Abstract
Proteomics is fast becoming one of the most interdisciplinary fields, bridging many chemical and biological disciplines. Major challenges, however, can limit the reach of proteomics to studies of model organisms. Challenges include the adequate preservation of field samples, and the reliance of in-depth proteomics on sequenced genomes. Seeking to better establish the evolutionary relationships of hornets and yellowjackets comprising the subfamily Vespinae, we are combining classical morphological and genomic information with a functional genomics trait using proteomics. Vespine species form highly social colonies and exhibit division of labor in almost all aspects of colony life. An extreme digestive division of labor has been reported in Vespa orientalis, in which larvae but not adult workers exhibit the capacity to digest proteins fully. This makes the colony dependent upon the amino acid-rich trophallactic fluid released to adults by larvae, and implies that the V. orientalis superorganism possesses larval-specific proteases. Identifying the proteases and the species exhibiting such extreme partitioning of digestive labor will allow for tracing the phylogenetic origins and elaboration of that digestive partitioning in the Vespinae. Herein we describe methods, generally applicable to field samples, showing the preservation of proteins and proteolytic activity from adult and larval vespine trophallactic fluid.
Keywords: mass spectrometry, proteomics, functional genomics, phylogenetics, Vespinae, hornet, yellowjacket, de novo sequencing, protein preservation, division of labor
Introduction
Model organisms have facilitated impressive progress in biological understanding. However, many lines of biological inquiry can only be advanced by direct examination of non-model organisms. In the field of systematics, phylogenetic relationships within a given clade can only be constructed by direct analysis of the diversity of taxa within the clade, and not by simple characterizations of one or two model organisms that might exist therein. A holistic approach to systematics seeks to determine evolutionary relationships via multiple data types, including genomic, morphological and behavioral traits1–3. Proteomic data can also be applied to systematics, offering “functional genomics” traits by indentifying and quantifying enzymatic activities, protein composition and post-translational modifications. Like all characters suitable for phylogenetic analysis, proteomic data can either be involved in the selection of an optimal tree topology, or those characters can mapped onto phylogenies post hoc. In either circumstance, this suite of character types will be useful in phylogenetic investigations of the complex social structures of the Vespinae.
Proteomic analysis of non-model organisms, however, is not trivial as it faces two primary challenges. First, non-model organisms by definition are rarely if ever cultured in laboratory settings. Thus simply obtaining and preserving the protein sample for enzymatic assays or mass spectrometry (MS) analysis could prove prohibitive. Second, the most useful MS-based proteomics is heavily dependent on species-specific protein databases to facilitate in-depth coverage and reliable peptide and protein identification. Even when manual or in silico de novo sequencing 4 methods accurately determine a peptide’s amino acid sequence, assembling the typically non-overlapping peptides into parent proteins remains problematic, in spite of some homology-driven computational assistance 5–8.
This study primarily addresses the first challenge as applied to the enzymatic and proteomic characterization of trophallactic fluid from wasps of the subfamily Vespinae. The Vespinae (hornets and yellowjackets) are comprised of 68 known species within four genera (J.M. Carpenter, J. Kojima; http://www.ipc.ibaraki.ac.jp/~jkrte/wasp/vespinae/top.html). Vespine species create sophisticated societies in which labor is divided among colony members 9, analogous to the partitioning of physiological activities in the tissues and organs of multicellular organisms. While most vespine species remain to be fully characterized, early work found that one species of hornet, Vespa orientalis, may have evolved an extreme digestive division of labor: Larvae but not adult workers (hereafter, workers) exhibit the capacity to fully digest and metabolize proteins 10. Survival of the V. orientalis colony therefore requires that workers forage for prey and provide protein to the larvae. In turn the larvae function as an essential digestive organ for the colony. Stimulating the mouth of the larvae, workers request larval release of a trophallactic fluid that is rich in amino acids and sugars 9. Although both larvae and workers release trophallactic fluids, larval-specific enzymes in V. orientalis appear to be required for their dramatic digestive division of labor. Therefore, a proteomic approach that would characterize vespine larval and worker trophallactic fluid could identify digestive enzymatic activities and proteins that participate in their digestive division of labor. Once such proteomic investigations have been conduced across many vespine species, phylogenetic investigations will shed light on the origins and elaborations of this functional genomics trait.
In order to conduct these investigations it is imperative to establish techniques for the collection and preservation of vespine trophallactic fluids for enzymatic assays and protein identification. We demonstrate herein simple methodologies that enable biological samples to withstand the rigors of typical warm-weather field expeditions by preserving both enzymatic activity, and protein integrity for subsequent MS analysis. These methods are generally applicable to field samples from non-model organisms and should facilitate greater access to proteomic technologies, which are playing increasingly important roles throughout the biological disciplines.
