Abstract
To better understand the mechanisms that regulate stem cell identity and function we sought to identify genes that are preferentially expressed by stem cells and critical for their function in multiple tissues. Prdm16 is a transcription factor that regulates leukemogenesis1, palatogenesis2, and brown fat development3–5, but which was not known to be required for stem cell function. We demonstrate that Prdm16 is preferentially expressed by stem cells throughout the nervous and hematopoietic systems and required for their maintenance. Prdm16 deficiency led to changes in reactive oxygen species (ROS) levels, increased cell death, altered cell cycle distribution, and stem cell depletion in the hematopoietic and nervous systems. In neural stem/progenitor cells, Prdm16 bound the Hgf promoter and in the absence of Prdm16 Hgf expression declined. Addition of recombinant HGF to culture partially rescued the increase in ROS levels and the depletion of Prdm16 deficient neural stem cells. Administration of the anti-oxidant, N-acetyl-cysteine, to Prdm16 deficient mice partially rescued defects in neural stem/progenitor cell function and neural development. Prdm16 therefore promotes stem cell maintenance in multiple tissues, partly by modulating oxidative stress.
The development and maintenance of vertebrate tissues depends upon diverse stem cells. Stem cells are functionally distinct from downstream progenitors and likely depend upon regulatory mechanisms that are conserved among stem cells from different tissues but are absent in most downstream progenitors6. Yet few such mechanisms have been identified. FoxO family transcription factors7–10, polycomb family (e.g. Bmi-1) epigenetic regulators11, and DNA repair genes12 are required for the maintenance of stem cells in multiple tissues but are widely expressed within tissues and likely to regulate many cells. Other genes, like Evi113 and Sox1714, are preferentially expressed by stem cells and required for stem cell maintenance in one tissue (in this case the hematopoietic system) but are not known to regulate stem cells in other tissues. Other genes, like Lgr515, 16, are preferentially expressed by stem cells in multiple tissues but it is unknown whether they regulate stem cell function. The identification of genes that are preferentially expressed by stem cells and required to maintain stem cells in multiple tissues would therefore provide important new insights into stem cell identity and function.
To identify genes that regulate the self-renewal of diverse stem cells we performed a Bmi-1 suppressor screen by transposon mutagenesis (data not shown). We reasoned that by screening for suppressors of the Bmi-1 deficiency phenotype we could identify genes that modulate a widely used self-renewal pathway and that might encode new self-renewal regulators required by diverse stem cells. Our screen revealed that over-expression of Prdm16 partially restored the ability of Bmi-1 deficient hematopoietic cells to reconstitute irradiated mice (data not shown). Prdm16 over-expression can contribute to leukemogenesis1, 17 and can increase the ability of cultured hematopoietic stem cells (HSCs) to reconstitute irradiated mice18, though no study has addressed whether Prdm16 is required for stem cell function in any tissue. We decided to test whether Prdm16, like Bmi-1, is physiologically required for stem cell function in multiple tissues.
We reanalyzed previously published gene expression profile data19 and found that Prdm16 was expressed by highly purified HSCs and transiently reconstituting multipotent hematopoietic progenitors (MPPs) but was not detectable in unfractionated hematopoietic cells (Fig. 1a). Quantitative RT-PCR (qPCR) in independent samples yielded similar results as Prdm16 was expressed at 100-fold higher levels in highly purified CD150+CD41−CD48−Sca1+c-kit+ HSCs19 as compared to unfractionated bone marrow cells (Fig. 1b).
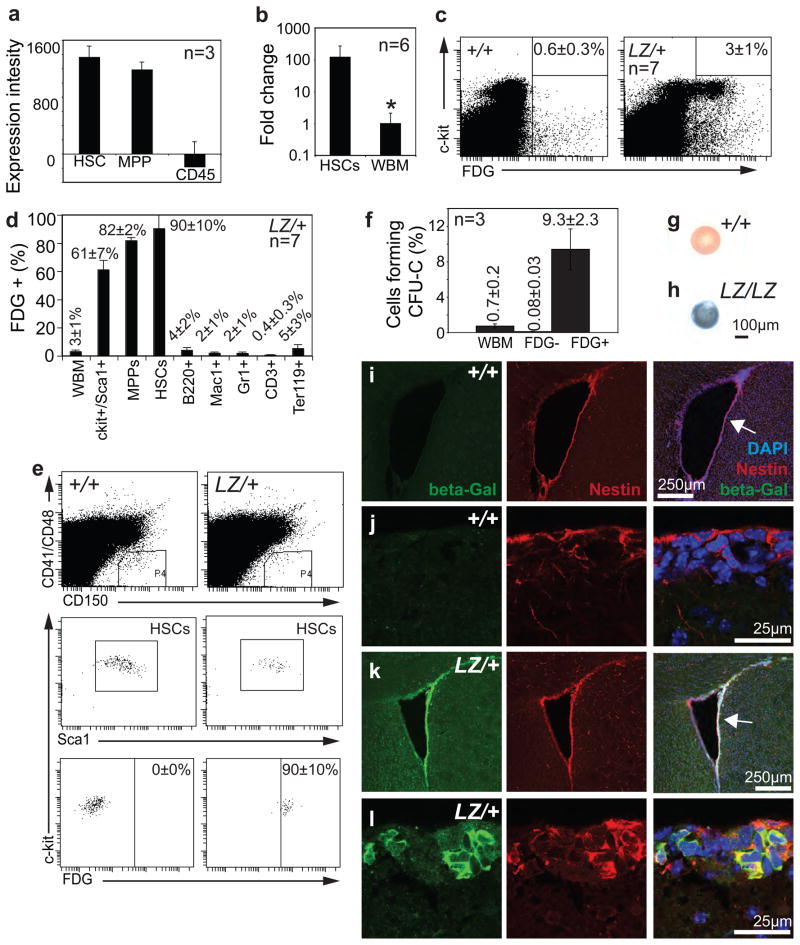
Figure 1. Prdm16 is preferentially expressed by stem cells and primitive progenitors in the hematopoietic and nervous systems.
a) Expression intensity for Prdm16 in HSCs, non-self-renewing multipotent progenitors (MPPs) and CD45+ bone marrow cells from young adult mice (data were extracted from a microarray analysis in an earlier study19). b) Quantitative RT-PCR of cDNA from CD150+CD48−CD41−Sca1+c-kit+ HSCs and unfractionated bone marrow cells from young adult mice in three experiments confirmed Prdm16 is expressed at 100-fold higher levels in HSCs. Samples were normalized using β-actin (*, P=0.029). c) FDG staining for β-galactosidase activity in cells from 2–6 month-old Prdm16LacZ/+ mice indicated that only 3±1% (mean±SD) of bone marrow cells expressed Prdm16. Background staining was observed in 0.6±0.3% of control bone marrow cells. Most of the bone marrow cells from Prdm16LacZ/+ mice that had β-galactosidase activity were c-kit+. d, e) The vast majority of CD150+CD48−CD41−Sca1+c-kit+ HSCs (90±10%; the same surface markers were used to isolate HSCs in subsequent figures) and CD150−CD48−CD41−Sca1+c-kit+ MPPs (82±2%; the same surface markers were used to isolate MPPs in subsequent figures) but few differentiated hematopoietic cells had β-galactosidase activity in Prdm16LacZ/+ mice. f) FDG+ bone marrow cells from Prdm16LacZ/+ adult mice were significantly (p<0.00005) enriched for colony-forming cells (CFU-C) in methylcellulose cultures and contained nearly all colony-forming cells in the bone marrow. The number of independent replicates is indicated in each panel that includes data from multiple independent experiments, and all error bars represent SD. Statistical significance was always assessed by Student’s t-test. g–h) Neurospheres cultured from lateral ventricle VZ cells from newborn Prdm16+/+ (g) or Prdm16LacZ/LacZ (h) mice and stained with X-gal (blue) revealed that virtually all Prdm16LacZ/LacZ neurospheres expressed Prdm16. i–l) Antibody staining for β-galactosidase indicated Prdm16 was expressed in the adult VZ in a pattern that overlaps with the stem/progenitor cell marker Nestin.
