panel; data not shown). Very weak unwinding by isolated 43S/DHX29 complexes (Figure 6A, right panel, lane 2) may in fact be attributable to ribosomal scanning after attachment of 43S complexes to the 5′-overhang. Unwinding by DHX29 of 10 nt-long duplexes with overhanging ends was marginal (<5%), and blunt duplexes were not unwound (Figure 6B, lanes 2, 3; data not shown). DHX29 could unwind Stem-2 duplex (Figure 6C, lane 3), although we note that this duplex was intrinsically unstable under experimental conditions, resulting in a noticeable background of RNA monomers (Figure 6C, lanes 2). Unwinding of Stem-3 duplex by DHX29 was marginal (Figure 6C, lane 6), and Stem-4 duplex was not unwound (Figure 6C, lane 9), whereas eIFs 4A/4F unwound ~5-10% of Stem-4 duplex (Figure 6C, lane 10). Thus, like some other DExH-box proteins (Tanaka and Schwer, 2005), DHX29 is not a processive RNA helicase.
Figure 6. Helicase activity of DHX29.
(A, B, C) Non-denaturing PAGE showing unwinding of (A) 13-bp, (B) 10-bp RNA duplexes with 25 nt-long single-stranded overhanging 5′-regions, and (C) RNA duplexes resembling Stems 2-4 by DHX29, 43S complexes, 43S/DHX29 complexes and eIF4A/eIF4F, as indicated. 1 nM duplex was incubated with 0.15 μM DHX29, 50 nM 43S complexes, 50 nM 43S/DHX29 complexes or 0.15 μM eIF4A/eIF4F and 0.2 mM NTPs, as indicated, at 37°C for 40 minutes. Mobilities of duplex and single-standed RNAs are indicated schematically on the left. 95°C represents the control for denatured strands. (D) SDG-purified 43S complexes containing different amounts of DHX29 and analyzed by SDS-PAGE and fluorescent SYPRO staining. (E, F) Toe-printing analysis of 48S complex formation on CAA-GUS Stem-1 mRNA in the presence of SDG-purified free 43S complexes and 43S complexes containing different amounts of DHX29 (shown in panel D). The positions of the initiation codon and assembled 48S complexes are indicated. Lanes C/T/A/G depict corresponding DNA sequence. (G) Association of DHX29 (and eIF2, as a loading control) with 43S complexes and 48S complexes assembled on native globin mRNA assayed by SDG centrifugation and western blotting.
DHX29 can participate in multiple rounds of 48S complex formation
DXH29 stimulated 48S complex formation most strongly when it was present in substoichiometric amounts relative to 43S complexes. Thus, SDG-purified 43S/DXH29 complexes with a 43S:DHX29 ratio of 10:1 (Figure 6D, lane 3) were most active in 48S complex assembly on CAA-GUS Stem-1 mRNA (Figure 6E, lane 2), whereas complexes with 43S:DHX29 ratios of 2:1 and 1:1 (Figure 6D, lanes 4, 5) were progressively less active (Figure 6E, lanes 3, 4). Importantly, a mixture of DHX29-free and DHX29-saturated 43S complexes that individually had the lowest activities (Figure 6E, lanes 4, 5), together promoted very efficient 48S complex formation (Figure 6E, lane 6). These results suggest that a proportion of DHX29 might be inactive, but that DHX29 from active 43S/DHX29 complexes could dissociate from ribosomal complexes and participate in new rounds of initiation. Alternatively, stimulation of 48S complex formation by DHX29 might require its dissociation from the 40S subunit at some point in the process before the 48S complex is formed, in which case the excess of free 43S complexes would ensure rebinding of dissociated DHX29 to a new 43S complex. To investigate this possibility, DHX29-saturated 43S complexes were mixed with purified 40S/eIF3/(CUUU)9 complexes, which themselves could not participate in 48S complex formation but could potentially provide a “trap” for dissociated DHX29 thereby stimulating 48S complex formation by 43S/DHX29 complexes. 40S/eIF3/(CUUU)9 complexes did not stimulate 48S complex formation by 43S/DHX29 complexes (Figure 6F, compare lanes 3, 5). Although this could be because DHX29’s affinity to 40S/eIF3/(CUUU)9 complexes is lower than to 43S complexes, the possibility that a proportion of DHX29 might be inactive cannot be excluded. The stage at which DHX29 dissociates from ribosomal complexes is not known, but we note that consistently less DHX29 was bound to 48S than to 43S complexes (Figure 6G).