Experimental Procedures
Whole cell extract preparation by hypotonic lysis
E1A-transformed human embryonic kidney (HEK) 293 cells were originally obtained from Ed Harlow (Harvard Medical School). Cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT) and 50units/ml penicillin and 50μg/ml streptomycin (Invitrogen, Carlsbad, CA). One confluent 10cm dish was washed twice with ice-cold phosphate-buffered saline and then subjected to hypotonic lysis on ice using 2ml of 25mM Tris, pH 7.2. The plate was then gently scraped and the lysate was transferred to microcentrifuge tubes and vortexed briefly. Lysates were cleared by spinning for 10min at 14,000 rpm in a VWR Galaxy 16 microcentrifuge at 4°C, and the supernatant was aliquoted into multiple tubes for further analysis. The supernatant was stored on ice for no longer than one hour before desiccation (see below) or freezing at −80°C.
Wasp trophallactic fluid collection and preparation
Trophallactic fluid was collected from workers caught with a net while in flight, and workers and larvae from active nests. Specimens and samples were collected in Singapore, and the following states in the USA: Colorado, New Mexico and Vermont. Samples from Japan were generously provided by Junichi Kojima. All appropriate collection permits were obtained. Samples were collected immediately from individuals caught with a net while in flight, and within three hours after nest disturbance.
Trophallactic fluid was harvested largely as described 10, 11. Briefly, a 5μl glass capillary (Drummond Scientific, Broomall, PA) was used to prod gently the larval mouth or mandibles of the workers, which stimulated the release of trophallactic fluid that was immediately drawn into the glass capillary. Samples were released from the glass capillary by applying air pressure facilitated by the manufacturer-supplied aspiration device. Samples were immediately frozen on dry ice, and stored at −80°C until future use. Alternatively, samples were desiccated as described below and processed immediately after returning from field expeditions of up to three week durations.
Sample desiccation
Samples were aliquoted into 0.7ml microcentrifuge tubes, the caps of which were punctured three times with a 21 gauge sterile needle. The tubes were stored upright in a storage box used for freezing tubes in liquid nitrogen. Such boxes have long open slits on the bottom running the length of each tube row. The tube lids were approximately 2cm below the top of the box allowing for ample aeration but preventing dislodgement in the field. This box was double-bagged in re-sealable, strong plastic storage bags containing either silica beads (BioQuip Productions, Rancho Dominguez, CA) or Drierite (W.A. Hammond Drierite, Xenia, OH) as a desiccant. When simulating high temperatures that may be encountered in the field, the bag was incubated at 37°C for three weeks. In some cases samples were added to tubes containing dried SDS (25μg or 50μg) or dried protease inhibitors. Dried protease inhibitors were prepared by dissolving one Mini-complete protease inhibitor tablet (Roche, Indianapolis, IN) in 7 mls of water and 10μl aliquots were dried in a Speed Vac to be used for each 5μl of sample (~3X final concentration). We also tested the desiccation method on up to 50μl of HEK 293 cell extract with no noticeable difference compared to a 10μl volume (not shown). However, the lowest suitable sample volume for a given field sample is recommended as it will dry more readily.
Proteolytic activity assay and myosin dialysis
Wasp trophallactic fluid was desiccated for three weeks as described above in 10μl, and 5μl, quantities respectively. Equal volumes of water were also desiccated for use as negative controls. All tubes were reconstituted to their original volumes in Milli-Q deionized water. To each sample was added either 1μl of a dialyzed myosin solution (see below), or 1ul of Milli-Q water. Samples were incubated overnight at 37°C. Only myosin was added to control (10μl water) tubes. Degradation of myosin indicated the sample contained proteolytic activity. As a positive control 1μg of trypsin was added to 10μl of water with either 1μl of myosin or 1μl of water. For myosin dialysis, a 250μl solution of porcine myosin purchased in a buffered glycerol solution (Sigma-Aldrich, St. Louis, MO) was dialyzed overnight at 4°C against 1L Milli-Q water using a Tube-O-Dialyzer with a 4,000 dalton molecular weight cutoff (G-Biosciences, Maryland Heights, MO). Aliquots were stored at −20°C until use.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)
SDS-PAGE was performed using hand-cast 4.2% stacking, 10% separating gels (37.5 acrylamide:1 bis-acrylamide). It is important to note that in experiments using SDS as a preservative, the final concentration of SDS in the sample buffer was normalized to 2% (the final concentration for all samples) prior to SDS-PAGE. Gels were stained with Coomassie blue for visualization and scanned.
In-gel tryptic digestion
Bands or regions were cut from stained SDS-PAGE gels and diced into 1mm cubes. Gel pieces were destained with 50mM ammonium bicarbonate (Sigma-Aldrich, St. Louis, MO), 50% acetonitrile (MeCN; Honewell Burdick & Jackson, Morristown, New Jersey), then dehydrated with 100% MeCN and stored at −20°C until future use. The trypsin in-gel digest was performed by adding 30–60μl of 6ng/μl of trypsin in 50mM ammonium bicarbonate and allowing the gel pieces to rehydrate on ice (about 30min), followed by an overnight incubation at 37°C. Digests were centrifuged briefly and the liquid was collected. Peptides were further extracted from the gel pieces by adding 100–200μl of 50% MeCN, 2.5% formic acid (F.A.) and spinning for ten minutes in a microcentrifuge at full speed. The extraction was combined with the previously collected peptides. A final rinse/extraction using 30–60 μl of 100% acetonitrile was performed for five minutes at room temperature. The resulting extracted peptide solution was dried to completion in a Speed Vac, and resuspended in 2.5% MeCN, 2.5% F.A. prior to MS.
