Summary
Objective
We describe semantic relation (SR) classification on medical discharge summaries. We focus on relations targeted to the creation of problem-oriented records. Thus, we define relations that involve the medical problems of patients.
Methods and Materials
We represent patients’ medical problems with their diseases and symptoms. We study the relations of patients’ problems with each other and with concepts that are identified as tests and treatments. We present an SR classifier that studies a corpus of patient records one sentence at a time. For all pairs of concepts that appear in a sentence, this SR classifier determines the relations between them. In doing so, the SR classifier takes advantage of surface, lexical, and syntactic features and uses these features as input to a support vector machine. We apply our SR classifier to two sets of medical discharge summaries, one obtained from the Beth Israel-Deaconess Medical Center (BIDMC), Boston, MA and the other from Partners Healthcare, Boston, MA.
Results
On the BIDMC corpus, our SR classifier achieves micro-averaged F-measures that range from 74% to 95% on the various relation types. On the Partners corpus, the micro-averaged F-measures on the various relation types range from 68% to 91%. Our experiments show that lexical features (in particular, tokens that occur between candidate concepts, which we refer to as inter-concept tokens) are very informative for relation classification in medical discharge summaries. Using only the inter-concept tokens in the corpus, our SR classifier can recognize 84% of the relations in the BIDMC corpus and 72% of the relations in the Partners corpus.
Conclusion
These results are promising for semantic indexing of medical records. They imply that we can take advantage of lexical patterns in discharge summaries for relation classification at a sentence level.
Keywords: Lexical context, support vector machines, relation classification for the problem-oriented record, medical language processing
1. Introduction
Clinical records contain important medical information. However, they can lack consistent structure and content. Weed argues for organizing medical records according to the patient’s medical and social problems [1]. He proposes the use of a “problem-oriented medical record” for documenting the information that guides the diagnosis and the plan for care, as well as the results of actions taken in response to the problems of the patient [2]. The medical and social problems of the patient lie in the center of a problem-oriented record. These problems are collected in a “problem list” which is constantly updated based on the changes observed in the patient. This list serves as a problem-oriented summary index for the information that is detailed in the rest of the medical record of the patient.
Figure 1 shows a template problem-oriented record which marks for each problem whether it is known to be present or possible in the patient, the actions taken, treatments administered, and the outcomes of actions taken (if known). When applied to the medical problem “pneumonia”, the problem-oriented record indicates that “pneumonia” was present in the patient, was identified by a “chest x-ray”, and was successfully treated with “antibiotics”. Additionally, the cause of “pneumonia” was not identified; however, this problem co-occurred with another medical problem, “respiratory distress”.
Figure 1.
Skeleton of a sample problem-oriented record. It shows that “pneumonia” was asserted to be “present” in the patient. “Pneumonia” was identified via a “chest x-ray”; its cause is not mentioned in the text. “Respiratory distress” was reported as a problem associated with it. Finally, it was successfully treated with an “antibiotic”.
In this article, we focus on the medical problems of patients. We observe that part of the information summarized in Figure 1 represents the relations of patients’ medical problems with each other, with tests, and with treatments. We study these relations in the context of semantic relation (SR) classification and we present a system that extracts relations that can populate a problem-oriented record.
We define patients’ medical problems, tests, and treatments based on the Unified Medical Language System (UMLS) [3]. The UMLS Metathesaurus includes vocabulary that corresponds to biomedical terms and health-related concepts1. The UMLS organizes concepts by meaning. It consolidates lexically-disparate terms corresponding to the same concept and maps concepts to semantic types.
We represent patients’ medical problems as diseases and symptoms. We define diseases as the following UMLS semantic types: pathologic functions, disease or syndrome, mental or behavioral dysfunction, cell or molecular dysfunction, congenital abnormality, acquired abnormality, injury or poisoning, anatomic abnormality, neoplastic process, and virus/bacterium. We define symptoms as the UMLS semantic type sign or symptom. For our purposes, tests correspond to UMLS semantic types laboratory procedure, diagnostic procedure, clinical attribute, and organism attribute. Treatments correspond to UMLS semantic types therapeutic or preventive procedure, medical device, steroid, pharmacologic substance, biomedical or dental material, antibiotic, clinical drug, and drug delivery device. We refer to diseases, symptoms, tests, and treatments as semantic categories [4].
We assume that the concepts corresponding to patients’ medical problems, treatments, and tests have already been tagged. Studying the relations of these concepts requires that we differentiate the medical problems that are asserted to be absent in the patient from those that are asserted to be present or possible. We assume that our input includes annotations that mark the patient’s medical problems as present (the patient has the medical problem), possible (the patient may have the medical problem), or absent (the patient does not have the medical problem) in the patient as determined by the assertions made in the narrative text [5].
Given already identified and assertion-classified concepts, we present an SR classifier based on support vector machines (SVMs) [6] and apply this classifier to medical discharge summaries in order to determine the physician-stated relations between concepts. This limits our scope to relations that have been stated in narrative text.
Our SR classifier studies the data one sentence at a time and a pair of concepts at a time. We refer to the pair of concepts under study as the candidate pair. Each candidate pair includes a medical problem that is asserted to be present or possible in a patient. The SR classifier determines the relation of each medical problem with other present or possible medical problems, treatments, and tests mentioned in the same sentence. These relations are fine grained, i.e., there are multiple relations that can hold for a given candidate pair. Successful classification of fine-grained relations requires features that reveal the difference between such relations. We use our SR classifier to explore the contribution of various features to the automatic extraction of fine-grained relations. We show that lexical cues capture most of the information that is necessary for extracting these relations.
2. Related Work
Relation extraction builds on entity extraction and is the next step in the endeavor to create a structured representation of the contents of unstructured natural language text. Below, we review some representative works performed in this area.
Our goal in SR classification is to determine the relations that a sentence states about the pairs of concepts it contains. Lexical cues found in sentences can be informative for this purpose [7]. For example, the verb “treats” indicates the treatment–disease relation [8]. Inflectional variants of the verb “bind” indicate a protein binding relation [9]. Such lexical patterns can be enriched with semantic patterns [10–12], syntactic patterns, and even syntacto-semantic patterns.
Rindflesch et al. developed the SemRep system and applied it to detect “branching” of anatomical structures as expressed in coronary catheterization reports [13], to extract molecular interactions from biomedical text [9], and to identify drug therapies in MEDLINE citations [14]. In order to determine the “branching” relations, they observed that these relations are expressed with various syntactic structures in coronary catheterization reports. For example, the syntactic structures “is a branch of”, “arises from”, and “takes off from” indicate “branching” relations between concepts that are marked as “body part, organ, or organ component” by UMLS. In simple declarative sentences, Rindflesch et al. identified pairs of UMLS concepts that fall on either side of the target syntactic structures and marked them as having the “branching” relation. They employed further rules to process sentences involving coordination and relativization.
Friedman et al. designed the MedLEE system for identifying relations in clinical records [11]. For them, pattern matching and semantic grammars determined the nature of the relations. For example, the semantic rule “degree + change + finding” when mapped to a phrase “drastic increase in temperature” directly indicates that “increase” describes the change in “temperature” and this “increase” is quantified as being “drastic”. Such semantic rules map to syntactic variants and even syntactically incorrect variants of these phrases, as they are based on the semantic categories of concepts rather than the exact linguistic context surrounding them. Various parsing and linguistic regularization steps minimize the ambiguity and complexity that is present in the linguistic context of the identified concepts as well as in the lexical variations in the concepts themselves. Friedman et al. adapted MedLEE to the biology domain for the extraction of relations indicating biomolecular interactions [15] and to the bioscience domain for the extraction of gene–phenotype relations [16].
Fundel et al. applied rules to dependency parse trees and chunked sentences. They have thus identified candidate pairs of proteins which can be in activation and interaction relations. After identifying the candidates, they filtered those that are negated. On a corpus containing 92 relation instances extracted from of 50 MEDLINE articles, they achieved an F-measure of 89% [17].