To evaluate Prdm16 expression at the single cell level we used Prdm16Gt(OST67423)Lex (further referred to as Prdm16LacZ) genetrap mice in which LacZ was inserted into the first intron of Prdm162. This allele allowed us to assess Prdm16 expression based on β-galactosidase activity but terminated Prdm16 translation after the first exon, causing a loss of Prdm16 function2. Based on staining with the fluorogenic β-galactosidase substrate fluorescein di-β-D-galactopyranoside (FDG), less than 3% of Prdm16LacZ/+ bone marrow cells had β-galactosidase activity (Fig. 1c–d). Most β-galactosidase+ cells were c-kit+ and Sca-1+, markers of HSCs and other primitive progenitors. 61±7% of all c-kit+Sca1+ bone marrow cells were β-galactosidase+ in Prdm16LacZ/+ mice (Fig. 1d). Almost all CD150+CD41−CD48−Sca1+c-kit+ HSCs19 (90±10%) and CD150−CD41−CD48−Sca1+c-kit+ MPPs20 (82±2%) from 2 month-old Prdm16LacZ/+ mice expressed β-galactosidase by flow-cytometry (Fig. 1d, e). In contrast, differentiated B, myeloid, T, and erythroid cells rarely expressed β-galactosidase (Fig. 1d). β-galactosidase+ cells from the bone marrow of 2 month-old Prdm16LacZ/+ mice contained nearly all of the colony-forming cells (CFU-Cs; 0.7±0.2% of bone marrow cells) in Prdm16LacZ/+ bone marrow (Fig. 1f). Prdm16 is thus preferentially expressed by HSCs and other primitive progenitors in the hematopoietic system.
Prdm16 was also preferentially expressed by stem cells in the nervous system. In forebrain sections from adult Prdm16LacZ/+ mice, β-galactosidase strongly overlapped with the stem/progenitor cell marker, Nestin, in the lateral ventricle subventricular zone (SVZ) but much less β-galactosidase staining was observed among the differentiated cells in the striatum and cortex (Fig. 1i–l). Similar results were observed in the newborn forebrain where β-galactosidase overlapped with Nestin in the lateral ventricle ventricular zone (VZ) (Suppl. Fig. 1). Virtually all neurospheres cultured from the Prdm16LacZ/+, but not Prdm16+/+, forebrain exhibited strong β-galactosidase staining (Fig. 1g–h). Cultured and uncultured neural crest stem cells from the enteric nervous system also expressed Prdm16 (Suppl. Fig. 2). Prdm16 is therefore expressed by neural stem/progenitor cells in the central and peripheral nervous systems but expression declines as these cells differentiate.
We next tested whether Prdm16 is required to regulate HSCs. Prdm16LacZ/LacZ mice were born at Mendelian frequency but died soon after birth (Suppl. Fig. 3a, b). Hematopoiesis was grossly normal in newborn Prdm16LacZ/LacZ mice as liver and spleen cellularity were normal (Fig. 2a). No significant differences were detected in the frequency of myeloid, B, or T cells in the livers or spleens of newborn Prdm16LacZ/LacZ, Prdm16LacZ/+, and Prdm16+/+ mice (Fig 2b, Suppl. Fig. 3c). Most colony-forming progenitors were present at normal frequencies in the neonatal liver of Prdm16LacZ/LacZ mice (Fig. 2c). However, mixed myeloerythroid (CFU-GEMM) and mixed myeloid (CFU-GM) progenitors were significantly depleted in the livers of neonatal Prdm16LacZ/LacZ mice (Fig. 2c). The frequency of CD150+CD41−CD48−Sca1+c-kit+ HSCs in the embryonic day (E)14.5 liver (Fig. 2d), newborn (P0) liver (Fig. 2d–e), and newborn spleen (Suppl. Fig. 3d) was reduced by approximately 2-fold in Prdm16LacZ/+ mice and by approximately 20-fold in Prdm16LacZ/LacZ mice compared to Prdm16+/+ littermates. Loss of Prdm16 therefore profoundly depletes HSCs without depleting most downstream hematopoietic progenitors.
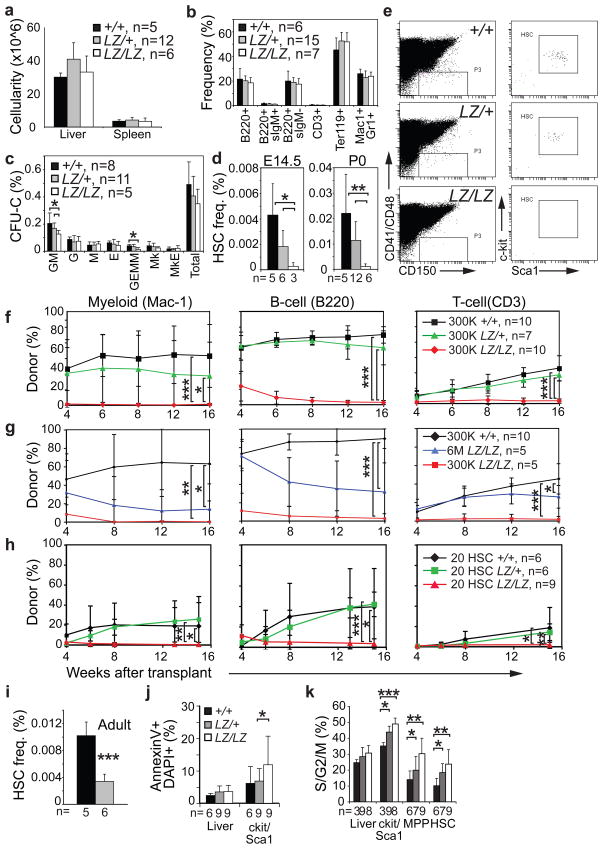
Figure 2. Prdm16 is required for survival, cell cycle regulation, and maintenance in fetal and adult HSCs.