Liquid chromatography tandem MS (LC-MS/MS) analysis
LC-MS/MS analysis was carried out largely as described previously 12, 13. Briefly, MS was performed on a linear ion trap (LTQ)-orbitrap hybrid mass spectrometer fitted with a Finnigan Surveyor MS Pump Plus HPLC and a Finnigan Micro AS auto sampler (all instruments from Thermo Fisher Scientific, San Jose, CA). A micro-capillary column was prepared by torching and stretching the tip of a length of fused silica (Polymicro Technologies, Pheonix, AZ) from an original inner diameter of 100μm to an approximate diameter of 4μm (outer diameter was 360μm). The column was then packed with ~14cm of C18 reversed-phase resin (Magic C18AQ; particle size, 5μm; pore size, 200Å; Michrom Bioresources, Auburn, CA). After 15 minutes of isocratic loading at 100% solvent A (2.5% MeCN, 0.15% F.A.) elution was achieved by a steady increase in solvent B (99.85% MeCN, 0.15% F.A.) from 5% to 35% over the course of the following 33 minutes, and ending with five minutes of 100% solvent B. The electrospray voltage was 2.1 kV, and the heated capillary temperature was 225°C. Data acquisition proceeded for the entire run. All spectra were acquired in the orbitrap at 30,000 resolution, using a precursor full scan of 360–1600 m/z followed by 10 data dependent MS/MS scans. Dynamic exclusion was enabled with a repeat count of 3 and a repeat cycle of 180 seconds. Lock mass was enabled and set to calibrate on the mass of a polydimethylcyclosiloxane ion [(Si(CH3)2O)5 +H+]+, m/z = 371.10120.
Data analysis
For HEK 293E cell proteomic analyses, .RAW data files were converted to .mzXML files and were subjected to SEQUEST search analysis as described 13 using a concatenated forward and reverse 14 human IPI database, a 30ppm precursor ion search window, a static modification of cysteine of 71.0371 for acrylamide adduction, and permitting differential oxidation of methionine residues. Search data were filtered using Xcorr cutoff values of 1.8, 2.2 and 2.5 for singly-, doubly- and triply-charged ions respectively; a ΔCn cutoff value of 0.18; and a requirement of two or more peptides identified for each protein. These filters gave a less than 0.01% false peptide discovery rate 14 for each analysis. Prior to data comparison between gel regions, contaminating peptides from trypsin and human skin (e.g. keratin) were removed. For V. vulgaris proteomics analyses .RAW files were converted to .dta.tgz files and were uploaded for PepNovo 4 analysis (website: http://proteomics.ucsd.edu/LiveSearch) with the following specifications: parent ion tolerance of 0.01, MS/MS ion tolerance of 0.01, maximum of 3 PTMs allowed per peptide (with methionine and tryptophan oxidation as the only dynamic PTMs), cysteine was set with a 71.0371 static modification for acrylamide adduction. PepNovo output was converted into fasta format, and a batch BLAST search was performed against Apis mellifera, Nasonia vitripennis, and a non-redundant (NR) database. The peptides with the top 200 PepNovo scores were examined manually to determine the proteins to which the highest number of peptides matched. Once the top five protein hits were identified, the remaining 763 peptides with a PepNovo score over 6 were examined to determine if any additional peptides could be mapped back to one of the top five best proteins matches from the top 200 peptides. MS/MS spectra for peptides mapping back to the top protein were subjected to manual de novo sequencing.
