Abstract
Studies over the past half-century have made it clear that environmental influences in development, particularly stress and traumatic experiences, can remain pervasive across the lifespan. Though it has been hypothesized for some time that the long-term consequences of early-life adversity represent epigenetic influences, it has not been until recently that studies have begun to provide empirical support of experience-driven epigenetic modifications to the genome. Here we focus on this theme, and review current knowledge pertaining to the epigenetics of behavioral development. At the center of our discussion is the brain-derived neurotrophic factor (BDNF) gene, as abnormal BDNF gene activity is a leading etiological hypothesis by which early-life adverse experiences persistently modify brain and behavioral plasticity.
Keywords: early-life experience, abuse, stress, epigenetic, DNA methylation, histone modification, BDNF gene
Introduction
Early-life stress and traumatic experiences are known to promote long-term neurobiological changes. For example, the experiences of childhood abuse and neglect are associated with elevated rates of anxiety, depression, and psychosis (e.g. Bremner, 2003; Heim and Nemeroff, 2001; Kaufman, et al., 2000; Schore, 2002). Imaging studies on adults who report such experiences have identified a number of lasting neural consequences, and suggest that aberrant function and responsiveness of the prefrontal cortex, amygdala, hippocampus, and hypothalamic-pituitary adrenal (HPA) axis likely have some role in the cognitive dysfunction associated with childhood maltreatment (e.g. De Bellis, 2005; Gunnar and Quevedo, 2007; Lupien et al., 2009; Perry, et al., 1995; Teicher, et al., 2003).
Adverse experiences (i.e. social interactions and environmental stressors) in developing rodents and non-human primates are equally associated with behavioral dysfunction, and common behavioral abnormalities include deficits in information processing, impaired memory, heightened fear- and anxiety-like behaviors, altered drug-seeking behavior, and social withdrawal (e.g. Gunnar and Quevedo, 2007; Kaffman and Meaney, 2007; Korosi and Baram, 2009; Pryce and Feldon, 2003; Sanchez, 2006). While rodent and non-human primate studies replicate the vulnerability of the prefrontal cortex, amygdala, hippocampus, and HPA axis to early-life adversity, they also highlight the lasting negative impact manifest at the cellular and molecular levels. A number of structural and functional consequences have been identified, and include aberrant synaptic density and structure, altered NMDA-receptor mediated signaling, attenuated neurogenesis, and deficits in synaptic long-term potentiation (Aisa, et al., 2009; Bock, et al., 2005; Brunson, et al., 2005; Fenoglio, et al., 2006; Gruss, et al., 2008; Huot, et al., 2002; Korosi, et al., 2010; Mirescue, et al., 2004).
Altogether, clinical and basic research efforts have made it clear that the developing brain is extraordinarily sensitive to environmental influences and that early-life experiences, particularly those occurring during heightened periods of brain plasticity, help determine life-long structural and functional aspects of brain and behavior. With the renewed interest in understanding the effects of early-life conditions on life-long health and behavior, this has prompted several recent investigations into whether the long-term consequences of early-life adverse conditions reflect sustained CNS gene effects that occurred as result of epigenetic modifications (McGowan, et al., 2008, 2009; Mueller and Bale, 2008; Murgatroyd, et al., 2009; Roth, et al., 2009).
Epigenetics refers to the chemical modifications made to chromatin (DNA and the associated histone proteins) that help regulate transcription of the genome, and at present, DNA methylation has been the most studied epigenetic mechanism in regard to understanding early-life experiences and neurobiological outcomes. DNA methylation is increasingly being recognized for its role in mediating gene-environment interplay throughout the lifespan, as studies have now documented both dynamic (Lubin, et al., 2008; Miller and Sweatt, 2007; Miller, et al., 2008; Penner, et al., in press; Westberry, et al., 2008; Yossifoff, et al., 2008) and stable (Abdomaleky, et al., 2005; Champagne, et al., 2006; Grayson, et al., 2005; McGowan, et al., 2008, 2009; Mueller and Bale, 2008; Murgatroyd, et al., 2009; Onishchenko, et al., 2008; Roth, et al., 2009; Weaver, et al., 2004) changes in CNS DNA methylation during early development and in adulthood. The stable nature of DNA methylation renders it an ideal substrate for mediating sustained gene effects controlling brain function and behavior. Thus we and others have proposed that the brain and behavioral dysfunction associated with early-life adverse experiences reflects the lasting imprint of such experiences on gene DNA methylation (McGowan, et al., 2008; 2009; Murgatroyd, et al., 2009; Roth, et al., 2009). Here we discuss data from these studies, including our own data demonstrating epigenetic marking of the brain-derived neurotrophic factor (BDNF) gene by adverse social interactions and environmental conditions in early infant development, and the hypothesized neurobiological consequences.