The influence of DHX29 on 48S complex formation during IRES-mediated initiation
The genomes of several families of RNA viruses contain internal ribosomal entry sites (IRESs), which mediate end-independent initiation, enabling viral mRNAs to bypass the canonical cap-dependent mechanism. IRESs are classified into structurally unrelated groups that mediate initiation by distinct mechanisms that require fewer eIFs than canonical initiation. Three mechanisms of IRES-mediated initiation have been identified, and they are all based on specific non-canonical interactions of IRESs with canonical components of the translation apparatus. The first is exemplified by type 2 picornavirus IRESs (e.g. encephalomyocarditis virus (EMCV)), which promote initiation at their 3′ border by a mechanism that relies on specific interaction of the IRES’ J-K domain upstream of the initiation codon with eIF4G and involves direct attachment of the 43S complex to the initiation codon, which is likely mediated by interaction of 43S-bound eIF3 with IRES-bound eIF4G (Pestova et al., 1996). Initiation on the IRESs of hepatitis C virus (HCV) and classical swine fever virus (CSFV) is determined by their ability to bind directly and independently to 40S subunits and eIF3 (Pestova et al., 1998b). These interactions enable 43S complexes to attach directly to the initiation codon of HCV-like IRESs without scanning or local unwinding of mRNA. Initiation on the intercistronic region (IGR) IRESs of dicistroviruses (e.g. cricket paralysis virus (CrPV)) is also determined by their ability to bind directly to 40S subunits, but unlike HCV-like IRESs, does not use eIFs or initiator tRNA: the ribosomal P-site is occupied by a domain of the IRES, which mimics the codon-anticodon interaction (Wilson et al., 2000).
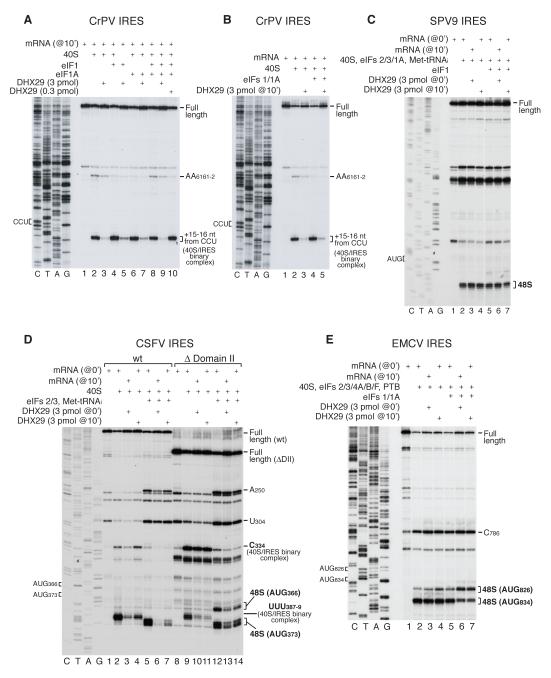
Binding of the CrPV IRES to 40S subunits yields two sets of toe-prints: at AG6228-9, corresponding to the leading edge of the 40S subunit +15-16 nt from the P-site CCU codon, and AA6161-2, corresponding to a second IRES-40S subunit interaction. When present in stoichiometric amounts relative to 40S subunits, DHX29 almost abrogated the toe-prints at AG6228-9 irrespective of whether DHX29 was added before CrPV IRES mRNA (Figure 7A) or to preassembled IRES/40S complexes (Figure 7B). eIF1, which stabilizes CrPV IRES/40S complexes (Pestova et al., 2004), only slightly mitigated the effect of DHX29 (Figure 7A, lanes 5, 7; Figure 7B, lane 5). Binding of the CSFV IRES to 40S subunits also yields two sets of toe-prints: at UUU387-9, corresponding to the leading edge of the 40S subunit +15-17 nt from the P-site AUG codon, and at C334, corresponding to a contact of the 40S subunit with the pseudoknot of the IRES (Pestova et al., 1998b). As with the CrPV IRES, DHX29 strongly reduced the toe-prints at UUU387-9 in 40S/CSFV IRES complexes irrespective of when it was added (Figure 7D, lanes 2-4). Interestingly, for both IRESs, DHX29 had less effect on toe-prints corresponding to 40S/IRES contacts outside the mRNA-binding cleft (AA6161-2 and C334 toe-prints) than on toe-prints at the leading edge of the bound 40S subunit (Figures 7A, B, D). Moreover, when assayed by SDG centrifugation, 40S/IRES complex formation in the presence of DHX29 was reduced by only ~30% for both IRESs (data not shown). Thus, binding of DHX29 to 40S subunits strongly affects fixation of the IRES in the area of the mRNA-binding cleft, but has a weaker effect on the overall affinity of IRESs to 40S subunits.