Results and Discussion
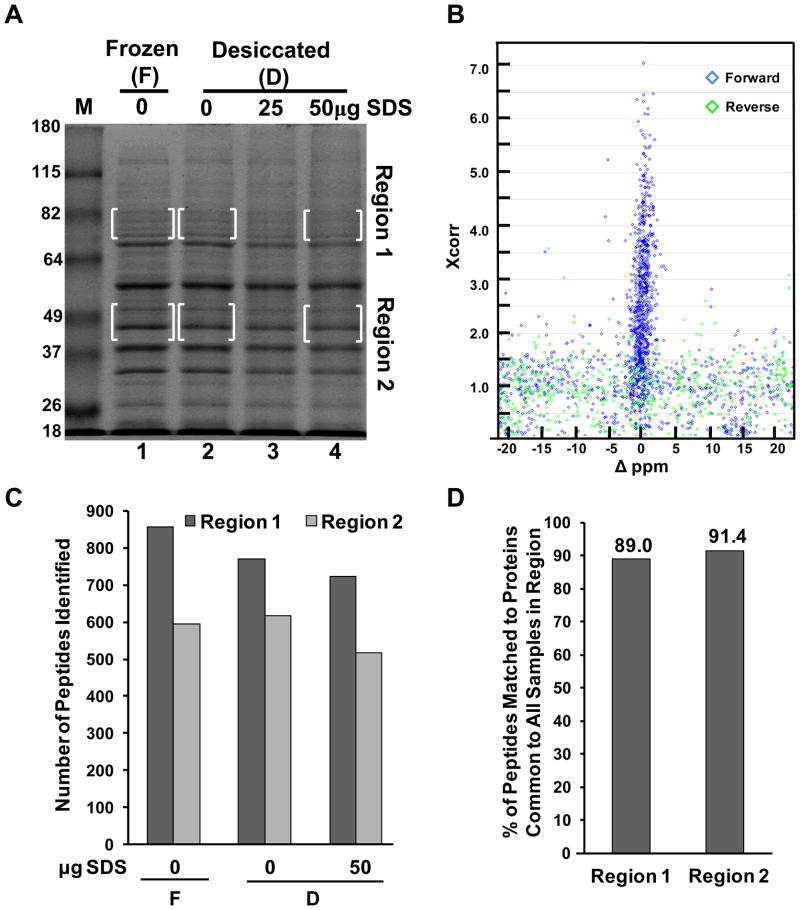
When access to freezing or refrigeration is not possible, desiccation is a simple mechanism to preserve protein samples collected during field expeditions. Toward developing a protein preservation method specifically for vespine trophallactic fluid, which could have applicability to field samples generally, we began with a protein preservation trial in the laboratory. We first generated a complex protein mixture with which to test methodologies that would assess protein preservation for subsequent mass spectrometry analysis. We made a hypotonic extract of HEK 293 cells. Cleared extracts were then aliquoted in 10μl volumes into multiple 0.7ml microcentrifuge tubes, some containing 25 μg or 50μg of dried SDS. We reasoned that SDS could more rapidly desolvate protein constituents and would remain compatible with SDS-PAGE upon return to the laboratory. Aliquots were then either frozen at −80°C directly, or placed into a simple hand-made, portable desiccation chamber (see methods) and left for 3 weeks at a constant temperature of 37°C to mimic the potentially warm temperatures and duration of an extended field expedition. The samples were then resuspended in protein sample loading buffer of different SDS concentrations such that the final SDS concentration for each sample was equal and at 2%. The samples were heated to 95°C for ten minutes and then subjected to SDS-PAGE and the results are shown in Figure 1A. Using the frozen sample as reference, all desiccated samples showed a similar banding pattern when stained with Coomassie blue, although the SDS-containing samples showed a slight reduction in total protein. To determine if proteomic identification would be compromised in the desiccated samples, two regions of the gel were chosen where some bands appeared diminished, particularly in the 50μg SDS lane. Region 1 was chosen for its complexity, higher molecular weight, and also because it was not dominated by a major protein species. Together these attributes would make for a fair challenge to test the efficacy of protein preservation in terms of peptide/protein identification and one might expect some reduction in peptide/protein identification in the SDS-containing samples as lower protein abundance could result in proteins falling below the level of detection of the mass spectrometer. Region 2 was chosen as a less complicated sample and one with largely high abundance proteins. Thus, a small drop in protein abundance would have little effect on protein or peptide identification as the analyte concentrations would be sufficiently above detection levels.
Figure 1.
Desiccation methods for protein preservation of a complex cell extract for MS-based identification. (A) SDS-PAGE and analysis of HEK 293 hypotonic whole cell extract preserved by freezing or desiccated in the presence and absence of SDS as indicated. Bracketed areas indicate gel regions subjected to further analysis via LC-MS/MS. Molecular weight markers in lane “M” are in kilodaltons. The gel was stained with Coomasie. (B) Scatter plot of SEQUEST Xcorr scores vs. Δppm values for each top peptide assignment matching to the forward (blue), or reverse (green) human IPI protein database. Scatter plot is for only one of the six regions analyzed and represents visually (along with Supporting Information Figure 1) the establishment of criteria to filter the data to a less than 0.01% false discovery rate. (C) Summary of the number of peptides identified in each boxed region from Figure 1A. (D) Percent of peptides matching to proteins identified in all three samples for a given region.