Early-life adversity, BDNF gene expression, and behavioral outcome
Since BDNF’s neurotrophic actions are vital for both brain development and plasticity and because BDNF exhibits activity-regulated release in the CNS (e.g. Conner, et al., 1997; Greenberg, et al., 2009; Hennigan, et al., 2007), the BDNF gene has been the focus of numerous developmental studies aimed at understanding the relationship between early-life stress, brain responses, and behavioral outcome. Regardless of the animal model of early-life adversity, studies have consistently indicated that altered behavioral outcomes are well-correlated with stable changes in both BDNF gene transcription and protein expression.
For example, infant mice raised in communal nests where they experience higher levels of maternal care have an increased propensity for social interaction in adulthood that is correlated with increased hippocampal BDNF protein levels (Branchi, 2009; Branchi, et al. 2006). Bouts of infant isolation from the mother and or nest have been shown to alter an array of behaviors, and in regard to BDNF, most studies indicate that isolation reduces levels of both mRNA and protein in the prefrontal cortex, amygdala, and hippocampus (e.g. Branchi, et al., 2004; Chatterjee, et al., 2007; Choy, et al., 2008; Fumagalli, et al., 2007; Lippman, et al., 2007; Nair, et al., 2007; Zimmerberg, et al., 2009). Other recent studies have demonstrated that adults who were weaned at an early age (at PN14) are more anxious and have increased stress responsivity, effects that are correlated with decreased hippocampal BDNF synthesis (Kikusui, et al., 2009), and that adolescent and adult rats who received less care from the mother have increased anxiety-related behavior that is accompanied by abnormal hippocampal and amygdala BDNF protein levels (Macri, et al., 2009).
Overall, data demonstrate that early-life events influence the BDNF gene and behavioral outcome. Hence aberrant BDNF gene activity continues to receive attention as a candidate molecular mechanism through which early-life adversity is able to produce stable modifications in brain and behavioral plasticity (Alleva and Francia, 2009; Branchi, et al., 2004; Calabrese, et al., 2009; Casey, et al., 2009; Cirulli, et al., 2009; Fumagalli, et al., 2007). It is interesting to note that other neurotrophins, such as nerve growth factor (NGF), may play similar roles (Alleva and Branchi, 2006; Cirulli and Alleva, 2009; Cirulli, et al., 2009). To provide an explanation as to how experiences so early in development are able to successfully influence transcriptional regulation of the BDNF gene and behavior into adulthood, we explored whether epigenetic modifications are involved (Roth, et al., 2009).
Epigenetic modifications regulate CNS gene activity
DNA methylation is the direct covalent modification of DNA, where at least three encoded enzymes known as DNA methyltransferases (DNMTs) are known to catalyze the addition of a -CH3 group to cytosine residues at the 5-position of the pyrimidine ring (Bird, 2002; Miranda and Jones, 2007). DNA methylation has been recognized for some time for its role in a number of developmental processes and neurodevelopmental disorders that are associated with long-lasting phenotypic changes. These include cellular differentiation, X-chromosome inactivation, Rett syndrome, and Fragile X mental retardation (Amir, et al., 1999; Das, et al., 1997; Yang and Kuroda 2007). The phenotypic outcomes in these examples are due to patterns of DNA methylation that are set very early in development and that remain stable throughout the lifespan. From this, DNA methylation has traditionally been viewed as a static process following neural development and cell differentiation.