Bui et al. wrote grammatical rules to extract causal relations between HIV and medications from PubMed abstracts. After post processing for negated relations and resolving contradictions between the assertions made on the same concepts, their system achieved an F-measure of 84.5% on 500 sentences [18].
In contrast to the above-mentioned rule-based systems, machine learning approaches to relation extraction can automatically learn the information salient for semantic relation classification [19, 20]. Niu and Hirst compile lists of lexical cues that mark relations between medications. Their lexical cues include “and”, “or”, etc. These cues help determine the combination, alternative, comparison, specification, substitute, and preference relations that hold between medications [21]. Roberts et al. study the has_target, has_finding, has_indication, has_location, negation_modifies, literality_modifies, and has_location_modifies relations between pairs of entities found in oncology narratives. Only one kind of relation can hold between a specific pair of entity types; for example, has_location relation always relates a condition entity to a locus entity. The only exception to this is the investigation and condition entity pairs which can relate either through has_finding or has_indication relations. Roberts et al. apply SVMs with features that mark the part of speech and surface information from a six-token window, the distance of the entities, the presence of intervening entities or events, and the information from a dependency parse of the text in order to determine which of their seven relations holds between a given entity pair. On a small set of 77 oncology narratives, their F-measure over all relations is at 70% [22].
Guiliano et al. extract the interactions between genes and proteins on two bioscience corpora. They use kernel methods with information from the sentence containing the concepts. They represent sentential context using bag of words and n-grams that capture patterns such as “binding of p1 and p2”, “p1 binding to p2”, “p1 and p2 interact”, etc. They then enrich this representation with information on ordering of the tokens, part-of-speech tags, lemmas, and orthographic features, achieving an F-measure of 61.7% [23].
Bundschus et al. extracted disease–treatment relations from the corpus prepared by Rosario and Hearst [19] using conditional random fields (CRFs) with part-of-speech tags. For their work, the relations of diseases and treatments can be any one of: cures, only disease, only treatment, prevents, side effect, vague, does not cure. On a corpus of 3570 sentences gathered from MEDLINE 2001 abstracts, Bundschus et al. report an F-measure of 72% [24].
In contrast to the relations targeted by related work (with the exception of Bundschus et al.), the relations described in our data are fine grained. Between any two kinds of entities, three or more relations are possible. Our task is to identify the exact fine-grained relation that is stated by doctors to hold between pairs of concepts. We focus on the relations that encompass concepts occurring within the same sentence and support the creation of a problem-oriented record. Our findings improve upon the very limited number of studies that extract physician-stated relations of concepts discussed in clinical records. In conducting our studies we focus on the evaluation of various features for the extraction of such fine-grained relations and determine that lexical cues contribute the most to this task.
3. Semantic Relations
We utilize semantic categories of concepts and the assertions made about these concepts to define the following semantic relation types: the present disease–treatment relation type; the possible disease–treatment relation type; the disease (possible and present)–test relation type; the disease–symptom relation type; the present symptom–treatment relation type; and the possible symptom–treatment relation type. Below, we explain the present disease–treatment relation type in detail and provide examples for each relation within this type. We list the relations under the rest of the relation types without much commentary as the relations in these types parallel those provided under the present disease–treatment relation type.
Present disease–treatment relation type
We define the following relations between a present disease and a treatment that are mentioned in the same sentence:
Treatment administered for disease (TADP): e.g., “[Solu-Medrol]treat was given for [tracheal edema]dis”.
Treatment causes disease (TCDP): e.g., “The patient experienced [femoral neuropathy]dis secondary to [radiation therapy]treat”.
Treatment cures disease (TXDP): e.g., “The patient had resolution of her [acute renal failure]dis, with [hydration]treat only”.
Treatment does not cure/worsens disease (TNDP): e.g., “The patient had also been treated for a [pneumonia]dis that was believed to have progressed despite treatment with [azithromycin ]treat”.
Treatment discontinued in response to disease (TDDP): e.g., “[ACE inhibitor]treat was held because of [renal failure]dis”.
None of the above-defined present disease–treatment relations holds (None).
Possible disease–treatment relation type
Treatment administered for possible disease (TAD); treatment causes possible disease (TCD); treatment discontinued in response to possible disease (TDD); and none of the above-listed possible disease–treatment relations holds (None).
Present symptom–treatment relation type
Treatment administered for symptom (TASP); treatment cures symptom (TXSP); treatment does not cure/worsens symptom (TNSP); treatment causes symptom (TCSP); treatment discontinued for symptom (TDSP); and none of the above-listed present symptom–treatment relations holds (None).
Possible symptom–treatment relation type
Treatment administered for possible symptom (TAS); treatment causes possible symptom (TCS); treatment discontinued in response to possible symptom (TDS); and none of the above-listed possible symptom–treatment relations holds (None). .
Disease–test relation type
test reveals disease (TRD); test conducted to investigate disease (TID); and none of the above-listed disease–test relations holds (None). .
Disease–symptom relation type
Disease causes symptom (DCS); symptom suggests presence of disease (SSD); and none of the above-listed disease–symptom relations holds (None). .
Now we can explain the contents of Figure 1 in terms of a subset of these relations. Given a medical problem, the problem and certainty fields correspond to the problem itself and its assertion class respectively. The treatments field lists treatments administered for that problem along with their outcome as captured, for example, by TAD, TADP, TAS, TASP, TXSP, TNSP, TXDP, or TNDP relations. The causes field lists possible causes of the problem, for example, as captured by the TCS, TCSP, TCDP, TCD, or DCS relations. The associated problems section refers to problems caused by the target problem. This field is only applicable if the target is a disease and it lists symptoms that relate to the target by DCS relation. Finally, the proof field lists tests and symptoms that relate to a disease by a TRD or a SSD relation.
4. Annotation
We conducted semantic relation classification on two corpora. One of these corpora consisted of 50 medical discharge summaries containing 11,619 sentences from the Beth Israel Deaconess Medical Center (BIDMC), Boston, MA. The other consisted of 13,443 sentences from 142 discharge summaries from various departments of the hospitals in Partners Healthcare. We manually annotated these corpora with semantic categories, assertions, and relations.
Two undergraduate computer science students were given the mapping of semantic categories to UMLS semantic types (from Section 1) and the definitions of these semantic types. These students independently marked the semantic categories of concepts in the BIDMC corpus on completely unannotated text. Two other undergraduate computer science students followed the same definitions and independently marked the semantic categories of the concepts on the completely unannotated text of the Partners corpus. The pairs of annotators discussed and resolved their disagreements on their respective corpora, providing us with semantic category annotations that they agreed on. Details of semantic category annotation and the agreement evaluation can be found in Sibanda et al. [4].
The manual semantic category annotations were then presented to an information studies doctoral student and a nurse librarian for manual assertion annotation. These annotators were asked to mark each mention of a medical problem as present if the text asserted that the problem was present in the patient; as uncertain (possible) if the text asserted that the problem could be present in the patient; as absent if the text asserted that the patient did not have the problem; and as alter-association if the discussed problem was not associated with the patient. After annotating the assertions made on medical problems, the two annotators discussed and resolved their disagreements, and provided us with gold standard assertion annotations that they agreed on. Details of the assertion annotation and agreement evaluation can be found in Uzuner et al. [5].
Having semantic categories and their assertions manually annotated allowed us to study relation classification without having to worry about the noise that could have been introduced to the data by automatic approaches to semantic category recognition and assertion classification. Given the semantic categories and assertions, and given our interest in extracting relations of concepts mentioned in the same sentence, we selected from our corpora those sentences containing pairs of concepts encompassed by our relation types. This selection process gave us 530 sentences from the BIDMC corpus and 1626 sentences from the Partners corpus. These sentences were annotated for relations by a computer science graduate student and a nurse librarian, under the supervision of a medical professional. The two annotators achieved a Kappa [25] agreement of 0.89 and 0.83 on the BIDMC and Partners corpora, respectively (see Hripcsak et al, [26] for a discussion of Kappa). The two annotators discussed their disagreements and provided us with gold standard annotations on the two corpora. Table 1 shows the breakdown of relations in our corpora. Relevant institutional review boards approved this study.