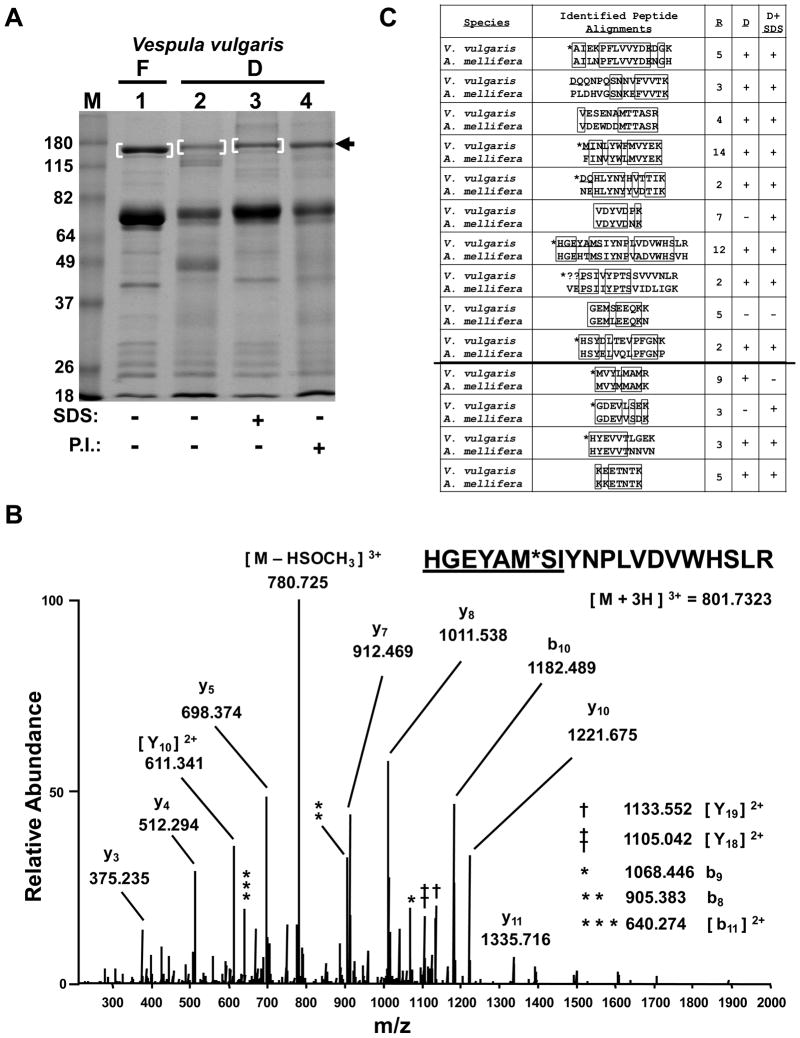
The indicated regions were cut from the gel, cubed and subjected to in-gel tryptic digestion and LC-MS/MS analysis in a linear ion trap-orbitrap hybrid mass spectrometer. Mass spectra were searched against a concatenated forward and reverse human IPI protein database 14. Top SEQUEST hits were plotted using Xcorr values and the difference in ppm of the observed and theoretical precursor masses (Fig. 1B). These parameters as well as the ΔCn scores (Supporting Information Fig. 1) were used to filter the data to a less than 0.01% false discovery rate 14. The total number of peptides identified in each region is indicated in Figure 1C and the percentage of peptides mapping back to proteins common across samples for each region is indicated in Figure 1D. These data suggested that there was an apparent small decrease in peptide identification in the SDS-containing samples from Region 1. However, the number of protein identifications across sample conditions was still rather constant. Taken together these desiccation methods were sufficiently strong at protein preservation for proteomic identification so as to warrant a test of our method on a true field sample. Larval trophallactic fluid of the yellowjacket Vespula vulgaris, a species common in Vermont, was collected as described in the methods. Aliquots were made into empty tubes or tubes containing SDS as was done for the HEK 293 cell extract, except some aliquots were placed directly on dry ice that had been carried into the field. Additionally, other aliquots were placed into tubes containing a dried cocktail of protease inhibitors as another possible preservation method. Aliquots that were not frozen were placed into the portable desiccation chamber, returned to the lab, and the chamber was placed at 37°C for three weeks. Samples were then resuspended in sample buffer and subjected to SDS-PAGE as for Figure 1A. Coomassie staining revealed that each protein preservation method afforded significant protection from proteolysis (Fig. 2A). However, some measure of degradation was observed particularly in the lane without additives. Given that larval trophallactic fluid is known to contain proteolytic activity 11, 15, 16 this was not unexpected. The sample preserved with SDS provided the strongest protection because in addition to more rapid desiccation, it was likely inhibitory to enzymatic activity. Although not as strong as the SDS, the protease inhibitors also afforded protection. In a separate experiment we determined that protease inhibitors kept in the desiccation chamber for three weeks at 37°C provided similar protection to freshly dried protease inhibitors (Supporting Information Fig. 2), but that dried protease inhibitors at room temperature for six months had significantly reduced protection (not shown) consistent with the manufacturer’s product details.
Figure 2.
Preservation of field samples for MS-based protein identification. (A) SDS-PAGE analysis of V. vulgaris larval trophallactic fluid following a 3 week preservation period by either freezing or desiccation, with or without SDS or protease inhibitors (P.I.) as indicated. Bracketed regions and arrow indicate bands chosen for in-gel digestion and LC-MS/MS analysis. “F” and “D” indicate frozen and desiccated respectively. Molecular weight markers in lane “M” are in kilodaltons. The gel was stained with Coomasie. (B) Low energy, high resolution MS/MS spectrum acquired in the orbitrap for the indicated peptide as a triply-charged precursor. “*” indicates an oxidized methionine. The underlined sequence was manually extended from PepNovo results. See Supporting Information Table 1 for theoretical and observed m/z values for b- and y-type ions and (C) for alignment of this peptide with its ortholgous sequence in A. mellifera. (C) PepNovo-identified and manually-validated V. vulgaris peptides matches to the top protein identified, Larval-Specific Very High Density Lipoprotein) from (A) band 1, are aligned with orthologous sequences from A. mellifera. “R” indicates the number of times a given peptide was identified redundantly by PepNovo. “D” and “D + SDS” indicate whether the given peptide was also identified from (A) lanes 2 and 3 respectively. The line separating the final four peptides indicates additional PepNovo sequences not found in the 200 scoring peptides with a PepNovo score > 6 and mapping back to the top protein hit. “*” indicates that the isobaric isoleucine and leucine residues were designated to match the A. mellifera sequence. Boxes indicate amino acid identity. “??” indicates this sequence had an unsolved N-terminal extension of likely two residues.