If however, DNA methylation is a mechanism contributing to the effects of early-life adversity, DNA methylation and its enzymatic machinery would need to remain labile and capable of responding to environmental factors. Indeed studies continue to challenge the static view of DNA methylation by providing evidence that DNA methylation remains an active process in post-mitotic cells (i.e. neurons). For example, robust levels of DNMTs are present in CNS neurons in the adult brain (Brooks, et al., 1996; Brown, et al., 2008; Goto, et al., 1994). Additionally, evidence continues to mount that changes in gene activity throughout the lifespan as a result of exposure to a variety of environmental factors, including toxins, diet, and stress, involve epigenetic mechanisms (e.g Jirtle and Skinner, 2007; Liu, et al., 2009; Onishchenko, et al., 2008). Furthermore, work continues to indicate that DNA methylation and demethylation can be rapidly and transiently induced in order to dynamically regulate gene transcription even in the adult brain (Levenson, et al., 2006; Lubin, et al., 2008; Miller and Sweatt, 2007; Miller, et al., 2008; Nelson, et al., 2008; Penner, et al., in press; Tian, et al., 2009 Westberry, et al., 2008).
DNA methylation in concert with histone post-translational modifications and their associated enzymatic machinery are increasingly being recognized for their important role in regulating gene transcription in the CNS. Our current understanding is that transcriptionally active genes are characterized by unmethylated cytosines and both histone acetylation and phosphorylation. Acetylation of the lysine residues of histone tails is catalyzed by enzymes known as histone acetyltransferases (HATs). This modification effectively decreases the affinity between the protein tail and DNA, relaxes chromatin, and thus promotes gene transcription (Marmorstein and Trievel, 2009).
Conversely, transcriptionally inactive chromatin is characterized by methylated cytosines and both histone deacetylation and histone methylation (Bird, 2002; Miranda and Jones, 2007). Histone deacetylases (HDACs) catalyze the reversal of histone acetylation (Haberland, et al., 2009), while DNMTs catalyze cytosine methylation. Methylated cytosines in turn bind repressor proteins, including the methyl-binding domain protein MeCP2, as well as HDAC1 (Bird, 2002; Miranda and Jones, 2007). This effectively condenses chromatin and thus suppresses transcription. However, recent studies have suggested that DNA methylation might also be associated with transcriptional activation (Chahrour et al., 2008; Cohen, et al., 2008; Yasui, et al., 2007).
Early-life adversity and epigenetic modifications to the BDNF gene
To determine whether DNA methylation could be a mechanism by which adverse infant experiences render some of their neurobiological consequences, we exposed infant rats to a stressed, “abusive” caregiver for 30 min daily during the first seven days of life (Roth, et al., 2009). We potentiated the maladaptive behaviors from mothers, such as pup dragging and rough handling, by placing them in an unfamiliar environment with limited bedding material. A limited bedding regimen has been used in other laboratories to produce stressful early-life environmental conditions that evoke changes in later behavior, increase levels of the stress hormone corticosterone, and elicit amygdala responsivity (Gilles, et al., 1996, Ivy, et al., 2008; Moriceau, et al., 2009; Roth and Sullivan, 2005). For control conditions, we exposed littermates to either a non-stressed, positive caregiver or kept littermates in the homecage.
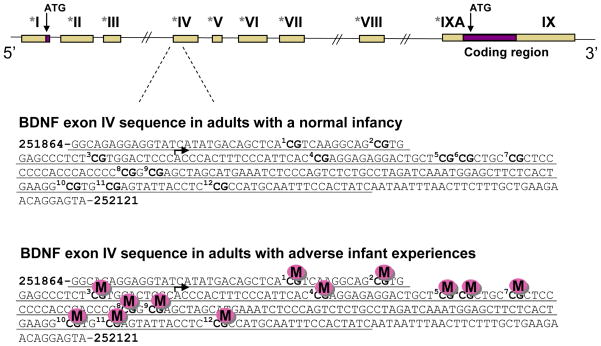
At least 3 months following these adverse conditions, rat had significantly lower levels of BDNF mRNA in their prefrontal cortex, an observation well in-line with previous findings where early-life experiences are known to having a lasting impact on this gene. Using a variety of techniques to assess DNA methylation within the prefrontal cortex, adults with adverse experiences during infancy were also found to have hypermethylated BDNF DNA, an effect that for the most part arose in infancy and persisted through adolescence and into adulthood. As depicted in Figure 1, sequencing of an important regulatory region of the BDNF gene (exon IV), revealed that across 12 CG dinucleotide sites within that regulatory region, normal adults (i.e. history of normal infancy) had either no or very little cytosine methylation. This is in sharp contrast to the adults who had experienced the adverse conditions during infancy, where sequencing revealed that those same CG sites were all highly methylated. Overall, our data illustrate that adverse experiences in early development can induce a lasting change in the methylation status of BDNF DNA. Variations in the level of normal maternal behaviors during the first week of life have also been shown to stably modify DNA methylation of the glucocorticoid receptor (GR) and estrogen receptor alpha (ER-alpha) genes such that it alters their gene transcription throughout the lifespan (Champagne, et al., 2006; Weaver, et al., 2004).