a) The cellularity of the liver and spleen and b) the frequencies of mature hematopoietic cells in the liver were normal in newborn Prdm16LacZ/LacZ mice (3 independent experiments with a total of 5–12 mice/genotype; the exact number of mice is indicated in the panel legend). c) The frequencies of most colony-forming progenitors were normal in the liver of Prdm16LacZ/LacZ mice, but CFU-GEMM and CFU-GM were significantly depleted. d) HSCs were depleted in the fetal (E14.5) and newborn (P0) liver of Prdm16LacZ/LacZ mice (4 independent experiments with a total of 3–12 mice/genotype; the exact number of mice is indicated under each bar). e) c-kit and Sca-1 staining is shown in the right column for CD150+CD48−CD41− cells gated in the left column for representative mice of each genotype. f) Irradiated CD45.1+ recipient mice were competitively reconstituted with 3×105 CD45.2+ neonatal liver cells from Prdm16+/+ (black lines), Prdm16LacZ/+ (green lines) or Prdm16LacZ/LacZ mice (red lines) along with 3×105 CD45.1+ recipient bone marrow cells. Each line represents average donor cell reconstitution levels (mean±SD; 3 independent experiments with 2–5 recipients/treatment/experiment; the total number of mice transplanted with cells of each genotype is inducted in panel legends for f–h). g) Donor cell reconstitution in an independent experiment in which recipients were transplanted with 3×105 CD45.2+ neonatal liver cells from a Prdm16+/+ donor (black lines), or 3×105 cells from a Prdm16LacZ/LacZ donor (red lines), or 6×106 cells from Prdm16LacZ/LacZ donor (blue lines) along with 3×105 young adult CD45.1+ bone marrow cells (1 experiment with at least 5 recipients/treatment). h) Donor cell reconstitution in 2 experiments in which recipients were transplanted with 20 CD45.2+ HSCs from Prdm16+/+ (black lines), Prdm16LacZ/+ (green lines), or Prdm16LacZ/LacZ donors (red lines) along with 3×105 CD45.1+ bone marrow cells (2 independent experiments). i) Bone marrow HSC frequency in adult Prdm16+/+ and Prdm16LacZ/+ mice (2 experiments with 5 or 6 mice/genotype). j) The frequency of annexin V+/DAPI+ unfractionated newborn liver cells or c-kit+Sca-1+ cells (5 independent experiments with 6 or 9 mice/genotype). k) Cell cycle distribution of cells from newborn liver (3 independent experiments with 3–9 mice/genotype). In panels i–k, the exact number of mice of each genotype is indicated under each bar in each panel (*, P<0.05; **, P<0.01; ***, P<0.001, error bars always represent SD).
To assess HSC function we performed competitive long-term reconstitution assays using neonatal liver cells from Prdm16LacZ/LacZ mice and littermate controls. 300,000 donor (CD45.2+) Prdm16+/+ cells or Prdm16LacZ/+ cells gave long-term multilineage reconstitution by myeloid, B, and T cells in all irradiated recipient (CD45.1+) mice (Fig. 2f). In contrast, 300,000 Prdm16LacZ/LacZ cells did not give long-term multilineage reconstitution in any recipients. All recipients of Prdm16LacZ/LacZ cells had low levels of transient reconstitution by donor B cells, and most had very low levels of transient myeloid or T cell reconstitution (Fig. 2f).
Since Prdm16LacZ/LacZ mice exhibited a 20-fold depletion of HSCs (Fig. 2d), we performed an additional experiment in which a 20-fold excess of donor (CD45.2+) cells from newborn Prdm16LacZ/LacZ mice was transplanted into irradiated wild-type recipients (CD45.1+) along with 300,000 recipient bone marrow cells. As in prior experiments, 300,000 Prdm16LacZ/LacZ cells gave poor reconstitution in all lineages while 300,000 Prdm16+/+ cells gave long-term multilineage reconstitution in all recipients (Fig. 2g). 6×106 neonatal Prdm16LacZ/LacZ liver cells gave long-term multilineage reconstitution in all recipients, but the levels of donor cell reconstitution were significantly lower than from 300,000 Prdm16+/+ control cells and were declining by 16 weeks after transplantation (Fig. 2g). Some HSC activity thus remains in newborn Prdm16LacZ/LacZ mice, and can sustain hematopoiesis, but these HSCs are greatly depleted.
To further test whether the residual Prdm16LacZ/LacZ HSCs were defective we transplanted 20 CD150+CD41−CD48−Sca1+c-kit+ HSCs from Prdm16LacZ/LacZ, Prdm16LacZ/+, or Prdm16+/+ neonates into irradiated recipients along with 300,000 recipient bone marrow cells. Four of 10 recipients of Prdm16+/+ HSCs and 6 of 10 recipients of Prdm16LacZ/+ HSCs, but none of 9 recipients of Prdm16Lacz/LacZ HSCs, were long-term multilineage reconstituted (Fig. 2h). The levels of donor cell reconstitution by Prdm16Lacz/LacZ HSCs were significantly lower than from control HSCs in all lineages (Fig. 2h). Even highly enriched HSC populations therefore have little reconstituting capacity in the absence of Prdm16. Furthermore, the frequency of HSCs in adult Prdm16LacZ/+ bone marrow was significantly reduced (3-fold) relative to Prdm16+/+ bone marrow (Fig. 2i). Prdm16 is therefore required for the maintenance of fetal and adult HSCs.
Prdm16 is required for normal cell cycle regulation and survival in HSCs and other primitive hematopoietic progenitors. Prdm16 deficiency significantly increased the frequency of c-kit+Sca-1+ cells, but not unfractionated liver cells, that stained positively for AnnexinV and DAPI (Fig. 2j; we could not evaluate highly purified HSCs in this assay because they were too rare in Prdm16Lacz/LacZ mice). Prdm16 deficiency also significantly increased the frequency of c-kit+Sca-1+ cells, but not unfractionated liver cells, that stained positively for activated caspase-3 (data not shown). Prdm16 deficiency significantly increased the frequency of HSCs, MPPs, and c-kit+Sca-1+ cells, but not unfractionated liver cells, in S/G2/M phase of the cell cycle (Fig. 2k).
Prdm16 also regulates neural development. Brain mass was significantly reduced in Prdm16LacZ/LacZ mice compared to littermate controls (Fig. 3a). Prdm16LacZ/LacZ forebrains had a thinner cortex (see brackets in Fig. 3c versus 3b), narrower ventricles (see arrowheads in Fig. 3c versus 3b), and agenesis of the corpus callosum (see arrows in Fig. 3c versus 3b, confirmed by serial sections). Axons in Prdm16LacZ/LacZ mice formed Probst bundles21 instead of crossing the midline (see * in Fig. 3e; see arrows in Fig. 3d versus 3e).