We next sought to determine if protein identification could be made using the field samples that had been subjected to our preservation regimes. The gel bands indicated in Figure 2A were cut from the gel and subjected to in-gel tryptic digestion. LC-MS/MS analysis of recovered peptides was performed as for the HEK 293 cell proteins. Given that V. vulgaris is a non-model organism without a sequenced genome, it was not surprising that SEQUEST searches against the Apis mellifera (honey bee), Drosophila melanogaster, Nasonia vitripennis (jewel wasp) and a non-redundant database did not reveal a confident identification. We then first analyzed the data from the most preserved, control sample (Fig. 2A, lane 1) using the de novo sequencing software program PepNovo 4. PepNovo results were trimmed to remove contaminant sequences such as trypsin and keratins, and peptide sequences achieving the top 200 PepNovo scores were subjected to BLAST searched in batch format against the A. mellifera or the N. vitripennis protein databases. The results of the BLAST searches, each giving the top ten best matches, were curated manually. The A. mellifera or N. vitripennis protein databases exhibited five proteins that had multiple unique peptide hits. The remaining PepNovo peptides with a score greater than six were then batch BLASTed and examined for the presence of one of the top five proteins identified in the top 200 batch BLASTs. The protein with the greatest number of unique hits had 14, while the four remaining proteins had four or less. This top protein was “larval-specific very high density lipoprotein,” an A. mellifera protein with a predicted molecular weight of 150 kDa, consistent with the molecular weight of the protein excised from the gel as determined by SDS-PAGE (Fig. 2A). The 14 unique peptide hits found for this protein were redundantly found, at least in part, a total of 76 times. We manually inspected each MS/MS spectrum for these 14 unique tryptic peptides to validate the PepNovo assignments and in some cases to extend the PepNovo sequences when PepNovo left an amino- or carboxyl-terminal mass deficiency (gap) off the precursor mass. Figure 2B shows an example MS/MS spectrum for one such peptide where we were able confidently to extend the PepNovo-identified sequence six additional amino acids. Manual validation was greatly aided by having acquired high resolution mass measurements during the MS/MS scans as can be appreciated by examining the theoretical and observed singly- and doubly-charged daughter ions for the MS/MS spectrum shown in Figure 2B (Supporting Information Table 1). Furthermore, the charge state of each fragment ion was readily determined owing to the high resolution measurements in the orbitrap. Importantly, similar results were obtained when analyzing the mass spectra generated from the protein bands from lanes two and three shown in Figure 2A. Indeed most of the 14 unique tryptic peptides found from the peptides derived from the band in lane one were found amongst the peptides originating from the bands in lanes two and three. The 14 V. vulgaris tryptic peptides are aligned with homologous sequences in A. mellifera and shown in Figure 2C. Supporting Information Figure 3 shows the complete amino acid sequence of the larval-specific very high density lipoprotein from A. mellifera and the location of the identified orthologous tryptic peptides from V. vulgaris. Supporting Information Figure 4 shows a second manually-validated and annotated MS/MS spectrum with theoretical and measured mass values shown in Supporting Information Table 2. Of note, despite the high degree of similarity between the V. vulgaris and A. mellifera sequences, none of the tryptic peptides was identical between these species, thus precluding the identification of these peptides using a SEQUEST search and the A. mellifera protein database. Taken together, these results describe methodology capable of preserving field samples for many weeks in warm temperatures. Furthermore, we have shown that using these methods, protein preservation is sufficient for protein identification using LC-MS/MS and subsequent database searching or via de novo sequencing methods when protein databases are not available.