Figure 1.
Epigenetic marking of the BDNF gene by early-life adverse experiences. Top panel – Schematic describing the organization of the rodent BDNF gene, which contains nine non-coding exons and a common coding exon (Aid, et al., 2007; Timmusk, et al., 1993). Asterisks designate promoters, tan boxes represent untranslated regions, and purple boxes represent protein coding regions. Bottom panels – Sequencing analysis in the prefrontal cortex of adults that had experienced favorable social interactions and environmental conditions during infancy revealed little or no cytosine methylation across 12 CG dinucleotide sites examined for BDNF exon IV (sites are numbered and in bold). Conversely, sequencing analysis of adult animals that had been maltreated as infants revealed significant cytosine methylation across the targeted region. Data recreated from Roth, et al., 2009.
In light of our BDNF methylation results, we reasoned that if the observed gene deficits were due to DNA methylation, then a drug capable of reversing DNA methylation levels should reverse the deficits in BDNF gene expression. To address this hypothesis, we took adult rats that had experienced adverse conditions during infancy and treated them with a demethylating agent called zebularine, a drug recognized for its remarkable ability to reverse DNA methylation in the treatment of cancer (Cheng, et al., 2003; Marquez, et al., 2005). We found that after a 7-day treatment with zebularine, both the aberrant DNA methylation and gene expression patterns incited by early-life adversity had been reversed. Interestingly, we found no effects of the treatment in control animals, indicating a specific interaction with early-life experience.
As discussed in an earlier section, there is ample evidence that early-life adversity produces aberrant BDNF gene activity that is correlated with lasting changes in behavior. We thus aimed to determine whether our early-life experience regimen had likewise affected behavioral outcome. We found that females with a history of infant adversity (maltreated-females) showed the same types of abusive behaviors toward offspring that they themselves had experienced as infants. Lastly, in one final series of experiments, we sought to determine whether the epigenetic modifications could be transmitted across a generation. We found that indeed eight-day-old offspring (both males and females) derived from the maltreated-females had significant methylation of BDNF DNA in their prefrontal cortex and hippocampus in comparison to offspring derived from normal-treated females. Strikingly, cross-fostering experiments (where offspring born to maltreated-females were cross-fostered to normal females and vice versa) allowed us to determine that the transgenerational inheritance was not simply a product of the postnatal experience, but likely reflected some unidentified prenatal component.
In sum, our results demonstrate a remarkable robustness to the experience-driven changes in BDNF DNA methylation by early-life adverse experiences. First, our data indicate that adverse social interactions and environmental conditions during the first week of life can alter cortical BDNF gene expression through epigenetic mechanisms. Second, and perhaps most intriguingly, our results have highlighted an epigenetic molecular mechanism potentially underlying not only lifelong but transgenerational effects incited by early-life adverse conditions. Finally, our results with zebularine indicate that the effects of early-life adversity are potentially modifiable. These latter results dovetail those where HDAC inhibitors have been used successfully to modify epigenetic effects of maternal care on the GR gene (Weaver, et al., 2004, 2005, 2006).