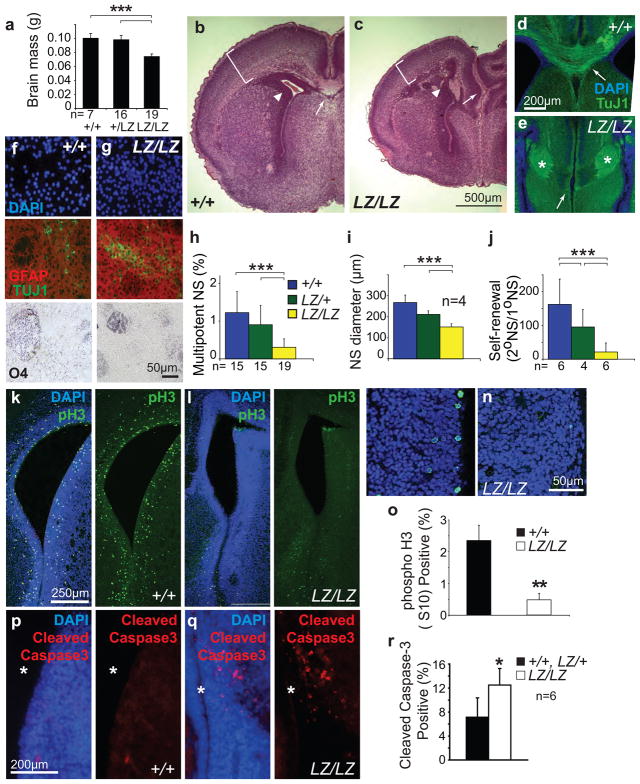
Figure 3. Prdm16 is required for survival, cell cycle regulation, and self-renewal in neural stem cells.
a) The brains of neonatal Prdm16LacZ/LacZ mice were significantly smaller than those of Prdm16+/+ or Prdm16LacZ/+ mice (the number of mice of each genotype is shown under each bar). b–c) Hematoxylin and eosin staining of coronal sections showed that Prdm16LacZ/LacZ brains (c) were smaller and that morphology was disrupted relative to control brains (b). Brackets show reduced cortical thickness, arrowheads show narrower lateral ventricle, and arrows point to the lack of a corpus callosum in the Prdm16LacZ/LacZ brain. d–e) Agenesis of the corpus callosum was confirmed by staining with the neuronal marker TuJ1 which revealed axon tracts that crossed the midline in wild-type (arrow, d) but Probst bundles that did not cross the midline in Prdm16LacZ/LacZ brains (*, e). f–j) Some primary neurospheres of all genotypes underwent multilineage differentiation, forming neurons (TuJ1+), astrocytes (GFAP+), and oligodendrocytes (O4+); however, a significantly lower percentage of VZ cells from newborn Prdm16LacZ/LacZ mice formed multipotent neurospheres compared to littermate controls (h) and the diameter (i) and self-renewal potential (j) of Prdm16LacZ/LacZ neurospheres was significantly less than control neurospheres (the number of mice per genotype is indicated in each panel; panels h, i, and j reflect 6, 4, and 3 independent experiments). Self-renewal was quantified as the number of multipotent secondary neurospheres generated upon the subcloning of individual primary neurospheres. k–o) Significantly fewer dividing cells were observed in the VZ of newborn Prdm16LacZ/LacZ mice compared to littermate controls based on phospho-histone3 (pH3) staining (3 mice per genotype and 4 sections per mouse). p–r) Significantly more cells underwent cell death in the lateral ventricle VZ of newborn Prdm16LacZ/LacZ mice compared to littermate controls (* marks lateral ventricle) in sections (p–q), and by flow cytometry (6 mice/genotype from 3 independent experiments). (*, P<0.05; **, P<0.01; ***, P<0.001, error bars always represent SD).
To test whether Prdm16 regulates neural stem cell function we cultured VZ cells from newborn Prdm16LacZ/LacZ mice and littermate controls at clonal densities (<1 cell/μl). Prdm16LacZ/LacZ VZ cells formed neurospheres that underwent multilineage differentiation (Fig. 3f, g). However, the frequency of VZ cells that formed multilineage colonies in culture was significantly reduced in Prdm16LacZ/LacZ mice (Fig. 3h). Prdm16LacZ/LacZ neurospheres were also significantly smaller than Prdm16+/+ and Prdm16LacZ/+ neurospheres (Fig. 3i). The self-renewal potential of Prdm16LacZ/LacZ neural stem cells was significantly less than control stem cells based on the number of multipotent daughter cells that could be subcloned from individual primary neurospheres (Fig. 3j). Prdm16 was similarly required for stem cell function in the peripheral nervous system (Suppl. Fig. 2c,d). Prdm16 deficiency thus reduces self-renewal potential and depletes neural stem cells, similar to the defects observed in the hematopoietic system.
To test whether we could observe neural stem/progenitor cell defects in vivo we stained sections through the forebrain of newborn Prdm16LacZ/LacZ mice and littermate controls with an antibody against phospho-Histone3 (pH3) to identify mitotic cells. We observed significantly fewer pH3+ cells in the lateral ventricle VZ of Prdm16LacZ/LacZ mice compared to littermate controls (Fig. 3k–o). We also observed a significantly increased frequency of activated caspase-3+ cells among Prdm16LacZ/LacZ VZ cells (Fig. 3p–r). These effects were more pronounced in the dorsal VZ than in the ventral VZ. Prdm16 is therefore required for normal cell cycle regulation and survival in neural stem/progenitor cells.
To investigate the underlying mechanisms we compared the gene expression profiles of uncultured VZ cells from newborn Prdm16LacZ/LacZ, Prdm16LacZ/+, and Prdm16+/+ mice (3 independent samples per genotype). Thirteen genes were significantly reduced in expression (Fig 4a) and six were significantly increased in expression (Suppl. Fig. 3e) in the Prdm16LacZ/LacZ VZ (at least 2.2-fold different (p<0.05) between Prdm16LacZ/LacZ and Prdm16+/+ VZ and at least 1.8-fold different (p<0.05) between Prdm16LacZ/LacZ and Prdm16LacZ/+ VZ). These differences were confirmed by qPCR in 3 independent samples per genotype (Fig. 4a; Suppl. Fig. 3e). Several of these genes regulate the generation of ROS or the response to oxidative stress (see * in Fig. 4a). In particular, Hepatocyte growth factor (Hgf), and Metallothienin2 (Mt2) expression were reduced in Prdm16LacZ/LacZ VZ (Fig. 4a). The reduction in Hgf expression in the absence of Prdm16 was further confirmed in neurospheres cultured from the newborn VZ (Fig. 4b). These changes would be predicted to increase ROS levels as MT2 scavenges free radicals22 and HGF can protect cells from oxidative stress by reducing ROS levels23, 24. Metallothionein1 (Mt1) expression was also reduced in the Prdm16LacZ/LacZ VZ (1.7-fold by microarray and 3.7-fold by qPCR; data not shown). Consistent with these predictions, DCFDA staining (an indicator of ROS levels) significantly increased in newborn Prdm16LacZ/LacZ VZ cells as compared to littermate controls (Fig. 4c–d). Although Prdm16LacZ/LacZ VZ cells had increased ROS levels, we did not detect any defects in mitochondrial mass or membrane potential (Suppl. Fig. 4c, d). Prdm16 is therefore required to control the expression of genes that regulate ROS levels in neural stem/progenitor cells and to avoid oxidative stress.