Figure 7. Influence of DHX29 on 48S complex formation on viral IRESs.
Toe-printing analysis of 40S/IRES binary and 48S complexes assembled on (A, B) CrPV, (C) SPV9, (D) wt and ΔDomain II CSFV, and (E) EMCV IRESs in the presence of eIFs as indicated. Initiation codons and positions of assembled ribosomal complexes are indicated. Lanes C/T/A/G depict corresponding DNA sequences.
Importantly, even upon delayed addition, DHX29 abrogated toe-prints corresponding to 48S complexes assembled on the CSFV IRES in the presence of eIFs 2/3 and Met-tRNAMeti (Figure 7D, lanes 5-7). The dissociating effect of DHX29 on 48S complexes assembled on this IRES is reminiscent of the effect of eIF1 (Pestova et al., 2008). Deletion of IRES domain II, which is responsible for conformational changes induced in 40S subunits by IRES binding (Spahn et al., 2001), eliminates the sensitivity of 48S complexes to dissociation by eIF1 (Pestova et al., 2008). Although deletion of domain II did not completely suppress the dissociating effect of DHX29, 48S complexes assembled on the IRES lacking domain II were nevertheless less sensitive to DHX29 than complexes assembled on the wt IRES (Figure 7D: compare lanes 5-7 with lanes 12-14). Interestingly, 48S complexes assembled on the HCV-like IRES of Simian picornavirus type 9 (SPV9), which are much more resistant to dissociation by eIF1 (de Breyne et al., 2008), were also resistant to dissociation by DHX29 (Figure 7C). It is likely relevant that the predicted structure of the SPV9 IRES domain II differs significantly from that of HCV and CSFV IRESs.
Initiation on the EMCV IRES occurs predominantly at AUG834, and infrequently at AUG826 (Kaminski et al., 1990). Although DHX29 did not affect the overall level of 48S complex formation on this IRES, in the presence of eIF1, it significantly increased the proportion of 48S complexes formed on AUG826 even upon its delayed addition (Figure 7E, lanes 5-7).
Such distinct effects of DHX29 on 48S complex formation on different IRESs are consistent with DHX29 causing conformational changes in 40S subunits, which can or cannot be tolerated by IRES-bound complexes. We note that eIF1, which induces conformational changes in 40S subunits (Passmore et al., 2007), also has distinct effects on 48S complex formation on different IRESs (de Breyne et al., 2008; Pestova et al. 1998a, 2004, 2008).