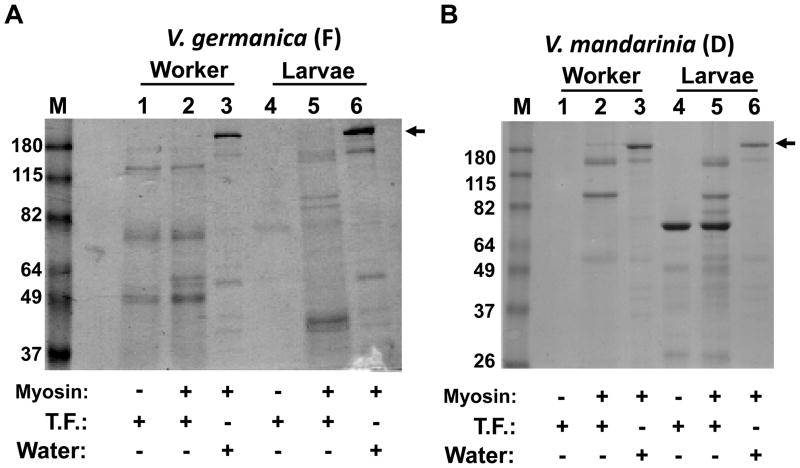
Having established preservation methods facilitating the identification of proteins from field samples, we next sought to determine if these methods were capable of preserving enzymatic activity so as to measure, at least qualitatively, the digestive capacity of larval and adult vespine trophallactic fluid. We reasoned that our methodologies to preserve enzymatic activity could not employ any of the additives used simply for protein preservation, as SDS or protease inhibitors would either fully inhibit or artificially alter proteolytic activity. We procured purified myosin heavy chain and asked if fresh vespine trophallactic fluid could degrade myosin, as myosin would be a major protein in vespine diets. Larval and worker trophallactic samples were collected from a colony of Vespula germancia located in Vermont. Samples were frozen in the field on dry ice and upon returning to the laboratory were challenged for the ability to digest myosin. Trophollactic fluid from both larvae and workers showed significant digestive capacity (Fig. 3A). We next tested for the presence of proteolytic activity in desiccated field samples. Larval and worker trophallactic fluid from Vespa mandarinia was collected in Japan, desiccated, and upon arrival at the US-based laboratory was subjected to the myosin digestion assay and results are shown in Figure 3B. Again, both worker and larval samples showed digestive capacity. Additional samples were collected and desiccated on field expeditions to Singapore, the Southwestern United States, Vermont and Japan. The results are shown in Table 1. To date, each species and animal type (workers and larvae) have tested positive for digestive capacity.
Figure 3.
SDS-PAGE analysis of myosin proteolytic activity assays performed on samples collected in the field. (A) Trophallactic fluid (T.F.) from V. germanica collected in Vermont, frozen (F) on dry ice, then stored at −80°C prior to myosin activity assay and SDS-PAGE. (B) V. mandarinia trophallactic fluid collected in Japan, and stored desiccated (D) until subjected to the myosin activity assay and SDS-PAGE. Arrow heads indicate full length myosin. Molecular weight markers in lane “M” are in kilodaltons. The gel was stained with Coomassie.
Table 1.
Summary of the results of myosin proteolytic activity assays from the 13 Vespine species analyzed. Collection sites are in parenthesis.
| Species | Workers | Larvae |
|---|---|---|
| Vespa ducalis (Japan) | + | + |
| Vespa annalis (Japan) | + | + |
| Vespa mandarinia (Japan) | + | + |
| Vespa affinis (Singapore) | + | ND |
| Vespa similima (Japan) | + | + |
| Provespa anomala (Singapore) | + | ND |
| Dolichovespula arenaria (VT, USA) | + | + |
| Dolichovespula maculata (VT, USA) | + | + |
| Vespula germanica (VT, USA) | + | + |
| Vespula flavapilosa (VT, USA) | + | + |
| Vespula vulgaris (VT, USA) | + | ND |
| Vespula pennsylvanica (NM, USA) | + | + |
| Vespula atropilosa (CO, USA) | + | ND |
“+” indicated the presence of myosin proteolytic activity. “ND” indicates Not Determined as larvae were not tested, because active nests for these species were not found.
While the purpose of this report is primarily to describe the methodology by which proteins from field samples can be successfully preserved for identification and enzymatic analysis, our results regarding vespine biology can be variably interpreted. First it is possible that V. orientalis has undergone such evolutionary divergence so as to become unique among vespines in exhibiting an extreme division of digestive labor. If such is the case, it is likely that specific proteolytic enzymes are sufficiently under-expressed in the workers of V. orientalis such that they are reliant on the larvae for digestion and survival. Given the arduous, but not impossible task of de novo peptide sequencing (Fig. 2B), a complete proteomic comparison of vespine trophallactic fluids will dramatically benefit by whole genome sequencing of vespine species, a goal we are pursuing. Second, it is possible that all vespine workers can digest proteins, at least to a degree, but some species such as V. orientalis may only be capable of limited digestion and therefore remain reliant on larvae for complete digestion. We are currently attempting to collect samples from V. orientalis workers and larvae to determine the degree to which they digest myosin in our assay. Should V. orientalis exhibit digestive capacity similar to what is observed in other vespines, our qualitative assay will be refined to take on a more quantitative approach. The development of a quantitative assay will be in consultation with phylogenetic relationships drawn from morphological and genomic analyses currently underway in our laboratory on a multitude of vespine species including those listed in Table 1. Once these relationships are better understood, we will quantify the digestive capacity of workers and larvae between samples of V. orientalis, our best approximation of its closest living relatives, and more distant living relatives—not hypothesized to exhibit any digestive division of labor—as outgroup controls.