Early-life adversity and epigenetic modifications to other genes
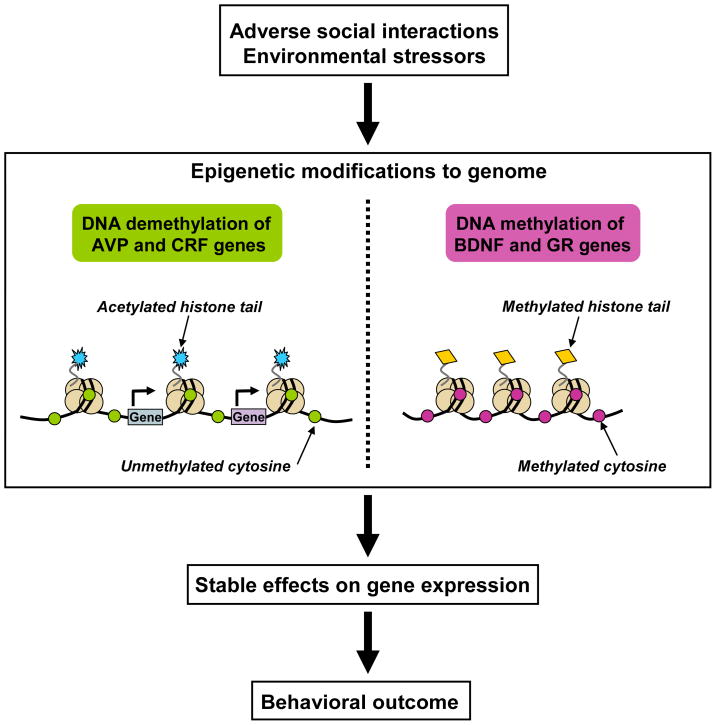
Recent studies have also indicated the ability of early-life adversity to epigenetically mark the DNA of other genes. For example, periodic separation of an infant from the caregiver (3 hr daily; ELS) during early-life induces hypomethylation of the arginine vasopressin (AVP) gene, an effect that coincided with increased corticosterone secretion both at basal conditions and in response to stress, as well as an attenuated memory capacity (Murgatroyd, et al., 2009). A chronic, variable daily stress regimen to pregnant mice during early gestation has been shown to produce a depressive-like phenotype in adult offspring that parallels hypomethylation of specific cytosines with the regulatory regions of the corticotropin-releasing factor (CRF) gene in both the hypothalamus and amygdala (Mueller and Bale, 2008). Finally, hippocampal samples derived from suicide victims with a history of childhood maltreatment (abuse and/or neglect) have decreased levels of GR mRNA that are correlated with increased cytosine methylation (McGowan, et al., 2009). Altogether, evidence continues to mount that both DNA methylation and demethylation at specific gene loci are involved in the neurobiological consequences of adverse early-life experiences (Figure 2). Though the effect of these experiences on histone modifications remains to be determined, it is likely the case that there is a combination of DNA methylation and histone modifications contributing to the long-term effects.
Figure 2.
Chromatin remodeling and its proposed role in governing the neurobiological consequences of early-life adverse experiences. Recent evidence indicates that adverse social interactions and stressful experiences early in development epigenetically mark genes in the CNS. Documented epigenetic changes include experience-driven DNA demethylation of the arginine vasopressin (AVP) and corticotropin-releasing factor (CRF) genes, and DNA methylation of the brain-derived neurotrophic factor (BDNF) and glucocorticoid receptor (GR) genes. These changes, along with those presumably occurring at histones, produce a unique epigenetic signature in the CNS that regulates transcription of the genome and influences behavioral output.
Concluding remarks
In the previous sections we discussed the data demonstrating that experiences in the immature animal produce lasting changes in behavior, BDNF gene activity, and epigenetic marking of the BDNF gene. The question remains unanswered whether these epigenetic alterations have directly caused the cognitive manifestations of early-life adversity. However, a growing body of literature continues to link epigenetic gene regulation, especially of the BDNF gene, with brain plasticity and cognitive function (Bredy, et al., 2007; Hunter et al., 2009; Levenson, et al., 2006; Lubin, et al., 2008; Tian, et al., 2009; Tsankova, et al., 2006). If epigenetic regulation of genes plays an active process in regulating an animal’s ability to respond to and form memories of its environment and experiences, then epigenetic modifications made to genes early in development could have the capacity to subsequently affect cognition. More studies are certainly necessary to provide a definitive link between early-experience-driven gene DNA methylation and behavioral outcome. In addition, as clinical work continues to indicate that there is an interaction between early-life stress, genotype (particularly for BDNF), and neurobehavioral outcome (e.g. Casey, et al., 2009; Caspi, et al., 2002, 2003; Gatt, et al., 2009), it is likely the case that to be in a position to fully understand how early experiences promote long-term changes in brain function and behavior, genetic polymorphisms with experience-driven epigenetic changes will need to be the focus of future work. A better understanding of their interaction holds promise of intervention strategies aimed at reducing the cognitive dysfunction and risk for psychiatric disorders associated with early-life stress and trauma. As a final point, since the effects of stressful and traumatic experiences on neurotrophins are not limited to early development (Alleva and Francia, 2009; Cirulli and Alleva, 2009; Duman and Monteggia, 2006), the characterization of stress-induced epigenetic modifications at these gene loci beyond infancy could provide valuable clues concerning the link between neurotrophins, later-life stress, and psychiatric disorders.