DISCUSSION
We have identified the DExH-box protein DHX29 as a factor that is required for efficient initiation on mammalian mRNAs with structured 5′-UTRs, which typically encode regulatory proteins. The extent of the requirement for DHX29 correlated with the stability of the secondary structure elements in the 5′-UTRs of mRNAs. Although isolated internal stems of ΔG=−13.1 kcal/mol or less could be efficiently overcome by scanning complexes in the presence of only eIF4A/4G/4B, efficient ribosomal movement through stems of ΔG>−19 kcal/mol required DHX29. Ribosomal scanning relies on the ATP-dependent helicase activity of eIF4A/4G/4B and is influenced by the conformation of scanning 43S complexes, which is modified by eIF1/1A. How does DHX29 stimulate 48S complex formation on mRNAs with structured 5′-UTRs? Does it participate directly in unwinding of mRNA or does it remodel 43S complexes to increase their scanning processivity? The answer to this question is linked to the mechanism by which eIF4A/4G/4B assist scanning, and elucidation of this requires knowledge of their location in ribosomal complexes. If eIF4A/4G/4B bind at the leading edge and unwind mRNA before it enters the 40S subunit, it is unlikely that another helicase, DHX29, would participate directly in the same process. In this case DHX29 could enhance the processivity of ribosomal movement by remodeling ribosomal complexes to ensure correct entry into and/or fixation of mRNA in the mRNA-binding cleft. But if eIF4A/4G/4B, as suggested (Siridechadilok et al., 2005), bind at the trailing edge near the E-site and assist scanning by helicase-mediated “ratcheting” of mRNA through the mRNA-binding channel, then DHX29 might directly unwind mRNA before it enters the 40S subunit. Although this possibility seems unlikely because DHX29 is not a processive helicase, it cannot be strictly excluded that binding of DHX29 to ribosomal complexes might enhance its helicase activity. However, even if eIF4A/4G/4B act at the trailing edge, DHX29 could still assist scanning not by direct unwinding of mRNA, but by remodeling ribosomal complexes and influencing accommodation of mRNA in the mRNA-binding channel, in which case mRNA secondary structure would be unwound by the scanning 40S subunit itself. In this hypothetical situation, correct positioning of mRNA at the entrance to the mRNA-binding channel would be particularly important. We note that the bacterial ribosome has helicase activity, which involves ribosomal proteins S3, S4 and likely S5 (Takyar et al., 2005).
Suppression of the aberrant +8-9 nt toe-print, which most likely represents a 48S complex with the 3′-portion of mRNA not firmly fixed in the mRNA-binding cleft of the 40S subunit, by DHX29 even on delayed addition to pre-assembled 48S complexes indicates that DHX29 does induce conformational changes in these complexes that influence ribosomal accommodation of the 3′-portion of mRNA. The appearance of aberrant +8-9 nt toe-prints depended on the presence of eIF1/1A. Binding of eIF1/1A to yeast 40S subunits causes the entry ‘latch’ between h18 in the body and h34/rpS5 in the neck to open and establishes a new connection between rpS3 and h16 (Passmore et al., 2007). Such opening of the entry ‘latch’ might weaken fixation of the 3′-portion of mRNA in the mRNA-binding cleft that could account for appearance of the +8-9 nt toe-print. It is likely that the conformation of the 40S subunit with the open latch is more conducive to attachment of 43S complexes to mRNA, whereas processive scanning might require firm fixation of mRNA in the mRNA-binding cleft. In this case the conformation of ribosomal complexes would require further modification, which could be promoted by DHX29.
Another indication that DHX29 causes conformational changes in 40S subunits comes from its influence on ribosomal complexes assembled on viral IRESs. Thus, even on delayed addition, DHX29 affected 40S-ribosomal binding and proper fixation in the mRNA-binding cleft of CrPV and CSFV IRESs. The CrPV and CSFV-like HCV IRESs both induce similar conformational changes in 40S subunits, which were suggested to facilitate fixation of these IRESs in the mRNA-binding cleft (Spahn et al., 2001, 2004). It is therefore likely that binding of DHX29 to 40S subunits either does not allow such IRES-induced changes to occur, and/or causes other conformational changes in 40S subunits that are not compatible with binding and proper positioning of the IRESs on 40S subunits. Moreover, DHX29 dissociated 48S complexes assembled on the CSFV IRES and influenced the ratio of 48S complexes assembled on AUG826 and AUG834 of the EMCV IRES. Although the dissociating effect of DHX29 on 48S complexes assembled on the CSFV IRES is similar to that reported for eIF1 (Pestova et al., 2008), the conformational changes induced in 40S subunits by eIF1 (Passmore et al., 2007) and the potential conformational changes induced by DHX29 are likely not identical because these factors have opposite effects on 40S/CrPV IRES complexes (Pestova et al., 2004; this study) and on the ratio of 48S complexes assembled on two AUGs of the EMCV IRES (Pestova et al., 1998a; this study).