Conclusion
In this report we demonstrate simple, inexpensive, and non-toxic desiccation methods capable of preserving proteins and enzymatic activity during field expeditions of up to three weeks at 37°C. Specifically these methods preserved protein for MS-based identification and for the determination of the presence of proteolytic activity. We have applied these methods to characterize the trophallactic fluid of 13 species of hornets and yellowjackets collected from across the world. While some protein samples collected by investigators in the field may have unique preservation requirements, the methods employed herein should be generally applicable, and enable biologists studying non-model organisms a greater access to proteomic technology which is increasingly bridging diverse biological disciplines.
Supplementary Material
Acknowledgments
We thank Junichi Kojima from Ibaraki University in Mito, Japan for generously providing us with trophallactic fluid from four different species of Vespa. We also thank Rudolf Meier for hosting us in his lab at the National University of Singapore, and we thank Heather Axen at the University of Vermont for help with field collection. This work was supported by National Science Foundation Award DEB0843505 (to K.M.P., J.M.C. and B.A.B) and the Vermont Genetics Network through National Institutes of Health grant P20 RR16462 from the Institutional Development Award Networks of Biomedical Research Excellence (INBRE) Program of the National Center for Research Resources (B.A.B.).
Footnotes
Supporting Information Available
Four supporting information figures and two supporting information tables are available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Freudenstein JV, Pickett KM, Simmons MP, Wenzel JW. From basepairs to birdsongs: phylogenetic data in the age of genomics. Cladistics. 2003;19(4):333–347. [Google Scholar]
- 2.Kluge AG. A concern for evidence and a phylogenetic hypothesis for relationships among Epicrates (Boidae, Serpentes) Syst Biol. 1989;38(1):7–25. [Google Scholar]
- 3.Nixon KC, Carpenter JM. On Simultaneous Analysis. Cladistics. 1996;12(3):221–241. doi: 10.1111/j.1096-0031.1996.tb00010.x. [DOI] [PubMed] [Google Scholar]
- 4.Frank A, Pevzner P. PepNovo: de novo peptide sequencing via probabilistic network modeling. Anal Chem. 2005;77(4):964–73. doi: 10.1021/ac048788h. [DOI] [PubMed] [Google Scholar]
- 5.Shevchenko A, Sunyaev S, Loboda A, Bork P, Ens W, Standing KG. Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time-of-flight mass spectrometry and BLAST homology searching. Anal Chem. 2001;73(9):1917–26. doi: 10.1021/ac0013709. [DOI] [PubMed] [Google Scholar]
- 6.Waridel P, Frank A, Thomas H, Surendranath V, Sunyaev S, Pevzner P, Shevchenko A. Sequence similarity-driven proteomics in organisms with unknown genomes by LC-MS/MS and automated de novo sequencing. Proteomics. 2007;7(14):2318–29. doi: 10.1002/pmic.200700003. [DOI] [PubMed] [Google Scholar]
- 7.Junqueira M, Spirin V, Balbuena TS, Thomas H, Adzhubei I, Sunyaev S, Shevchenko A. Protein identification pipeline for the homology-driven proteomics. J Proteomics. 2008;71(3):346–56. doi: 10.1016/j.jprot.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shevchenko A, Valcu CM, Junqueira M. Tools for exploring the proteomosphere. J Proteomics. 2009;72(2):137–44. doi: 10.1016/j.jprot.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spradbery JP. Wasps; an account of the biology and natural history of solitary and social wasps. University of Washington Press; Seattle: 1973. p. xvi.p. 408. [Google Scholar]
- 10.Ishay J, Ikan R. Food exchange between adults and larvae in Vespa orientalis F. Anim Behav. 1968;16(2):298–303. doi: 10.1016/0003-3472(68)90013-4. [DOI] [PubMed] [Google Scholar]
- 11.Ikan R, Bergmann ED, Ishay J, Gitter S. Proteolytic enzyme activity in the various colony members of the Oriental hornet, Vespa orientalis F. Life Sci. 1968;7(18):929–34. doi: 10.1016/0024-3205(68)90168-9. [DOI] [PubMed] [Google Scholar]
- 12.Haas W, Faherty BK, Gerber SA, Elias JE, Beausoleil SA, Bakalarski CE, Li X, Villen J, Gygi SP. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol Cell Proteomics. 2006;5(7):1326–37. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Ballif BA, Carey GR, Sunyaev SR, Gygi SP. Large-scale identification and evolution indexing of tyrosine phosphorylation sites from murine brain. J Proteome Res. 2008;7(1):311–8. doi: 10.1021/pr0701254. [DOI] [PubMed] [Google Scholar]
- 14.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 15.Sonneborn HH, Pfleiderer G. On the evolution of endopeptidases. VII. A protease of molecular weight 12500 with chymotryptic properties from larvae of Vespa orientalis. Hoppe Seylers Z Physiol Chem. 1969;350(3):389–95. [PubMed] [Google Scholar]
- 16.Hagenmaier HE. Proteolytic enzymes from the digestive tract of the hornet (Vespa orientalis F) Experientia. 1971;27(8):894–5. doi: 10.1007/BF02135728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.