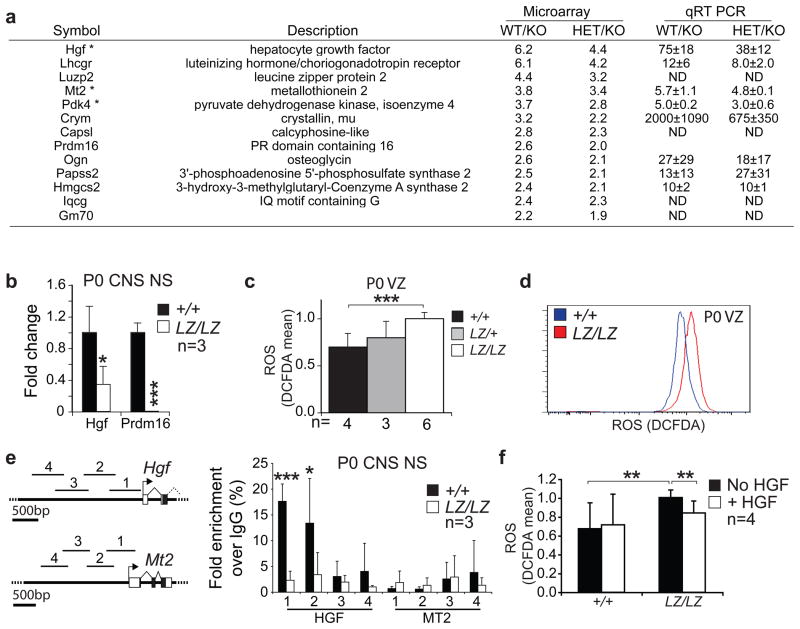
Figure 4. Prdm16 promotes the expression of Hgf and regulates ROS levels in neural stem/progenitor cells.
a) The gene expression profiles of VZ cells from newborn Prdm16+/+, Prdm16LacZ/+, and Prdm16LacZ/LacZ mice were compared by microarray (3 independent samples per genotype). The list shows all genes that were significantly (p<0.05) reduced in expression within Prdm16LacZ/LacZ VZ cells as compared to control cells (by at least 2.2 fold between Prdm16+/+ and Prdm16LacZ/LacZ VZ and at least 1.8 fold between Prdm16LacZ/+ and Prdm16LacZ/LacZ cells). Asterisks indicate genes associated with ROS regulation or response to oxidative stress. Differential expression was confirmed by qPCR in 3 independent samples/genotype. Genes that increased in expression in Prdm16LacZ/LacZ VZ cells are shown in Suppl. Fig. 3e. d) Hgf expression was confirmed to decline by qPCR in neurospheres cultured from Prdm16LacZ/LacZ mice. c–d) Newborn Prdm16LacZ/LacZ VZ cells had significantly and uniformly increased ROS levels based on DCFDA staining. e) Prdm16 bound to the promoter of Hgf but not Mt2. Chromatin immunoprecipitation was conducted using anti-Prdm16 antibodies and primary neurospheres from Prdm16+/+ and Prdm16LacZ/LacZ mice. qPCR was used to quantify the immunoprecipitated Hgf and Mt2 promoter regions indicated on the schematics. Data are shown as fold enrichment over control IgG immunoprecipitation. Statistical significance was determined by paired T-tests comparing fold enrichment of target sequences immunoprecipitated by anti-Prdm16/MT2 antibody versus input DNA compared to control IgG versus input DNA. f) HGF treatment reduced ROS levels in neurospheres grown adherently for 5–6 days. HGF was added to the cultures 20 hours before ROS measurement. The number of independent experiments is indicated in each panel, or on each bar in panel c to indicate the number of experiments in which mice of the indicated genotype were used. *, P<0.05; **, P<0.01; ***, P<0.001, error bars always represent SD.
Chromatin immunoprecipitation experiments were conducted to test if Prdm16 directly binds the promoters of Hgf and Mt2. Two of four sequences 5′ of the Hgf start codon were immunoprecipitated from Prdm16+/+, but not Prdm16LacZ/LacZ, neurospheres using anti-Prdm16 antibodies (Fig 4e). In contrast, no Prdm16 binding was detected at the Mt2 promoter. These data suggest that Hgf, but not Mt2, is a direct target of Prdm16.
To test whether HGF regulates oxidative stress in neural stem/progenitor cells we added recombinant HGF to neurospheres cultured from the VZ of newborn Prdm16LacZ/LacZ mice and littermate controls. The Prdm16LacZ/LacZ cells exhibited significantly higher levels of DCFDA staining but HGF treatment significantly reduced DCFDA staining in these cells (Fig. 4f). These data suggest that HGF regulates ROS levels in neural stem/progenitor cells and that Prdm16 regulates oxidative stress in neural stem/progenitor cells partly by regulating Hgf expression.
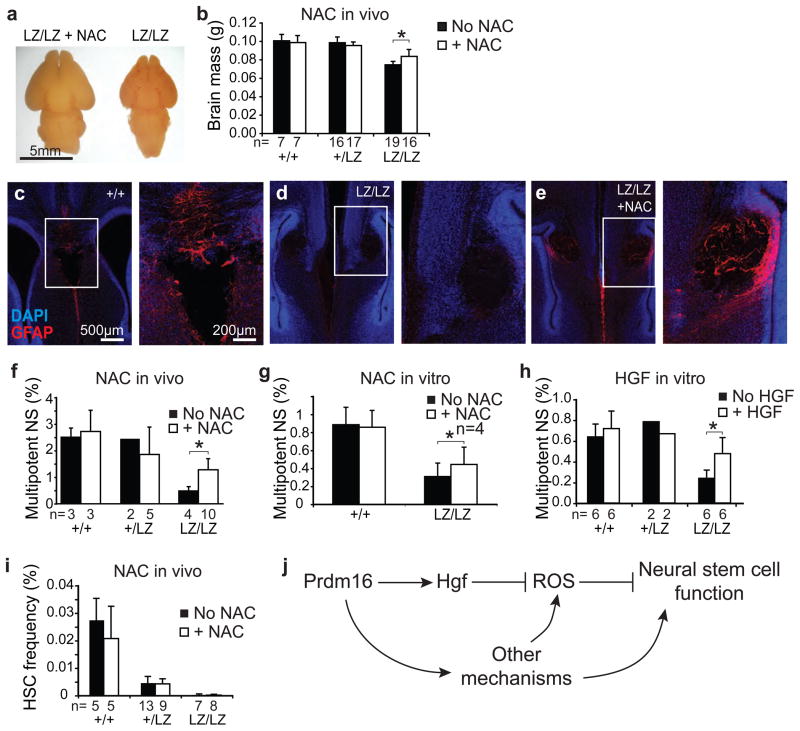
To test whether oxidative stress contributed to the defects in neural stem/progenitor cell function and neural development we administered the anti-oxidant N-acetyl-cysteine (NAC) to pregnant Prdm16LacZ/+ mice. NAC significantly increased brain size in newborn Prdm16LacZ/LacZ mice (Fig. 5a, b) but did not affect brain size in Prdm16LacZ/+ or Prdm16+/+ mice (Fig 5b). In wild-type mice, glial fibrillary acidic protein (GFAP) expressing astrocytes were evident around the midline and the corpus callosum at birth (Fig. 5c) but many fewer GFAP+ cells were observed around the midline of Prdm16LacZ/LacZ mice (Fig. 5d). Prdm16LacZ/LacZ mice treated with NAC in utero had more GFAP+ cells around the midline in the forebrain (Fig. 5e), though the corpus callosum still did not develop in these mice. NAC treatment in utero (Fig 5f) or in culture (Fig. 5g) also significantly increased the frequency of newborn Prdm16LacZ/LacZ VZ cells, but not Prdm16LacZ/+ or Prdm16+/+ VZ cells, that formed multipotent neurospheres in culture. Oxidative stress therefore contributes to the defects in neural development in Prdm16LacZ/LacZ mice.