Table 1.
Breakdown of semantic relations in our corpora.
| Relation | Count in BIDMC corpus |
Count in Partners corpus |
Relation | Count in BIDMC corpus |
Count in Partners corpus |
|---|---|---|---|---|---|
| Present disease–treatment type | Present symptom–treatment type | ||||
| None | 208 | 483 | None | 88 | 302 |
| TADP | 157 | 557 | TASP | 38 | 247 |
| TXDP | 14 | 7 | TXSP | 20 | 27 |
| TNDP | 11 | 28 | TNSP | 4 | 47 |
| TCDP | 10 | 131 | TCSP | 8 | 176 |
| TDDP | 16 | 22 | TDSP | 6 | 29 |
| Possible disease–treatment type | Possible symptom–treatment type | ||||
| None | 34 | 28 | None | 74 | 312 |
| TAD | 17 | 40 | TAS | 22 | 64 |
| TCD | 4 | 5 | TCS | 0 | 29 |
| TDD | 2 | 10 | TDS | 1 | 2 |
| Disease–test type | Disease–symptom type | ||||
| None | 346 | 575 | None | 204 | 649 |
| TRD | 285 | 1007 | SSD | 9 | 303 |
| TID | 35 | 221 | DCS | 13 | 251 |
5. The SR Classifier
Our SR classifier consists of six different multi-class SVM classifiers corresponding to the six relation types listed in Section 3. We generate separate SVM models for each relation type. For example, an SVM is trained to recognize the relations gathered under the disease–symptom relation type, another SVM is trained for the disease–test relation type, etc.
For each sentence in the text, the SR classifier uses information on the semantic categories and assertion classes of the candidate pair of concepts in order to determine the relation type for the pair. More specifically, the relation type of a candidate pair of concepts comes from their semantic categories and the assertions made on them. As a result, the first step in our SR classifier is a switch operating on the semantic categories and assertions. According to this switch, for example, a candidate pair that consists of a present disease and a treatment belongs to a present disease–treatment relation type; a candidate pair that consists of a disease and a symptom belongs to a disease–symptom relation type, etc. The SR classifier uses the relation type of the candidate pair in order to invoke only the SVM classifier for that relation type. The SVM for the relation type of the candidate concept pair is responsible for identifying the relation that holds between the concepts. For example, in the sentence “antibiotics were given for his pneumonia” where the disease “pneumonia” is asserted (by the possessive pronoun) to be present in the patient, only the SVM for the present disease–treatment relation type is invoked, which determines the relation between these concepts as TADP.
Each sentence in our data can contain one or more pairs of candidate concepts2. When a sentence contains multiple pairs of candidate concepts, each candidate concept pair of the sentence is matched to its own relation type and is processed by the SVM for that relation type. This requires some sentences to be processed by more than one SVM in our SR classifier. For example, the sentence “antibiotics were given for his pneumonia that was identified by a chest x-ray” needs to be processed by the SVM for the present disease–test relation type (for the concept pair “pneumonia” and “chest x-ray”) as well as the SVM for the present disease–treatment relation type (for the candidate concept pair “pneumonia” and “antibiotics”).
The SVM for each relation type is trained on examples representing the relations specific to that type. These examples are represented in the form of a feature vector. In this vector, we define a feature as a set of columns that collectively describe a given characteristic, e.g., “words” is a feature that corresponds to vector columns “disease”, “anemia”, “treat”, etc., that are extracted from the patient record. In this feature vector, for each candidate concept pair to be classified (row in the feature vector), we mark the values of features by setting the columns observed to 1, leaving the rest at zero [5]. If the candidate pair has no value for a feature, then all columns representing this feature will be set to zero.
5.1 Support vector machines
Given a collection of input samples represented by multi-dimensional vectors and class labels, (Xi, yi), i = 1, …, l where Xi Є Rn and y Є {1, −1}l, SVMs optimize [27, 28]:
| (1) |
| (2) |
where C>0 is the penalty parameter and ϕ is a function that maps Xi to a higher dimensional space. SVMs capture the hyperplane that best separates the input samples according to their class. The chosen hyperplane maximizes the distance to the closest samples from each class. The kernel function
| (3) |
determines the form of the hyperplane.
In this article, we explore a high dimensional feature space which can be prone to over-fitting. To minimize this risk, we employ a linear kernel
| (4) |
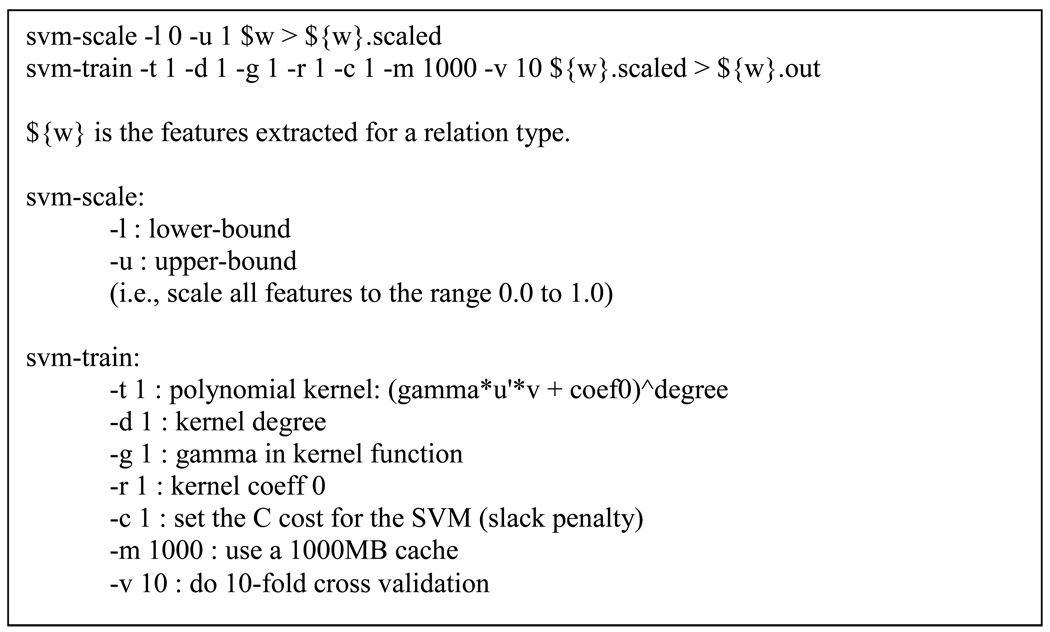
SVMs robustly handle large feature sets and the noise that may be present in them. We employ the multi-class SVM implementation of LibSVM as is. This implementation, referred to as C-SVC by LibSVM, builds a multi-class classifier out of binary classifiers using one-versus-one voting [6]. The choice of linear kernels for our classifier reduces the need for parameter tuning drastically, i.e., only C can be tuned (see Figure 2). Although parameter tuning usually improves performance, tuning needs to be repeated for every classifier and for every subset of the features. Given our interest in the evaluation and comparison of various features rather than the best possible performance in classification, we use the default C value of LibSVM and keep the classifier (and its parameters) exactly the same when experimenting with various feature sets. Figure 2 shows the exact invocation.
Figure 2.
Exact parameters used with LibSVM.
5.2 Features
The features for our SVMs are as follows:
5.2.1 Surface Features
5.2.1.1 Relative ordering of the candidate concepts
In most of the sentences in our corpus, diseases and symptoms are mentioned before treatments and tests; diseases are mentioned before symptoms. We consider this order to be the default order of the concepts. We observe that the default ordering of the concepts (and deviations from the default) can be indicative of (presence or absence of) some relations. Therefore, for each pair of candidate concepts, we mark whether or not they occur in their default order.
The relative ordering feature is a single column in our feature vector. The value of the column is zero if the concepts appear in the default ordering. Otherwise, it is one.