Foot-printing experiments revealed that in 43S complexes, DHX29 protects h16 of 18S rRNA. We cannot conclude unambiguously whether such protection is caused by direct contact of DHX29 with h16 or reflects conformational changes in the 40S subunit induced by DHX29. If DHX29 indeed binds h16 near the mRNA entrance, this position of DHX29 would be consistent with both hypothetical modes of action (remodeling of 43S complexes or mRNA unwinding). However, if the observed protections correspond to conformational changes in ribosomal complexes, then the region of such changes is entirely consistent with remodeling of 40S subunits near the mRNA entrance, which would likely affect accommodation of the 3′-portion of mRNA in the mRNA-binding cleft. Moreover, it is exactly the area of the 40S subunit that undergoes conformational changes upon binding of eIF1 and eIF1A. It is not known whether stable secondary structures in 5′-UTRs only slow ribosomal scanning or also increase drop-off of 43S complexes. If drop-off can occur, then proper fixation of mRNA in the mRNA-binding cleft would also stabilize ribosomal association with mRNA and increase the processivity of scanning complexes. Moreover, the potential influence of the conformation of ribosomal complexes on the processivity of the ribosome-bound eIF4A/4G/4B helicase complex could strictly also not be excluded. Although we are not yet in a position to discriminate between the remodeling and unwinding mechanisms by which DHX29 might stimulate 48S complex formation, it is worth noting that it has become apparent that many DExH/D proteins function primarily in remodeling of RNA and RNP complexes rather than in processive unwinding of RNA duplexes (reviewed by Pyle, 2008). Thus, many DExH/D proteins have additional RNA-binding domains that contribute to strong ATP-independent RNA annealing activity, which in conjunction with their ATP-dependent unwinding activity suggest that such proteins can induce alternating conformational rearrangements in RNA and RNP complexes upon their transition between ATP-bound and ATP-free states. In addition, DExH/D proteins can also function as RNPases, displacing proteins from RNA in ATP-dependent manner.
Interestingly, although DHX29 and eIF4F/4A/4B acted synergistically in 48S complex formation on mRNAs with 5′-UTRs containing stable hairpins, DHX29 alone also promoted relatively efficient 48S complex formation on mRNAs with 5′-UTRs containing less stable stems and unstructured 5′-terminal regions that could promote eIF4F/4A/4B-independent attachment of 43S complexes, and even mediated low-level 48S complex formation on mRNAs with 5′-UTRs containing stems of high stability. DHX29 might therefore be responsible for translation of at least a sub-class of mRNAs in conditions when eIF4G is depleted (Ramírez-Valle et al., 2008).
In RRL, DHX29 was wholly associated with 40S-ribosomal complexes, but DHX29-bound 40S-ribosomal complexes nevertheless constitute only ~10% of all 40S-ribosomal complexes. In this respect, it is particularly important that DHX29 can participate in multiple rounds of 48S complex formation. Although it would be most logical to suggest that DHX29 remains associated with ribosomal complexes during the entire scanning process and dissociates from assembled 48S complexes as a result of conformational changes that likely occur upon establishment of codon-anticodon base-pairing, we cannot exclude that DHX29 might dissociate earlier, particularly if it functions by remodeling ribosomal complexes rather than by unwinding mRNA. Our experiments also indicate that a proportion of purified DHX29 might be inactive in stimulating 48S complex formation, even though it could still bind ribosomal complexes. Although DHX29 might have been partially inactivated during purification, phosphorylation of human DHX29 at Ser192, Ser200, Tyr811 and Tyr826 (www.phosphosite.org) could also influence its activity.