Acknowledgments
This work has been supported by grants from the National Institutes of Health, the National Alliance for Research on Schizophrenia and Depression, Civitan International, the Rotary Clubs CART fund, and the Evelyn F. McKnight Brain Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, Shafa R, Glatt SJ, Nguyen G, Ponte JF, Thiagalingam S, Tsuang MT. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet. 2005;134:60–6. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aisa B, Elizalde N, Tordera R, Lasheras B, Río JD, Ramírez MJ. Effects of neonatal stress on markers of synaptic plasticity in the hippocampus: Implications for spatial memory. Hippocampus. 2009;19:1222–1231. doi: 10.1002/hipo.20586. [DOI] [PubMed] [Google Scholar]
- Alleva E, Branchi I. NGF: A social molecule. Psychoneuroendocrinol. 2006;31:295–296. doi: 10.1016/j.psyneuen.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Alleva E, Francia N. Psychiatric vulnerability: Suggestions from animal models and role of neurotrophins. Neurosci Biobehav Rev. 2009;33:525–536. doi: 10.1016/j.neubiorev.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl- CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Bock J, Gruss M, Becker S, Braun K. Experience-induced changes of dendritic spine densities in the prefrontal and sensory cortex: correlation with developmental time windows. Cereb Cortex. 2005;15:802–808. doi: 10.1093/cercor/bhh181. [DOI] [PubMed] [Google Scholar]
- Branchi I. The mouse communal nest: Investigating the epigenetic influences of the early social environment on brain and behavior development. Neurosci Biobehav Rev. 2009;33:551–559. doi: 10.1016/j.neubiorev.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Branchi I, D’Andrea I, Fiore M, Di Fausto V, Aloe L, Alleva E. Early social enrichment shapes social behavior and nerve growth factor and brain-derived neurotrophic factor levels in the adult mouse brain. Biol Psychiatry. 2006;60:690–696. doi: 10.1016/j.biopsych.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Branchi I, Francia N, Alleva E. Epigenetic control of neurobehavioural plasticity: the role of neurotrophins. Behav Pharmacol. 2004;15:353–362. doi: 10.1097/00008877-200409000-00006. [DOI] [PubMed] [Google Scholar]
- Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem. 2007;14:268–276. doi: 10.1101/lm.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremner JD. Long-term effects of childhood abuse on brain and neurobiology. Child Adolesc Psychiatr Clin N Am. 2003;12:271–292. doi: 10.1016/s1056-4993(02)00098-6. [DOI] [PubMed] [Google Scholar]
- Brooks PJ, Marietta C, Goldman D. DNA mismatch repair and DNA methylation in adult brain neurons. J Neurosci. 1996;16:939–945. doi: 10.1523/JNEUROSCI.16-03-00939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SE, Weaver ICG, Meaney MJ, Szyf M. Regional-specific global cytosine methylation and DNA methyltransferase expression in the adult rat hippocampus. Neurosci Lett. 2008;440:49–53. doi: 10.1016/j.neulet.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Brunson KL, Kramar E, Lin B, Chen Y, Colgin LL, Yanagihara TK. Mechanisms of late-onset cognitive decline after early-life stress. J Neurosci. 2005;25:9328–9338. doi: 10.1523/JNEUROSCI.2281-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese F, Molteni R, Racagni G, Riva MA. Neuronal plasticity: A link between stress and mood disorders. Psychoneuroendocrinology. 2009;34:S208–S216. doi: 10.1016/j.psyneuen.2009.05.014. [DOI] [PubMed] [Google Scholar]