5.2.1.2 Distance between the candidate concepts
The greater the distance between the candidate concepts, the less likely that they are related. We measure the distance between the candidate concepts in terms of the number of tokens between them. We include both word tokens and punctuation tokens in our count.
Our feature vector includes a single column that marks the distance between the concepts. We represent the distance between two concepts by setting the value of this column to the number of tokens between the concepts.
5.2.1.3 Presence of intervening disease, symptom, test, and treatment concepts
Presence of mentions of other diseases, symptoms, tests, and treatments between the candidate concepts often implies that the candidate concepts are unrelated, e.g., “[Tylenol] was given for his pain, aspirin for his [headache]” where “Tylenol” and “headache” are intervened by “pain” and “aspirin”.
This information is represented in our feature vector with a single column that is set to one if there is an intervening concept and is left at zero otherwise.
5.2.2 Lexical Features
Both the tokens that constitute the candidate concepts and the tokens that make up the context of the candidates can play a role in semantic relation extraction. We therefore consider the following lexical features in our experiments. For all of our lexical features, we normalize [29] the text of the discharge summaries so that the morphological variants of a token can be treated as the same token.
5.2.2.1 Tokens in concepts
We hypothesize that the tokens in candidate phrases may be indicative of the relation that they are involved in. For example, in encountering the sentence “[Tylenol] was given for his [headache]”, we may know that in our corpus the word token “Tylenol” always appears in the relation TASP. We therefore represent each candidate concept with the tokens it contains.
In our feature vector, we list all the tokens of all concepts found in the training corpus as separate columns. Together, these columns represent the feature “Tokens in concepts”. The tokens that are observed in the candidate concepts are indicated by setting their columns to one. The rest are left at zero.
5.2.2.2 Lexical trigrams
We study the left and right lexical trigrams (of tokens corresponding to words and punctuation) of the candidate concepts. For example, in the sentence “[Tylenol] was given for his [headache]”, knowing that the trigram “was given for” follows the treatment “Tylenol”, and the trigram “given for his” precedes the symptom “headache”, suggests that the relation here is TASP.
Given a target concept at the nth position in the sentence, the lexical trigrams represent the (n−1)th, (n−2)th, (n−3)th, (n+1)th, (n+2)th, and (n+3)th positions of each of the candidate concepts in the sentence in two ways. The first representation lists all possible tokens for each of the positions (based on the training set) as columns and marks the tokens that occur at each of the positions by setting their columns to one. For some candidate pairs, one or more of the positions can have no tokens specified, e.g., the second token of the sentence will have no (n−3)th tokens specified, i.e., all columns for that position will be left at zero. This representation of lexical trigrams subsumes lexical bigrams and lexical unigrams which capture (n−2)th through (n+2)th and (n−1)th through (n+1)th positions, respectively.
The second representation of lexical trigrams creates two additional features, one representing the left lexical trigram string of the (n−1)th, (n−2)th, and (n−3)th positions together, and the other representing the right lexical trigram string of the (n+1)th, (n+2)th, and (n+3)th positions together. For example, in the sentence “[Tylenol] was given for his [headache]”, the column corresponding to “was given for” is marked with a one.
5.2.2.3 Inter-concept tokens
Inter-concept tokens include the words and punctuation between the candidate concepts. In the sentence “[Tylenol] was given for his [headache]”, the inter-concept tokens are “was”, “given”, “for”, and “his”. We hypothesize that certain inter-concept tokens are indicative of specific relations. Punctuation often suggests lack of a relation, e.g., “[Tylenol] was given for pain: his [headache] has not been treated”.
Our feature vector lists all inter-concept tokens of all concept pairs found in the training corpus as columns. The inter-concept tokens that are observed between the candidate concepts are indicated by setting their columns to one.
5.2.3 Shallow Syntactic Features
Lexical and surface features are limited in the information they capture. We augment these features with shallow syntactic information in the form of syntactic bigrams and verbs that appear between and surrounding the candidate concepts.
We extract the verbs from part-of-speech tagged text obtained from the output of the Brill tagger [30]. The Brill tagger is rule based. It assigns tags to words in three basic steps: First, it labels each word with the most popular part of speech observed for it in the training corpus. Second, for the words that are not found in the training corpus, it guesses a tag based on their orthographic properties. Third, it applies rules that check sequences of tags and modifies the already assigned tags based on their neighbors. In the absence of a clinical corpus which would allow us to train the Brill tagger, we utilize the Brill tagger as trained on the Wall Street Journal corpus, fully understanding the limitation caused by the mismatch [31] between our corpus and the training corpus of the Brill tagger. Even then, we use the output of this tagger only to identify verbs.
We determine additional syntactic features from the output of the Link Grammar Parser [32]. This parser treats words as blocks and utilizes a lexicon that imposes constraints on the syntactic links of blocks with each other. Given the blocks and their constraints, the Link Grammar Parser tries to construct the most complete parse for each sentence so that all blocks have been linked without violating the constraints that govern them. If the search for a complete linkage fails, the parser enters “panic mode” which allows it to create partial parses for phrases. Before running this parser on our corpora, we augment its lexicon [33].
5.2.3.1 Verbs
Verbs are often the strongest indicators of the relations between concepts. For example, in the sentence fragment, “[Levaquin] for [right lower lobe pneumonia], which has resolved”, the verb “resolved” suggests the relation TXDP. We capture the information provided by verbs that appear before, between, and after candidate concepts. We gather up to two verbs that occur before and up to two verbs that appear after the candidate concepts along with the verbs that occur between the candidate concepts.
Our feature vector lists verbs found in the training corpus as separate columns. Three sets of columns represent verbs. The first set represents verbs that occur between the candidate concepts. The other two represent verbs that occur before and after the candidate concept pair. The verbs that are observed for a candidate concept pair are indicated by setting their columns to one. The rest of the verbs in the vector are left at zero.
5.2.3.2 Headword
The headword of a concept is the head noun contained within the concept phrase. We heuristically extract the head from each concept by traversing the Link Grammar Parser [32] output for the sentence from the leftmost to rightmost word in each concept. We return the last word before the target of an 'M' link (modifier-preposition link), excluding adjectival 'Ma' links, or the word before the first 'J' link (preposition-object link) as the head.
We represent the headword feature with a set of columns corresponding to the headwords found in the training set. We mark the head of each candidate concept found in the test set by setting its column to one.
5.2.3.3 Syntactic bigrams
Syntactic bigrams are the tokens that are at most two links away from the candidate concepts in the parse provided by the Link Grammar Parser [32]. We study the left and right syntactic bigrams of the candidate concepts. This feature captures syntactic dependencies among tokens, even when the tokens are separated by relative clauses.
Our feature vector represents syntactic bigrams analogously to lexical trigrams. For each candidate concept, the values of (n−1)th, (n−2)th, (n+1)th, and (n+2)th syntactic positions (created from the training set) are marked.
5.2.3.4 Link path
This is the syntactic link path between the candidate concepts. We extract this feature from the Link Grammar parse of the text by following the links from one candidate concept to the other. If there is no link path between the concepts, then this feature has the value none. Ding et al. [34] showed that it is possible to determine whether two biochemicals are related by checking for a link path between them. We take this idea one step further by checking if the exact nature of the links in the path indicates the type of relation. For example, a subject-object (S-Os) path between a test (chest x-ray) and a disease (pneumonia) in Figure 3, could suggest the TRD relation.
Figure 3.
Sample link grammar output.
Our feature vector represents link paths in a similar manner to inter-concept tokens. For each candidate concept pair, the columns of the links are set to one when observed, left at zero otherwise.
5.2.3.5 The link path tokens
These are the tokens encountered on the link path between the candidate concepts. We predict that using just the link path between entities is susceptible to noise. Therefore, we also use the actual tokens encountered on the path as features.