The preceding discussion is based on the assumption that eIF4A is the only DEAD-box RNA helicase involved in initiation. However, biochemical and genetic analyses have implicated other DEAD/DExH-box proteins in initiation, including Ded1p and the homologous mammalian proteins DDX3/PL10, mammalian RNA helicase A (RHA) and Drosophila Vasa (Chuang et al., 1997; de la Cruz et al., 1997; Hartman et al., 2006; Johnstone and Lasko, 2004; Lee et al., 2008). The mechanism(s) by which Ded1p, DDX3, RHA and Vasa act in the initiation process are incompletely characterized but are likely distinct. Ded1p has been characterized in the greatest detail: it is a more processive helicase than eIF4A (Marsden et al., 2006), its function is not redundant with that of eIF4A and mutations in Ded1p are synthetic-lethal with mutations in TIF1 (eIF4A) and cdc33 (eIF4E) and deletion of TIF4631 (eIF4G) or STM1/TIF3 (eIF4B) (Chuang et al., 1997; de la Cruz et al., 1997). This has led to suggestions that eIF4A may, as a subunit of eIF4F, function in promoting recruitment of 43S complexes to the cap-proximal region of mRNA, whereas Ded1p assists ribosomal complexes during scanning, particularly on mRNAs with long 5′UTRs (e.g. Marsden et al., 2006). The molecular interactions that could couple Ded1p with scanning ribosomes are not known. It remains to be seen whether mammalian Ded1p homologues like DDX3 also function during scanning, after ribosomal loading, in which case they would likely unwind mRNA before it enters the mRNA-binding cleft, near its entrance. If this is indeed the case, it is even more likely that DHX29 functions in remodeling the 40S/mRNA/eIFs complex rather than in unwinding mRNA during initiation.
EXPERIMENTAL PROCEDURES
Plasmids
See Supplemental Data.
Purification of initiation factors, ribosomal subunits and aminoacylation of tRNA
40S and 60S subunits, eIFs 2/3/4F, eEF1H, eEF2 and total aminoacyl-tRNA synthetases were purified from RRL or HeLa cells, recombinant eIFs 1/1A/4A/4B/5/5B, PTB and E. coli methionyl tRNA synthetase were expressed and purified from E. coli, and native total tRNA (Novagen) and in vitro transcribed tRNAMeti were aminoacylated as described (Pisarev et al., 2007).
DHX29 purification
DHX29 was purified from RRL on the basis of activity in supporting 48S complex formation in the in vitro reconstituted system on mRNAs with structured 5′-UTRs, which was monitored by toe-printing. Purification involved preparation of ribosomal salt wash, fractionation by ammonium sulphate precipitation, chromatography on DEAE cellulose and on phosphocellulose, and FPLC on MonoS, MonoQ and hydroxyapatite columns. DHX29 was identifi by mass spectrometry of tryptic peptides.
Assembly and analysis of initiation complexes
48S complexes were assembled on capped in vitro transcribed (CAA)-GUS mRNA, its derivatives containing stems or two AUG codons, (CAA)-Stem-MVHC-STOP mRNAs, NCF2 mRNA, CDC25 mRNA, native β-globin mRNA, and CrPV, SPV9, EMCV and CSFV IRESs, and analyzed by primer extension using AMV-RT and [32P]-labeled primers as described (Pisarev et al., 2007). To assay elongation on (CAA)-Stem-MVHC-STOP mRNAs, 48S complexes were supplemented with eIF5, eIF5B, 60S subunits, tRNA aminoacylated with Met, Val, His and [35S]Cys, eEF1H and eEF2 and incubated at 37°C for 20 min. Elongation complexes were assayed by toe-printing, or by centrifugation through 10-30% SDGs in a Beckman SW55 rotor at 53,000 rpm for 75min with subsequent monitoring of [35S]MVHC formation.
To investigate the requirement for NTP hydrolysis by DHX29 for its activity in stimulating 48S complex formation and the ability of DHX29 to participate in multiple rounds of initiation, 48S complexes were assembled on CAA-GUS Stem-1 mRNA using SDG-purified 43S complexes.
For toe-printing analysis of 48S complexes assembled on β-globin mRNA in RRL, globin mRNA was incubated in RRL (Promega) in the presence of 2 mM GMPPNP for 10 min at 30°C.
Analysis of ribosomal binding of DHX29
DHX29 was incubated with 40S subunits, 60S subunits, 80S ribosomes, 40S/eIF3/(CUUU)9 or 43S complexes in the presence/absence of nucleotides and subjected to centrifugation through 10-30% SDGs. Fractions that corresponded to ribosomal complexes were analyzed by SDS-PAGE with subsequent fluorescent SYPRO staining or western blotting using DHX29 antibodies. To investigate the ribosomal association of DHX29 in RRL, RRL (Promega) was incubated with 1 mM GMPPNP and subjected to SDG centrifugation. Gradient fractions were analyzed by western blotting.