- Casey BJ, Glatt CE, Tottenham N, Soliman F, Bath K, Amso D, Altemus M, Pattwell S, Jones R, Levita L, McEwen B, Magariños AM, Gunnar M, Thomas KM, Mezey J, Clark AG, Hempstead BL, Lee FS. Brain-derived neurotrophic factor as a model system for examining gene by environment interactions across development. Neuroscience. 2009;164:108–120. doi: 10.1016/j.neuroscience.2009.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, McClay J, Moffitt TE, Mill J, Martin J, Craig IW, Taylor A, Poulton R. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–854. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R. Influence of life stress on depression: moderation by a polymorphism in the 5-HT gene. Science. 2003;301:386–400. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong STC, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne F, Weaver I, Diorio J, Dymov S, Szyf M, Meaney MJ. Maternal care associated with methylation of the estrogen receptor-alpha1b promoter and estrogen receptor-alpha expression in the medial preoptic area of female offspring. Endocrinology. 2006;147:2909–2915. doi: 10.1210/en.2005-1119. [DOI] [PubMed] [Google Scholar]
- Chatterjee D, Chatterjee-Chakraborty M, Rees S, Cauchi J, de Medeiros CB, Fleming AS. Maternal isolation alters the expression of neural proteins during development: ‘Stroking’ stimulation reverses these effects. Brain Res. 2007;1158:11–27. doi: 10.1016/j.brainres.2007.04.069. [DOI] [PubMed] [Google Scholar]
- Cheng JC, Matsen CB, Gonzales FA, Ye W, Greer S, Marquez VE, Jones PA, Selker EU. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J Natl Cancer Inst. 2003;95:399–409. doi: 10.1093/jnci/95.5.399. [DOI] [PubMed] [Google Scholar]
- Choy KH, de Visser Y, Nichols NR, van den Buuse M. Combined neonatal stress and young-adult glucocorticoid stimulation in rats reduce BDNF expression in hippocampus: Effects on learning and memory. Hippocampus. 2008;18:655–667. doi: 10.1002/hipo.20425. [DOI] [PubMed] [Google Scholar]
- Cirulli F, Alleva E. The NGF saga: From animal models of psychosocial stress to stress-related psychopathology. Front Neuroendocrinol. 2009;30:379–395. doi: 10.1016/j.yfrne.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Cirulli F, Francia N, Berry A, Aloe L, Alleva E, Suomi SJ. Early life stress as a risk factor for mental health: Role of neurotrophins from rodents to non-human primates. Neurosci Biobehav Rev. 2009;33:573–585. doi: 10.1016/j.neubiorev.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Zhou Z, Greenberg ME. Activating a repressor. Science. 2008;320:1172–1173. doi: 10.1126/science.1159146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner JM, Lauterborn JC, Yan Q, Gall CM, Varon S. Distribution of brain-derived neurotrophic factor (BDNF) protein and mRNA in the normal adult rat CNS: Evidence for anterograde axonal transport. J Neurosci. 1997;17:2295–2313. doi: 10.1523/JNEUROSCI.17-07-02295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Kubota T, Song M, Daniel R, Berry-Kravis EM, Prior TW, Popovich B, Rosser L, Arinami T, Ledbetter DH. Methylation analysis of the fragile X syndrome by PCR. Genetic Test. 1997;1:151–155. doi: 10.1089/gte.1997.1.151. [DOI] [PubMed] [Google Scholar]
- De Bellis MD. The psychobiology of neglect. Child Maltreatment. 2005;10:150–172. doi: 10.1177/1077559505275116. [DOI] [PubMed] [Google Scholar]
- Duman RS, Monteggia LM. A neurotrophic model for stress-related mood disorders. Biol Psychiatry. 2006;59:1116–1127. doi: 10.1016/j.biopsych.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Fenoglio KA, Brunson KL, Baram TZ. Hippocampal neuroplasticity induced by early-life stress: Functional and molecular aspects. Front Neuroendocrinol. 2006;27:180–192. doi: 10.1016/j.yfrne.2006.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli F, Molteni R, Racagni G, Riva MA. Stress during development: Impact on neuroplasticity and relevance to psychopathology. Prog Neurobiol. 2007;81:197–217. doi: 10.1016/j.pneurobio.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Gatt JM, Nemeroff CB, Dobson-Stone C, Paul RH, Bryant RA, Schofield PR, Gordon E, Kemp AH, Williams LM. Interactions between BDNF Val66Met polymorphism and early life stress predict brain and arousal pathways to syndromal depression and anxiety. Mol Psychiatry. 2009;14:681–695. doi: 10.1038/mp.2008.143. [DOI] [PubMed] [Google Scholar]