In our inter-concept tokens, we consider all of the tokens between the concepts. We hypothesize that by only including tokens on the link paths between concepts, we avoid spurious tokens, such as prepositions, and other modifiers that do not contribute to the semantic relation in the sentence. If there is no link path between the two concepts, we revert to using all of the tokens between the concepts.
Our feature vector represents link path tokens in a similar manner to link paths. For each candidate concept pair, the columns of the link path tokens are set to one when observed, left at zero otherwise.
6. Evaluation
We evaluated the SR classifier using 10-fold cross validation. At each round of cross-validation, we recreated the feature vector based on the training corpus used for that round. As a result, the features of the candidate concepts that appear only in the validation set of that round may not appear in the training feature vector. We computed precision (Equation 5), recall (Equation 6), and F-measures (Equation 7) for each relation separately. Precision measures the percentage of true positives (TP) among all the assignments (TP and false positive (FP)) made by the classifier. Recall measures the percentage of TPs in the samples that belong to the class (given by TPs and false negatives (FNs)). F-measure is the harmonic mean of precision and recall. It determines the relative weights of precision and recall using a constant β. For our task, precision and recall are equally important; therefore we set β to 1 and weight precision and recall equally.
| (5) |
| (6) |
| (7) |
We obtained system-level results from micro-averaged F-measures (Equation 8). Micro-averaged F-measure weights each relation by its relative frequency. It is computed from micro-averaged precision (Equation 9) and micro-averaged recall (Equation 10). We also compute macro-averaged F-measures (Equation 11) over all relations. Macro-averaged F-measure gives equal weight to each relation and presents the arithmetic mean of the F-measures of all relations. In the below equations, M stands for the number of relations.
| (8) |
| (9) |
| (10) |
| (11) |
Micro- and macro-averaged statistics can be computed for each relation type and also on the complete data consisting of various relation types. The micro- and macro-averaged performance of the SR classifier on a relation type is identical to the performance of the SVM specific to that relation type, i.e., compute micro-and macro-averaged precision, recall, and F-measure on only the data corresponding to that relation type. Micro- and macro-averaged performance of the SR classifier on the complete data are obtained by treating the SR classifier as a “black box” and counting the TP, FN, and FP predictions generated by the SR classifier as a whole without any regard to which relation type SVM each prediction came from, i.e., compute micro- and macro-averaged precision, recall, and F-measure of the complete system output on the complete data.
We use micro-averaged F-measure as the basis for our discussions and present macro-averaged results, without much commentary, for completeness. We test the significance of the difference in micro-averaged F-measures using a Z-test on two proportions [35, 36] with a z-value of ±1.645 which represents an alpha of 0.1 [37]. The z-value is computed from Equation 12 and Equation 13:
| (12) |
| (13) |
where n1 and n2 refer to sample sizes, and p1 and p2 refer to two compared F-measures.
7. Results and Discussion
We compare our SR classifier with two baseline feature sets. The first baseline uses only the tokens contained in concepts, i.e., only the “tokens in concepts” as defined in Lexical Features in Section 5.2.2., while the second uses all of the tokens in the complete sentence. We refer to these baselines as the tokens-in-concepts and the tokens-in-sentence baselines, respectively. Table 2 shows the performance of the baselines and of the SR classifier on individual relations on each of our corpora. Table 3 shows that the SR classifier significantly outperforms both baselines on most of the individual relation types and in aggregate (over all relation types) on both of our corpora. In aggregate, on the BIDMC corpus, the SR classifier scores micro- and macro-averaged F-measures of 86% and 63% respectively, compared to 75% and 49% of the tokens-in-concepts baseline and 75% and 54% of the tokens-in-sentence baseline (see Table 3). In aggregate, on the Partners corpus, the SR classifier scores micro- and macro-averaged F-measures of 76% and 62% respectively, compared to 67% and 49% of the tokens-in-concepts baseline and 71% and 54% of the tokens-in-sentence baseline (see Table 3).
Table 2.
| a: Results for individual relations on the BIDMC corpus. P stands for precision, R stands for recall, and F1 stands for F-measure. Bold indicates statistically significant difference from corresponding F1 of the tokens-in-concepts baseline. Italic indicates significant difference from the corresponding F1 of the tokens-in-sentence baseline. | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| BIDMC corpus | |||||||||
| P | R | F1 | P | R | F1 | P | R | F1 | |
| Tokens-in-concepts baseline | Tokens-in-sentence baseline | Semantic relation classifier | |||||||
| Relation | Present disease–treatment relation type | ||||||||
| None | 0.65 | 0.75 | 0.69 | 0.70 | 0.74 | 0.72 | 0.84 | 0.86 | 0.85 |
| TADP | 0.60 | 0.57 | 0.58 | 0.61 | 0.66 | 0.63 | 0.76 | 0.83 | 0.79 |
| TXDP | 0.75 | 0.43 | 0.55 | 0.40 | 0.29 | 0.33 | 0.89 | 0.57 | 0.70 |
| TNDP | 0.89 | 0.73 | 0.80 | 0.63 | 0.45 | 0.53 | 1.00 | 0.73 | 0.84 |
| TCDP | 1.00 | 0.30 | 0.46 | 1.00 | 0.30 | 0.46 | 0.75 | 0.40 | 0.52 |
| TDDP | 0.50 | 0.20 | 0.29 | 0.75 | 0.40 | 0.52 | 0.75 | 0.30 | 0.43 |
| Possible disease–treatment relation type | |||||||||
| None | 0.72 | 0.79 | 0.75 | 0.78 | 0.88 | 0.83 | 0.76 | 0.85 | 0.80 |
| TAD | 0.60 | 0.71 | 0.65 | 0.78 | 0.82 | 0.80 | 0.78 | 0.82 | 0.80 |
| TCD | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| TDD | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Present symptom–treatment relation type | |||||||||
| None | 0.71 | 0.83 | 0.76 | 0.80 | 0.84 | 0.82 | 0.87 | 0.89 | 0.88 |
| TASP | 0.35 | 0.29 | 0.32 | 0.39 | 0.47 | 0.43 | 0.64 | 0.76 | 0.70 |
| TXSP | 0.59 | 0.50 | 0.54 | 0.83 | 0.75 | 0.79 | 0.88 | 0.75 | 0.81 |
| TNSP | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| TCSP | 0.71 | 0.63 | 0.67 | 1.00 | 0.75 | 0.86 | 1.00 | 0.75 | 0.86 |
| TDSP | 0.50 | 0.33 | 0.40 | 1.00 | 0.17 | 0.29 | 0.50 | 0.50 | 0.50 |
| Possible symptom–treatment relation type | |||||||||