Chemical and enzymatic foot-printing
Ribosomal complexes were assembled by incubating 40S subunits, eIFs 2/3/1/1A and Met-tRNAMeti with or without DHX29, and then enzymatically digested by incubation with RNase V1 or RNase T1, or modified by incubation with CMCT or DMS. Cleavage/modification sites in 18S rRNA were identified by primer extension using AMV-RT.
NTPase assay
DHX29 was incubated with [α-32P]ATP, [α-32P]GTP, [α-32P]UTP, [α-32P]CTP or [γ-32P]ATP in the presence/absence of (CUUU)9 RNA, 18S rRNA, SDG-purified 43S complexes or 43S/(CUUU)9 complexes. Reaction mixtures were analyzed by chromatography on PEI cellulose.
Helicase assay
Short RNA oligonucleotides (Dharmacon) were 32P-labeled with T4 polynucleotide kinase and annealed with complementary long RNA oligonucleotides. RNA duplexes were purified on Superdex 75, incubated with DHX29, 43S complexes, 43S/DHX29 complexes, eIF4A/eIF4F and NTPs, and analyzed in 16% non-denaturing gels.
Supplementary Material
ACKNOWLEDGEMENTS
We thank K. Gauss for the NCF2 vector and A. Marintchev for helpful discussion. This work was supported by NIH Grant GM059660 and HFSP Grant RPG0055/2006-C to TVP, NIH Grant AI051340 to CUTH and National American Heart Association Grant 0730120N to AAK.
Footnotes
SUPPLEMENTAL DATA Supplemental data include detailed Experimental Procedures and Table 1.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Battiste JL, Pestova TV, Hellen CUT, Wagner G. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell. 2000;5:109–119. doi: 10.1016/s1097-2765(00)80407-4. [DOI] [PubMed] [Google Scholar]
- Chuang RY, Weaver PL, Liu Z, Chang TH. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- de Breyne S, Yu Y, Pestova TV, Hellen CU. Factor requirements for translation initiation on the Simian picornavirus internal ribosomal entry site. RNA. 2008;14:367–380. doi: 10.1261/rna.696508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Iost I, Kressler D, Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- Hartman TR, Qian S, Bolinger C, Fernandez S, Schoenberg DR, Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- Jackson RJ. The ATP requirement for initiation of eukaryotic translation varies according to the mRNA species. Eur. J. Biochem. 1991;200:285–294. doi: 10.1111/j.1432-1033.1991.tb16184.x. [DOI] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Interaction with eIF5B is essential for Vasa function during development. Development. 2004;131:4167–4178. doi: 10.1242/dev.01286. [DOI] [PubMed] [Google Scholar]
- Kaminski A, Howell MT, Jackson RJ. Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J. 1990;9:3753–3759. doi: 10.1002/j.1460-2075.1990.tb07588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CUT, Pestova TV. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- Lee CG, Chang KA, Kuroda MI, Hurwitz J. The NTPase/helicase activities of Drosophila maleless, an essential factor in dosage compensation. EMBO J. 1997;16:2671–2681. doi: 10.1093/emboj/16.10.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Dias AP, Jedrychowski M, Patel AH, Hsu JL, Reed R. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 2008;36:4708–4718. doi: 10.1093/nar/gkn454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden S, Nardelli M, Linder P, McCarthy JE. Unwinding single RNA molecules using helicases involved in eukaryotic translation initiation. J. Mol. Biol. 2006;361:327–335. doi: 10.1016/j.jmb.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CUT. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998a;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998b;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Hellen CU. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lorsch JR, Hellen CUT. In: Translational control in biology and medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2007. pp. 87–128. [Google Scholar]
- Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 2008;27:1060–1072. doi: 10.1038/emboj.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CUT, Pestova TV. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Unbehaun A, Hellen CUT, Pestova TV. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Jackson RJ. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- Ramírez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J. Cell. Biol. 2008;181:293–307. doi: 10.1083/jcb.200710215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Schwer B. Characterization of the NTPase, RNA-binding, and RNA helicase activities of the DEAH-box splicing factor Prp22. Biochemistry. 2005;44:9795–9803. doi: 10.1021/bi050407m. [DOI] [PubMed] [Google Scholar]
- Tanner NK, Cordin O, Banroques J, Doére M, Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CUT, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon–anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.