Figure 5. Prdm16 promotes neural stem/progenitor cell function by regulating Hgf expression and ROS levels.
NAC treatment significantly increased brain size (a) and mass (b) in Prdm16LacZ/LacZ mice but not in littermate controls (the number of mice per treatment is indicated under each bar; each panel reflects at least 3 independent experiments). c–e) Newborn Prdm16LacZ/LacZ mice (d) had reduced GFAP staining (red) in the midline relative to control littermates (c) but this phenotype was partially rescued by NAC treatment (e). Higher magnification images of the boxed regions are shown to the right. f) NAC treatment in utero significantly (by paired t-test) increased the percentage of newborn Prdm16LacZ/LacZ VZ cells that formed multipotent neurospheres in culture. Addition of NAC (g) or HGF (h) to culture significantly increased the percentage of newborn Prdm16LacZ/LacZ VZ cells that formed multipotent neurospheres. i) NAC treatment of pregnant mice did not rescue the depletion of HSCs in Prdm16LacZ/LacZ mice. (*, P<0.05; **, P<0.01; ***, P<0.001, error bars always represent SD). j) A model of Prdm16 function in neural stem cells.
Since Prdm16 promotes Hgf expression in neural stem/progenitor cells (Fig. 4a,b,e), HGF regulates ROS levels in these cells (Fig. 4f), and increased ROS levels contribute to the defects in neural stem/progenitor cell function in Prdm16LacZ/LacZ mice (Fig. 5), we tested whether addition of HGF to culture could partially rescue the defects in Prdm16LacZ/LacZ neural stem cell function. Addition of HGF significantly increased the frequency of Prdm16LacZ/LacZ, but not Prdm16LacZ/+ or Prdm16+/+, VZ cells that formed multipotent neurospheres (Fig. 5h).
To test whether loss of Prdm16 changes ROS levels within primitive hematopoietic progenitors, we stained c-kit+Sca-1+ stem/progenitor cells from newborn Prdm16LacZ/LacZ mice with DCFDA. Surprisingly, we observed a significant decline in ROS levels within Prdm16LacZ/LacZ c-kit+Sca-1+ liver cells compared to control cells while ROS levels remained unchanged in unfractionated liver cells (Suppl. Fig. 4a–b). Thus, Prdm16 is also required to regulate ROS levels in primitive hematopoietic progenitors, though loss of Prdm16 appeared to decrease ROS levels in these cells, in contrast to the nervous system. Furthermore, NAC treatment of pregnant mice did not rescue the depletion of CD150+CD41−CD48−Sca1+c-kit+ HSCs in Prdm16LacZ/+ or Prdm16LacZ/LacZ mice (Fig. 5i). Prdm16 might promote stem cell maintenance by different mechanisms in different tissues. Alternatively, there could be an increase in ROS levels in Prdm16LacZ/LacZ HSCs that is masked by other changes that occur among the heterogeneous c-kit+Sca-1+ cells. Unfortunately, this is impossible to test because too few HSCs can be recovered from Prdm16LacZ/LacZ mice (Fig. 2d) to assess DCFDA staining. In either case, the failure of NAC to rescue the depletion of Prdm16LacZ/LacZ HSCs suggests that Prdm16 has other critical functions in HSCs beyond regulating oxidative stress. Finally, compensatory changes might also be induced in response to oxidative stress25 in either HSCs or neural stem/progenitor cells that alter the observed effects of Prdm16 deficiency on ROS levels.
Our results demonstrate that Prdm16 is required for stem cell function in the nervous and hematopoietic systems (Fig. 2, 3). In the central nervous system, Prdm16 appears to promote neural stem/progenitor cell function partly by promoting HGF expression (Fig. 4a,b,e) and regulating ROS levels (Fig. 4f). However, Prdm16 likely regulates ROS levels and stem cell maintenance by other mechanisms as well. Our results emphasize the critical and diverse mechanisms required by stem cells to regulate oxidative stress. Bmi-1 also regulates ROS levels in stem cells26, but Prdm16 does not appear to regulate Bmi-1 expression or vice versa (data not shown). Considerable additional work will be required to fully elucidate the mechanisms by which Prdm16 regulates oxidative stress and stem cell maintenance.
METHODS
Mice
Prdm16Gt(OST67423)Lex genetrap mice were obtained from the NIH Mutant Mouse Regional Resource Center (http://www.mmrrc.org/). C57BL/Ka-CD45.1:Thy-1.2 mice were recipients in reconstitution experiments. All mice were backcrossed onto a C57BL/Ka background.
Reconstitution assay
Single cell suspensions of liver cells from neonatal mice were transplanted into recipient mice by retro-orbital venous sinus injection. Eight week-old recipient mice were irradiated in two doses at least two hours apart with a total of 1140 cGy using a Cesium137 GammaCell40 Irradiator (MDS Nordia, Kanata, ON, Canada). Primary transplants were performed with 300,000 unfractionated recipient bone marrow cells for radioprotection.
Flow cytometry
HSCs were analyzed and isolated as previously described 19. CD150+CD41−CD48−Sca1+c-kit+ HSCs were isolated using anti-CD150 (TC15-12F12.2-PE; BioLegend, San Diego, CA), anti-CD41 (MWReg30-FITC; BD Pharmingen, San Diego, CA), anti-CD48 (HM48-1-FITC; BioLegend), anti-Sca-1 (E13-6.7-APC), and anti-c-kit (2B8-biotin, eBiosciences, San Diego, CA), followed by Streptavidin APC-Cy7. HSCs were enriched using an AutoMacs (Miltenyi, Auburn, CA) with anti-biotin paramagnetic beads. The medium was supplemented with 200μM verapamil and 200μM chloroquine (Sigma) and incubated at 37°C for 10 minutes. An equal volume of pre-warmed deionized water with or without 200μM FDG was added to the cells and incubated at 37°C for 5 minutes. The reaction was terminated by addition of 5 volumes of ice cold staining medium with verapamil and chloroquine. All analyses excluded dead cells that stained positively for DAPI (1μg/ml) (Sigma, St. Louis, MO).
For staining of gut neural crest cells, outer muscle/plexus layer cells from the gut were dissected and dissociated as described below. Cells were stained with antibodies against p75 (Millipore, Billerica, MA) and CD49b (eBioscience, DX5). Cells were analyzed and sorted with FACS Vantage SE, FACS Aria, or FACS Canto II flow-cytometers (BD Biosciences).
Cell cycle status was determined in hematopoietic cells by using DAPI to stain DNA content. Stained hematopoietic cells were fixed in CytoFix/Cytoperm (BD Biosciences) and stained with 5 μg/ml DAPI. APC-Annexin V (BD biosciences) was used to quantify apoptotic cells following the manufacturer’s instructions.