| None | 0.81 | 0.92 | 0.86 | 0.91 | 0.97 | 0.94 | 0.99 | 0.97 | 0.98 |
| TAS | 0.46 | 0.27 | 0.34 | 0.82 | 0.64 | 0.72 | 0.87 | 0.91 | 0.89 |
| TCS | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| TDS | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Disease–test relation type | |||||||||
| None | 0.84 | 0.84 | 0.84 | 0.73 | 0.77 | 0.75 | 0.89 | 0.90 | 0.90 |
| TRD | 0.82 | 0.82 | 0.82 | 0.72 | 0.71 | 0.71 | 0.88 | 0.91 | 0.89 |
| TID | 0.38 | 0.34 | 0.36 | 0.68 | 0.49 | 0.57 | 0.78 | 0.51 | 0.62 |
| Disease–symptom relation type | |||||||||
| None | 0.94 | 0.99 | 0.96 | 0.96 | 0.98 | 0.97 | 0.97 | 0.99 | 0.98 |
| SSD | 0.88 | 0.78 | 0.82 | 0.63 | 0.56 | 0.59 | 0.88 | 0.78 | 0.82 |
| DCS | 0.60 | 0.23 | 0.33 | 0.82 | 0.69 | 0.75 | 0.89 | 0.62 | 0.73 |
| b: Results on individual relations on Partners corpus. P stands for precision, R stands for recall, and F1 stands for F-measure. Bold indicates statistically significant difference from corresponding F1 of the tokens-in-concepts baseline. Italic indicates significant difference from the corresponding F1 of the tokens-in-sentence baseline. | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Partners corpus | |||||||||
| P | R | F1 | P | R | F1 | P | R | F1 | |
| Tokens-in-concepts baseline | Tokens-in-sentence baseline | Semantic relation classifier | |||||||
| Relation | Present disease–treatment relation type | ||||||||
| None | 0.71 | 0.69 | 0.70 | 0.76 | 0.80 | 0.78 | 0.81 | 0.80 | 0.81 |
| TADP | 0.69 | 0.77 | 0.73 | 0.76 | 0.78 | 0.77 | 0.77 | 0.84 | 0.81 |
| TXDP | 0.00 | 0.00 | 0.00 | 0.67 | 0.29 | 0.40 | 0.00 | 0.00 | 0.00 |
| TNDP | 0.62 | 0.29 | 0.39 | 0.69 | 0.39 | 0.50 | 0.86 | 0.43 | 0.57 |
| TCDP | 0.67 | 0.53 | 0.59 | 0.66 | 0.56 | 0.61 | 0.75 | 0.54 | 0.63 |
| TDDP | 0.20 | 0.09 | 0.13 | 0.36 | 0.18 | 0.24 | 0.73 | 0.63 | 0.68 |
| Possible disease–treatment relation type | |||||||||
| None | 0.59 | 0.57 | 0.58 | 0.69 | 0.64 | 0.67 | 0.86 | 0.68 | 0.76 |
| TAD | 0.66 | 0.68 | 0.67 | 0.67 | 0.75 | 0.71 | 0.72 | 0.90 | 0.80 |
| TCD | 0.67 | 0.40 | 0.50 | 0.00 | 0.00 | 0.00 | 0.50 | 0.20 | 0.29 |
| TDD | 0.58 | 0.70 | 0.64 | 0.70 | 0.70 | 0.70 | 0.78 | 0.70 | 0.74 |
| Present symptom–treatment relation type | |||||||||
| None | 0.61 | 0.70 | 0.65 | 0.68 | 0.71 | 0.70 | 0.72 | 0.77 | 0.74 |
| TASP | 0.60 | 0.57 | 0.58 | 0.59 | 0.62 | 0.61 | 0.67 | 0.73 | 0.70 |
| TXSP | 0.20 | 0.07 | 0.11 | 0.22 | 0.07 | 0.11 | 0.33 | 0.11 | 0.16 |
| TNSP | 0.54 | 0.45 | 0.49 | 0.65 | 0.68 | 0.67 | 0.77 | 0.66 | 0.71 |
| TCSP | 0.54 | 0.56 | 0.55 | 0.56 | 0.59 | 0.58 | 0.60 | 0.59 | 0.59 |
| TDSP | 0.47 | 0.31 | 0.38 | 0.40 | 0.14 | 0.21 | 0.75 | 0.41 | 0.53 |
| Possible symptom–treatment relation type | |||||||||
| None | 0.81 | 0.94 | 0.87 | 0.84 | 0.93 | 0.88 | 0.94 | 0.96 | 0.95 |
| TAS | 0.48 | 0.23 | 0.32 | 0.43 | 0.25 | 0.32 | 0.77 | 0.75 | 0.76 |
| TCS | 0.71 | 0.34 | 0.47 | 0.72 | 0.62 | 0.67 | 0.89 | 0.86 | 0.88 |
| TDS | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Disease–test relation type | |||||||||
| None | 0.64 | 0.69 | 0.66 | 0.63 | 0.69 | 0.66 | 0.73 | 0.75 | 0.74 |
| TRD | 0.78 | 0.80 | 0.79 | 0.79 | 0.81 | 0.80 | 0.84 | 0.86 | 0.85 |
| TID | 0.41 | 0.28 | 0.33 | 0.59 | 0.43 | 0.49 | 0.63 | 0.52 | 0.57 |
| Disease–symptom relation type | |||||||||
| None | 0.66 | 0.75 | 0.70 | 0.76 | 0.79 | 0.78 | 0.78 | 0.81 | 0.79 |
| SSD | 0.57 | 0.49 | 0.52 | 0.64 | 0.65 | 0.65 | 0.62 | 0.62 | 0.62 |
| DCS | 0.51 | 0.43 | 0.47 | 0.57 | 0.51 | 0.54 | 0.58 | 0.53 | 0.55 |
Table 3.
Performance on relation types and overall. Bold indicates micro-averaged F-measures of SR classifier that are significantly different from the corresponding micro-averaged F-measure of the tokens-in-concepts baseline. Italic indicates micro-averaged F-measures of the SR classifier that are significantly different from the corresponding micro-averaged F-measure of the tokens-in-sentence baseline.
| BIDMC corpus | ||||||
|---|---|---|---|---|---|---|
| Relation type | Tokens-in-concepts baseline |
Tokens-in-sentence baseline |
SR classifier | |||
| Micro-avgd F-measure |
Macro- avgd F- measure |
Micro-avgd F-measure |
Macro- avgd F- measure |
Micro-avgd F-measure |
Macro- avgd F- measure |
|
| Present disease–treatment | 0.64 | 0.56 | 0.66 | 0.53 | 0.81 | 0.69 |
| Possible disease–treatment | 0.67 | 0.35 | 0.75 | 0.41 | 0.74 | 0.40 |
| Present symptom–treatment | 0.61 | 0.45 | 0.69 | 0.53 | 0.80 | 0.62 |
| Possible symptom–treatment | 0.76 | 0.30 | 0.89 | 0.41 | 0.95 | 0.47 |
| Disease–test | 0.81 | 0.67 | 0.73 | 0.68 | 0.88 | 0.80 |
| Disease–symptom | 0.93 | 0.71 | 0.94 | 0.77 | 0.96 | 0.84 |
| Overall performance | 0.75 | 0.49 | 0.75 | 0.54 | 0.86 | 0.63 |
| Partners corpus | ||||||
| Relation type |
Tokens-in-concepts baseline |
Tokens-in-sentence baseline |
SR classifier | |||
|
Micro-avgd F-measure |
Macro- avgd F- measure |
Micro-avgd F-measure |
Macro- avgd F- measure |
Micro- avgd F- measure |
Macro- avgd F- measure |
|
| Present disease–treatment | 0.69 | 0.42 | 0.75 | 0.55 | 0.78 | 0.58 |
| Possible disease–treatment | 0.63 | 0.60 | 0.66 | 0.52 | 0.76 | 0.65 |
| Present symptom–treatment | 0.58 | 0.46 | 0.62 | 0.48 | 0.68 | 0.57 |
| Possible symptom–treatment | 0.78 | 0.41 | 0.80 | 0.47 | 0.91 | 0.65 |
| Disease–test | 0.70 | 0.60 | 0.72 | 0.65 | 0.78 | 0.72 |
| Disease–symptom | 0.61 | 0.56 | 0.69 | 0.65 | 0.69 | 0.66 |
| Overall performance | 0.67 | 0.49 | 0.71 | 0.54 | 0.76 | 0.62 |
Table 3 shows that, on the BIDMC corpus, the SR classifier gives its best micro-averaged performance in possible symptom–treatment and disease–symptom relation types. On both corpora, the SR classifier significantly outperforms the baseline in recognizing the possible symptom–treatment relation type. Table 1 shows that two of the four relations in the possible symptom–treatment relation type are non-existent in the BIDMC corpus. In other words, the micro-averaged F-measure of 95% on this relation type is due to the fact that SR classifier mostly makes a decision between TAS and none relations. On the Partners corpus, the relatively improved sample size for this relation type changes the task to three-way classification and gives a micro-averaged F-measure of 91%.
On both corpora, the SR classifier consistently gains significantly over both baselines in present disease–treatment, present symptom–treatment, and disease-test relation types. The relations under these types are well represented in our corpora and they are characterized by repeating context. For example, the verb “show” occurring between a disease and a test is almost always indicative of the TRD relation.
The SR classifier fails to gain significantly over the baselines on the possible disease–treatment, possible symptom–-treatment, and disease–symptom relation types in the BIDMC corpus. The sample sizes limit the ability of our classifier to learn the relations in these types based on the BIDMC corpus. The improvement in the sample sizes of these relation types in the Partners corpus improves the significance of the gain of the SR classifier over the baselines.
To understand the strengths of the SR classifier, we re-evaluated it with each feature separately. Table 4 shows that most of the SR classifier’s gains come from inter-concept tokens, link path tokens, and lexical trigrams. The success of inter-concept tokens implies that key information regarding the nature of relations is captured by the context between the concepts. The lexical trigrams overlap in their content with inter-concept tokens; they capture trigrams of inter-concept tokens and add to these the left and right lexical trigrams of the candidate concepts. Link path tokens are similar in their content to lexical trigrams and inter-concept tokens; they represent a subset of the inter-concept tokens and lexical trigrams that are highlighted by the Link Grammar Parser.
Table 4.
Performance of SR classifier with different features over all relation types. Best micro-averaged results are in bold.
| BIDMC corpus | Partners corpus | |||
|---|---|---|---|---|
| Features | Overall micro-avgd F-measure |
Overall macro-avgd F-measure |
Overall micro-avgd F-measure |
Overall macro-avgd F-measure |
| Surface features | 0.68 | 0.27 | 0.58 | 0.25 |
| Tokens in concepts | 0.75 | 0.49 | 0.67 | 0.49 |
| Lexical trigrams | 0.81 | 0.58 | 0.70 | 0.56 |
| Inter-concept tokens | 0.84 | 0.56 | 0.72 | 0.58 |
| Verbs | 0.78 | 0.52 | 0.68 | 0.48 |
| Link path tokens | 0.83 | 0.55 | 0.71 | 0.54 |
| Link path | 0.67 | 0.27 | 0.55 | 0.26 |
| Syntactic bigrams | 0.80 | 0.55 | 0.70 | 0.53 |
| Head words | 0.75 | 0.47 | 0.65 | 0.48 |
| Inter-concept tokens + surface features | 0.84 | 0.57 | 0.73 | 0.58 |
| Inter-concept tokens + tokens in concepts | 0.85 | 0.60 | 0.75 | 0.63 |
| Inter-concept tokens + lexical trigrams | 0.86 | 0.63 | 0.74 | 0.59 |
| Inter-concept tokens + verbs | 0.85 | 0.61 | 0.74 | 0.60 |
| Inter-concept tokens + link path tokens | 0.84 | 0.58 | 0.72 | 0.58 |
| Inter-concept tokens + link path | 0.84 | 0.56 | 0.73 | 0.58 |
| Inter-concept tokens + syntactic bigrams | 0.85 | 0.60 | 0.73 | 0.59 |
| Inter-concept tokens + head words | 0.86 | 0.60 | 0.75 | 0.62 |
The second half of Table 4 shows the contribution of each of the individual feature sets to the performance of the SR classifier with inter-concept tokens. Inter-concept tokens by themselves achieve a micro-averaged F-measure of 84% on the BIDMC corpus and fail to gain significantly from any of the rest of the features; the difference between 84% and the maximum performance of 86% is not statistically significant. On the Partners corpus, the inter-concept tokens achieve a micro-averaged F-measure of 72%. These features obtain a significant contribution from tokens in concepts, lexical trigrams, verbs, and head words. In particular, the addition of either tokens in concepts or head words to the inter-concept tokens produces an F-measure of 75%, which is not significantly different from the performance of the SR classifier (with all of the features) on this corpus.
Due to the nature of our data, the Link Grammar’s contribution to our feature set is limited. Manual analysis of a sample of sentences from our BIDMC corpus reveal that of the 2,471 candidate pairs found in these sample sentences, only 35% (858) had link paths between them and 16% had complete linkages (386), i.e., do not contain null links. Despite the Link Grammar Parser’s inability to completely parse most of the analyzed sentences, the parser does make consistent decisions. Therefore, the link path tokens we extract from its output tend to exhibit some patterns. Link paths themselves tend to be less useful than link path tokens because of the granularity of information they contain. Sibanda et al. [4] showed that for semantic category recognition, even when sentences are incorrectly or only partially parsed, local links around most tokens tend to be correct and hence Link Grammar information is useful. However, for SR classification, we need long distance links (linking the candidate concepts) which are more susceptible to parsing errors. In the absence of valid parses, lexical information dominates the SR task.
8. Limitations
Our SR classifier builds on semantic categories and assertions. Therefore, its performance is affected by the quality of the annotations provided on categories and assertions. The experiments in this manuscript are run on ground truth semantic categories and assertions. This ground truth was generated mostly by non-medical annotators, and we expect that its quality could improve by more active involvement from annotators with medical training. Nonetheless, running the SR classifier on the ground truth for semantic categories and assertions shields the SR classifier from the noisy annotations that would have been created by automatic means of generating these annotations. Therefore, we expect that when incorporated into a system that works end-to-end and processes unannotated free text to first find semantic categories, then assertions, and then relations, the performance may degrade. The development of this end-to-end system is left for future work.
9. Conclusion
We have described a SR classifier that can effectively identify a set of fine-grained relations for providing a problem-oriented account of medical discharge summaries. This classifier uses semantic categories and assertions made on them as a starting point. In experiments conducted with ground truth semantic categories and assertions, our classifier significantly outperformed the baseline of classifying relations solely based on the tokens found in concepts. It also significantly outperformed the baseline of classifying relations based on the tokens in sentences.
This SR classifier uses lexical and syntactically-informed feature sets; however, the lexical features (in particular, tokens occurring between candidate concepts) are the most informative for identifying the relations studied. Using only the inter-concept tokens in the corpus, our SR classifier can recognize 84% of the relations in the BIDMC corpus and 72% of the relations in the Partners corpus.
This SR classifier is to be integrated into a system that can process raw text of discharge summaries to extract first semantic categories, then their assertions, and finally their relations. However, when run on the non-perfect output of automatic semantic categories and assertions, the performance of the SR classifier would most likely decrease. Nonetheless, the results presented in this manuscript imply that we can take advantage of lexical patterns in classifying physician-stated fine-grained relations of concepts found in clinical records. These relations provide high level summaries of the findings, observations, and the rationale for treatment as stated by the doctors and support the development of a problem-oriented account of the experiences of patients.
Acknowledgement
This work was supported in part by the NIH Road Map for Medical Research Grants U54LM008748. Institutional Review Board approval has been granted for the studies presented in this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
We define a concept as a phrase that can be mapped to a UMLS semantic type.
As mentioned in Section 4, we filter sentences with zero candidate concept pairs from our corpus.