ROS levels were measured by incubating 2×106 antibody stained hematopoietic cells with 5 μM 2′-7′-dichlorofluorescein diacetate (DCFDA, Molecular Probes, Eugene, OR) for 15 min at 37°C. Mitochondrial mass was measured by incubating 2×106 cells with 1 nM Mitotracker Deep Red (Molecular Probes) and 50μM verapamil (Sigma) for 15 min at 37 °C. Mitochondrial membrane potential was measured by incubating 2×106 cells with 25 nM TMRM for 15 min at 37°C. Freshly isolated VZ cells were stained for 5 min at 37°C with vital dyes to preserve viability.
Colony formation assays
Four hundred live neonatal liver or spleen cells, or single HSCs were sorted per well of a 96 well plate (Corning, Corning, NY) containing 100μl of MethoCult M3434 medium (StemCell Technologies). The medium was supplemented with 1% penicillin/streptomycin, 10ng/ml Flt-3, and 10ng/ml thrombopoietin (R&D Systems). Colonies were counted after 12 days incubation at 37°C in 6% CO2. We analyzed 32 wells per sample.
Quantitative (real-time) RT-PCR
Cells were harvested or sorted into trizol and RNA was isolated using an RNeazy Mini Kit (Qiagen Sciences, MD). cDNA was made with oligo dT primers and SuperScript II reverse transcriptase (Invitrogen). Quantitative PCR was performed with cDNA from using a SYBR Green Kit and a LightCycler 480 (Roche). Each sample was normalized to β-actin and amplification products were tested for specificity by analyzing melting curves, band size by gel electrophoresis, and amplicon sequencing.
CNS stem cell culture, differentiation and self-renewal assays
CNS and PNS cells were dissociated and cultured as previously described27. To isolate CNS VZ cells, ventricular zone cells were dissected from the lateral wall of the lateral ventricle of newborn Prdm16LacZ/LacZ, Prdm16LacZ/+ and Prdm16+/+ mice. The cells were dissociated in 1ml of a 1:4 mixture of 0.025% trypsin/EDTA (Calbiochem, San Diego, CA) and Ca-, Mg-free HBSS for 4 min at 37°C. Dissociation was quenched with 2 volumes of L15 medium containing 10 mM HEPES (pH 7.4), 1 mg/ml BSA, and 25 μg/ml DNAse1. For non-adherent cultures, cells were plated at 1000 cells per well in 1.5 ml of self-renewal medium in ultra low binding six-well plates (Corning). Self-renewal medium contained a 5:3 mixture of DMEM low glucose and neurobasal medium, 1 μg/ml penicillin/streptomycin (GIBCO), 10% chick embryo extract (CEE), 2% B27 supplement, 1% N2 supplement (GIBCO), 50 μM 2-mercaptoethanol, 20 ng/ml bFGF, and 20 ng/ml EGF (R&D Systems). After 9 days culture in self-renewal medium single neurospheres larger than 50 μm in diameter were counted and transferred into differentiation medium for another 8–10 days. The differentiation phase of the culture was performed adherently in 48 well plates (1 neurosphere per well) coated with 150 μg/ml poly-D-lysine (Biomedical Technologies, Sloughton, MA) and 20μg/ml laminin (Invitrogen). Differentiation medium was the same as self-renewal medium except that it contained 5% fetal bovine serum (FBS) instead of CEE, no EGF, and 10ng/ml of bFGF instead of 20 ng/ml. Differentiated colonies were tested for multipotency by staining with neuron (Tuj1, Covance, Princeton, NJ), astrocyte (GFAP, Sigma) and oligodendrocyte (O4) markers as described in the immunochemistry section below.
To assay self-renewal potential, individual primary neurospheres (>50μm in diameter) were dissociated and re-plated at clonal density in non-adherent secondary cultures. Secondary neurospheres were counted 7–9 days later and transferred to adherent cultures containing differentiation medium to assess their ability to undergo multilineage differentiation as described above. Self-renewal was quantified as the number of multipotent secondary colonies that were subcloned per primary neurosphere.
CNS tissue processing and immunochemistry
Newborn mouse brains were fixed in 4% paraformaldehyde, cryoprotected in 30% sucrose, embedded in Cryo-Gel embedding medium (Instrumedics, St. Louis, MO) and flash frozen. 40 μm floating sections were collected and stained with anti-β-galactosidase (ab9361, Abcam, Cambridge, MA), anti-Nestin (MAB353, Millipore, Billerica, MA) anti-phospho-Histone H3 (3H10, Millipore), anti-Tuj1 (Covance, Princeton, NJ), or anti-GFAP (DAKO) primary antibodies followed by Alexa-Fluor 488 Alexa-Fluor 594 secondary antibodies (Invitrogen). Sections were counterstained with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) then mounted with ProLong antifade reagent (Invitrogen).
To assess the differentiation of CNS cultures, live cells were stained with an antibody against O4 (mouse ascites, Developmental Studies Hybridoma Bank, University of Iowa), fixed with 5% acetic acid in ethanol, labeled with donkey anti-mouse IgM secondary antibody conjugated to horseradish peroxidase (HRP) (Jackson Immunoresearch) and visualized by nickel-diaminobenzidine staining. Neurons and astrocytes were labeled with the primary antibodies to Tuj1 (MMS-435P, Covance) and GFAP (G3893 Sigma), followed by Alexa-Fluor 488 goat anti-mouse IgG1 (Invitrogen) and Alexa-Fluor 555 goat anti-mouse IgG2a (Invitrogen). To assess the differentiation of PNS cultures, colonies were fixed in 5% acetic acid in ethanol for 20 min at −20°C, washed, blocked and labeled with antibodies against peripherin, (Chemicon International), GFAP and SMA (Sigma).
NAC and HGF administration
For in vitro experiments, N-acetyl-cysteine (NAC, Sigma) was added to the culture medium at a concentration of 50μM. For in vivo administration of NAC, freshly made NAC (10mg/ml in PBS, pH. 7.4) was subcutaneously injected daily into pregnant Prdm16LacZ/+ mice at a dose of 67mg/kg. Pregnant mice were treated with NAC for the last 5 to 8 days of their pregnancy in most experiments but for the entire pregnancy in some experiments. To assess the effect of HGF on cultured neurospheres, CNS cells were cultured with or without 25 ng/ml HGF (R&D Systems) added to culture medium lacking CEE. Neurospheres were counted after 9 or 10 days culture, photographed to measure neurosphere diameter, and differentiated in adherent cultures as described above to assess multipotency.
Microarray analysis
Total RNA was extracted from freshly isolated VZ cells from 3 Prdm16+/+, Prdm16LZ/+ and Prdm16LZ/LZ neonatal brains using Trizol with 25μg/ml linear acrylamide (Ambion, Austin, TX) and treated with 2 Units of RNase-free recombinant DNase I (Ambion) to remove any contaminating genomic DNA. Purified RNA was reverse transcribed and amplified using the WT-Ovation™ Applause WT/Amp RNA amplification system (NuGEN Technologies, San Carlos, CA) following the manufacturer’s instructions. Sense strand cDNA was fragmented and labeled using the FL-Ovation™ cDNA Biotin Module V2 (NuGEN). 2.5μg of labeled cDNA were hybridized to Affymetrix Mouse Gene ST 1.0 microarrays. The chips were hybridized and scanned according to the manufacturer’s instructions. Expression values for all genes were calculated using the robust multi-array average (RMA) method 28. For the identification of genes with differential expression levels between groups, the raw expression data were analyzed using Expander v5.1 software 29, and genes with fold changes greater than 2 and p-values less than 0.05 (t-test, using log2 transformed expression values) between sample groups were considered to be significantly changed.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed using the Magna ChIP (Millipore) kit according to the manufacturer’s protocol. Briefly, neurospheres from newborn Prdm16+/+ or Prdm16LacZ/LacZ littermates were grown for 9–10 days, cross-linked with 1% paraformaldehyde for 10 min, washed, and then lysed in buffer with protease inhibitor cocktail. Sonicated nuclear extracts were immunoprecipitated overnight at 4°C with anti-Prdm16 antibody (Abcam), IgG Isotype negative control antibody, or anti-Acetyl-H3-K9 (Cell Signaling) positive control antibody and Magna ChIP magnetic beads. Nuclear extracts from 3–5×105 cells were used per immunoprecipitation reaction. Eluted DNA and input control was purified and quantitated by qPCR. Primers were designed using Primer3 software as follow: Hgf_1: 5′-AGTCCAACGGGTCTCAAGTG-3′, 5′-AGTCAAGGCACAGGCAGAAC-3′; Hgf_2: 5′-TGCCTTCCCCTTCTCTTTCT-3′, 5′-CACTTTGCATCTCCCTGACA-3′; Hgf_3: 5′-TACGGGGAAACTGCTTCCTA-3′, 5′-GATCTCTTCACCCTCCTCCA-3′; Hgf_4: 5′-CATGTTGCTCTGTCTTATTTCAA-3′, 5′-CCCGCCCATTTTCCTATTAT-3′; Mt2_1: 5′-GAACACTCCAACCAGCGTTT-3′, 5′-GCGACCTTTATAGCGGAGAG-3′, Mt2_2: 5′-GTGGGGAAACACCATGTACC-3′, 5′-AGACCCTGCGTACAGGAAAA-3′; Mt2_3: 5′-CTGGCGGATACATCCAGTCT-3′, 5′-CTGAGGGGAAGGGTGGAG-3′; Mt2_4: 5′-CAGGATTTGTCTTTCTGGAAGC-3′, 5′-TCCCGTAAATTCATGGAGGT-3′.
Statistical analysis
All data were analyzed by homoscedastic t-tests except as noted.
Accession number
Microarray data files are available at the GEO repository (www.ncbi.nlm.nih.gov/geo/) under accession number GSE23406.
Supplementary Material
Acknowledgments
This work was supported by the Howard Hughes Medical Institute and by the National Institutes of Neurological Disease and Stroke (NS40750). SC was supported by a career development award from the Leukemia and Lymphoma Society. BPL was supported by an American Heart Association Postdoctoral Fellowship (0725726Z) and an Irvington Institute-Cancer Research Institute/Edmond J. Safra Memorial Fellowship. Flow-cytometry was partially supported by the UM-Comprehensive Cancer, NIH CA46592. We thank Martin White and David Adams for flow-cytometry assistance, George Wendt for technical assistance, and Elizabeth Smith (Hybridoma Core Facility) for antibody production, partially supported through the Rheumatic Core Disease Center (P30 AR48310).
Footnotes
AUTHOR CONTRIBUTION
SC and BPL characterized Prdm16 expression and function with assistance from MLS. SC, BPL, and SJM designed experiments, interpreted results, and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors are not aware of any competing financial interests.
References
- 1.Morishita K. Leukemogenesis of the EVI1/MEL1 gene family. International journal of hematology. 2007;85:279–286. doi: 10.1532/IJH97.06174. [DOI] [PubMed] [Google Scholar]
- 2.Bjork BC, Turbe-Doan A, Prysak M, Herron BJ, Beier DR. Prdm16 is required for normal palatogenesis in mice. Human molecular genetics. 2010;19:774–789. doi: 10.1093/hmg/ddp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seale P, et al. Transcriptional control of brown fat determination by PRDM16. Cell metabolism. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murholm M, et al. Dynamic regulation of genes involved in mitochondrial DNA replication and transcription during mouse brown fat cell differentiation and recruitment. PloS one. 2009;4:e8458. doi: 10.1371/journal.pone.0008458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annual review of cell and developmental biology. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- 7.Miyamoto K, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell stem cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Paik JH, et al. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell stem cell. 2009;5:540–553. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renault VM, et al. FoxO3 regulates neural stem cell homeostasis. Cell stem cell. 2009;5:527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Rossi DJ, Jamieson CH, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 13.Goyama S, et al. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell stem cell. 2008;3:207–220. doi: 10.1016/j.stem.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Kim I, Saunders TL, Morrison SJ. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell. 2007;130:470–483. doi: 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007 doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 16.Jaks V, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nature genetics. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 17.Du Y, Jenkins NA, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deneault E, et al. A functional screen to identify novel effectors of hematopoietic stem cell activity. Cell. 2009;137:369–379. doi: 10.1016/j.cell.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. SLAM Family Receptors Distinguish Hematopoietic Stem and Progenitor Cells and Reveal Endothelial Niches for Stem Cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 20.Kiel MJ, Yilmaz OH, Morrison SJ. CD150- cells are transiently reconstituting multipotent progenitors with little or no stem cell activity. Blood. 2008;111:4413–4414. doi: 10.1182/blood-2007-12-129601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren T, Zhang J, Plachez C, Mori S, Richards LJ. Diffusion tensor magnetic resonance imaging and tract-tracing analysis of Probst bundle structure in Netrin1- and DCC-deficient mice. J Neurosci. 2007;27:10345–10349. doi: 10.1523/JNEUROSCI.2787-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.West AK, Hidalgo J, Eddins D, Levin ED, Aschner M. Metallothionein in the central nervous system: Roles in protection, regeneration and cognition. Neurotoxicology. 2008;29:489–503. doi: 10.1016/j.neuro.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozaki M, Haga S, Zhang HQ, Irani K, Suzuki S. Inhibition of hypoxia/reoxygenation-induced oxidative stress in HGF-stimulated antiapoptotic signaling: role of PI3-K and Akt kinase upon rac1. Cell death and differentiation. 2003;10:508–515. doi: 10.1038/sj.cdd.4401172. [DOI] [PubMed] [Google Scholar]
- 24.Yoon YS, Lee JH, Hwang SC, Choi KS, Yoon G. TGF beta1 induces prolonged mitochondrial ROS generation through decreased complex IV activity with senescent arrest in Mv1Lu cells. Oncogene. 2005;24:1895–1903. doi: 10.1038/sj.onc.1208262. [DOI] [PubMed] [Google Scholar]
- 25.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, et al. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459:387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 29.Shamir R, et al. EXPANDER--an integrative program suite for microarray data analysis. BMC Bioinformatics. 2005;6:232. doi: 10.1186/1471-2105-6-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.