References
- 1.Weed L. Medical records that guide and teach. The New England Journal of Medicine. 1968;178(12) doi: 10.1056/NEJM196803212781204. [DOI] [PubMed] [Google Scholar]
- 2.Meystre S, Haug PJ. Automation of a problem list using natural language processing. BMC Medical Informatics and Decision Making. 2005;5(30) doi: 10.1186/1472-6947-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Unified Medical Language System. [accessed November 29, 2009];2009 http://www.nlm.nih.gov/research/umls/
- 4.Sibanda T, He T, Szolovits P, Uzuner Ö. Syntactically-informed semantic category recognizer for discharge summaries. In: Tang PC, editor. Proceedings of the American Medical Informatics Association; Hanley & Belfus, Inc; Philadelphia, PA, USA. 2006. pp. 714–718. [PMC free article] [PubMed] [Google Scholar]
- 5.Uzuner O, Zhang X, Sibanda T. Machine learning and rule-based approaches to assertion classification. J Am Med Inform Assoc. 2009;16:109–115. doi: 10.1197/jamia.M2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang C, Lin C. Manual, Department of Computer Science and Information Engineering. Taipei, Taiwan: National Taiwan University; 2001. [accessed 6.10.09]. LIBSVM: a library for support vector machines. http://www.csie.ntu.edu.tw/cjlin/libsvm/ [Google Scholar]
- 7.Malaisé V, Zweigenbaum P, Bachimont B. Detecting semantic relations between terms in definitions. In: Ananiadou S, Zweigenbaum P, editors. Proceedings of the 3rd International Workshop on Computational Terminology (CompuTerm 2004); Association for Computational Linguistics; Morristown, NJ, USA. 2004. pp. 55–62. [Google Scholar]
- 8.Delbecque T, Jacquemart P, Zweigenbaum P. Indexing UMLS semantic types for medical question-answering. Studies in Health Technology and Informatics. 2005;116:805–810. [PubMed] [Google Scholar]
- 9.Rindflesch TC, Rayan JV, Hunter L. Extracting molecular binding relationships from biomedical text. In: Nirenburg S, editor. Proceedings of the 6th Conference on Applied Natural Language Processing; Association for Computational Linguistics; Morristown, NJ, USA. 2000. pp. 188–195. [Google Scholar]
- 10.Feldman R, Regev Y, Finkelstein-Landau M, Hurvitz E, Kogan B. Mining biomedical literature using information extraction. Current Drug Discovery. 2002;2(10):19–23. [Google Scholar]
- 11.Friedman C, Alderson PO, Austin JHM, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1:161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leroy G, Hsinchun C, Martinez J, Eggers S, Falsey R, Kislin K, Huang Z, Li J, Xu J, McDonald D, Nq G. Genescene: Biomedical Text and Data Mining. In: Marshall CC, editor. JCDL '03: Proceedings of the 3rd ACM/IEEE-CS joint conference on Digital libraries; IEEE Computer Society; Washington, DC, USA. 2003. pp. 116–118. [Google Scholar]
- 13.Rindflesch TC, Bean CA, Sneiderman CA. Argument identification for arterial branching predications asserted in cardiac catheterization reports. In: Overhage JM, editor. Proceedings of the American Medical Informatics Association; Hanley & Belfus, Inc; Philadelphia, PA, USA. 2000. pp. 704–708. [PMC free article] [PubMed] [Google Scholar]
- 14.Srinivasan P, Rindflesch T. Exploring text mining from MEDLINE. In: Kohane IS, editor. Proceedings of the American Medical Informatics Association; Hanley & Belfus, Inc; Philadelphia, PA, USA. 2002. pp. 722–726. [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman C, Kra P, Yu H, Krauthammer M, Rzhetsky A. GENIES: a natural-language processing system for the extraction of molecular pathways from journal articles. Bioinformatics. 2001;17 Suppl 1:S74–S82. doi: 10.1093/bioinformatics/17.suppl_1.s74. [DOI] [PubMed] [Google Scholar]
- 16.Chen L, Friedman C. Extracting phenotypic information from the literature via natural language processing. Studies in Health Technology and Informatics. 2004;107(Pt 2):758–762. [PubMed] [Google Scholar]
- 17.Fundel K, Küffner R, Zimmer R. RelEx--Relation extraction using dependency parse trees. Bioinformatics. 2007 Feb 1;23(3):365–371. doi: 10.1093/bioinformatics/btl616. Epub 2006 Dec 1. [DOI] [PubMed] [Google Scholar]
- 18.Bui QC, Nualláin BO, Boucher CA, Sloot PM. Extracting causal relations on HIV drug resistance from literature. BMC Bioinformatics. 2010 Feb 23;11:101. doi: 10.1186/1471-2105-11-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosario B, Hearst M. Classifying semantic relations in Bioscience texts. In: Scott D, editor. ACL '04: Proceedings of the 42nd Annual Meeting on Association for Computational Linguistics; Association for Computational Linguistics; Morristown, NJ, USA. 2004. p. 430. [Google Scholar]
- 20.Craven M. Learning to extract relations from MEDLINE. In: Califf ME, editor. Proceedings of the AAAI Workshop on Machine Learning for Information Extraction; American Association for Artificial Intelligence; Menlo Park, California, USA. 1999. pp. 25–30. [Google Scholar]
- 21.Niu Y, Hirst G. Analysis of semantic classes in medical text for question answering. In: Molla D, Vicedo JL, editors. Proceedings of the Workshop on Question Answering in Restricted Domains; Association for Computational Linguistics; Morristown, NJ, USA. 2004. pp. 54–61. [Google Scholar]
- 22.Roberts A, Gaizauskas R, Hepple M, Guo Y. Mining clinical relationships from patient narratives. BMC Bioinformatics. 2008 Nov 19;9 Suppl 11:S3. doi: 10.1186/1471-2105-9-S11-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guiliano C, Lavelli A, Romano L. Exploiting shallow linguistic information for relation extraction from biomedical Literature. In: Pado S, Read J, Seretan V, editors. Proceedings of the 11th Conference of the European Chapter of the Association for Computational Linguistics (EACL06); Association for Computational Linguistics; Morristown, NJ, USA. 2006. pp. 401–408. [Google Scholar]
- 24.Bundschus M, Dejori M, Stetter M, Tresp V, Kriegel HP. Extraction of semantic biomedical relations from text using conditional random fields. BMC Bioinformatics. 2008 Apr 23;9:207. doi: 10.1186/1471-2105-9-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen J. A Coefficient of Agreement for Nominal Scales. Educational and Psychological Measurement. 1960;20(1):37–46. [Google Scholar]
- 26.Hripcsak G, Rothschild AS. Agreement, the F-measure, and reliability in information retrieval. J Am Med Inform Assoc. 2005 May–June;12(3):296–298. doi: 10.1197/jamia.M1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burges CJC. A tutorial on support vector machines for pattern recognition. Data Min Knowl Discov. 1998;2(2):121–167. [Google Scholar]
- 28.Cortes C, Vapnik V. Support-vector networks. Machine Learning, v. 20. 1995;no. 3:273–297. [Google Scholar]
- 29.The Specialist NLP tools. [accessed 28 July 2009];2009 http://lexsrv3.nlm.nih.gov/SPECIALIST/index.html.
- 30.Brill E. A simple rule-based part of speech tagger. In: Batest M, Stock O, editors. Proceedings of the 3rd Conference on Applied Natural Language Processing; Association for Computational Linguistics; Morristown, NJ, USA. 1992. pp. 152–155. [Google Scholar]
- 31.Bhooshan N. Master’s thesis, Electrical Engineering and Computer Science Department. Cambridge, MA, USA: Massachusetts Institute of Technology; 2005. Feb, Classification of semantic relations in different syntactic structures in medical text using the MeSH hierarchy. [Google Scholar]
- 32.Sleator D, Temperley D. Parsing English with a link grammar. Pittsburgh, PA, USA: Carnegie Mellon University; Technical Report CMU-CS-91-196, Computer Science Department. 1991
- 33.Szolovits P. Adding a medical lexicon to an English parser. In: Musen M, editor. Proceedings of the American Medical Informatics Association; Hanley & Belfus, Inc; Philadelphia, PA, USA. 2003. pp. 639–643. [PMC free article] [PubMed] [Google Scholar]
- 34.Ding J, Berleant D, Xu J, Fulmer A. Extracting biochemical interactions from MEDLINE using a Link Grammar Parser. In: Zhang D, editor. Proceedings of the IEEE International Conference on Tools with Artificial Intelligence; IEEE Computer Society; Los Alamitos, CA, USA. 2003. p. 467. [Google Scholar]
- 35.Z-test for two proportions. [accessed 6 August, 2009];2007 http://www.dimensionresearch.com/resources/calculators/ztest.html.
- 36.Osborn CE. Statistical applications for health information management. 2nd Ed. Boston, MA, USA: Jones & Bartlett Publishers; 2005. [Google Scholar]
- 37.Chinchor N. The Statistical Significance of the MUC-4 Results. MUC4 '92: Proceedings of the 4th Conference on Message Understanding; Association for Computational Linguistics; Morristown, NJ, USA. 1992. pp. 30–50. [Google Scholar]