- Gilles EE, Schultz L, Baram TZ. Abnormal corticosterone regulation in an immature rat model of continuous chronic stress. Pediatr Neurol. 1996;15:114–119. doi: 10.1016/0887-8994(96)00153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Numata M, Komura J, Ono T, Bestor TH, Kondo H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994;56:39–44. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- Grayson D, Jia X, Chen Y, Sharma RP, Mitchell C, Guidotti A, Costa E. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci U S A. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ME, Xu B, Lu B, Hempstead BL. New insights in the biology of BDNF synthesis and release: Implications in CNS function. J Neurosci. 2009;29:12764–12767. doi: 10.1523/JNEUROSCI.3566-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss M, Braun K, Frey JU, Korz V. Maternal separation during a specific postnatal time window prevents reinforcement of hippocampal long-term potentiation in adolescent rats. Neuroscience. 2008;152:1–7. doi: 10.1016/j.neuroscience.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Gunnar M, Quevedo K. The neurobiology of stress and development. Annu Rev Psychol. 2007;58:145–173. doi: 10.1146/annurev.psych.58.110405.085605. [DOI] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim C, Nemeroff CB. The role of childhood trauma in the neurobiology of mood and anxiety disorders: preclinical and clinical studies. Biol Psychiatry. 2001;49:1023–1039. doi: 10.1016/s0006-3223(01)01157-x. [DOI] [PubMed] [Google Scholar]
- Hennigan A, O’Callaghan RM, Kelly ÃM. Neurotrophins and their receptors: roles in plasticity, neurodegeneration and neuroprotection. Biochem Soc Trans. 2007;35:424–427. doi: 10.1042/BST0350424. [DOI] [PubMed] [Google Scholar]
- Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proceedings of the National Academy of Sciences. 2009;106:20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot RL, Plotsky PM, Lenox RH, McNamara RK. Neonatal maternal separation reduces hippocampal mossy fiber density in adult Long Evans rats. Brain Res. 2002;950:52–63. doi: 10.1016/s0006-8993(02)02985-2. [DOI] [PubMed] [Google Scholar]
- Ivy AS, Brunson KL, Sandman C, Baram TZ. Dysfunctional nurturing behavior in rat dams with limited access to nesting material: A clinically relevant model for early-life stress. Neuroscience. 2008;154:1132–1142. doi: 10.1016/j.neuroscience.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A, Meaney MJ. Neurodevelopmental sequelae of postnatal maternal care in rodents: clinical and research implications of molecular insights. J Child Psychol Psychiatry. 2007;48:224–244. doi: 10.1111/j.1469-7610.2007.01730.x. [DOI] [PubMed] [Google Scholar]
- Kaufman J, Plotsky PM, Nemeroff CB, Charney DS. Effects of early adverse experiences on brain structure and function: clinical implications. Biol Psychiatry. 2000;48:778–790. doi: 10.1016/s0006-3223(00)00998-7. [DOI] [PubMed] [Google Scholar]
- Kikusui T, Ichikawa S, Mori Y. Maternal deprivation by early weaning increases corticosterone and decreases hippocampal BDNF and neurogenesis in mice. Psychoneuroendocrinology. 2009;34:762–772. doi: 10.1016/j.psyneuen.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Korosi A, Baram TZ. The pathways from mother’s love to baby’s future. Front Behav Neurosci. 2009:3. doi: 10.3389/neuro.08.027.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korosi A, Shanabrough M, McClelland S, Liu ZW, Borok E, Gao XB, Horvath TL, Baram TZ. Early-Life experience reduces excitation to stress-responsive hypothalamic neurons and reprograms the expression of corticotropin-releasing hormone. J Neurosci. 2010;30:703–713. doi: 10.1523/JNEUROSCI.4214-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Lippmann M, Bress A, Nemeroff CB, Plotsky PM, Monteggia LM. Long-term behavioural and molecular alterations associated with maternal separation in rats. Eur J Neurosci. 2007;25:3091–3098. doi: 10.1111/j.1460-9568.2007.05522.x. [DOI] [PubMed] [Google Scholar]
- Liu L, van Groen T, Kadish I, Tollefsbol TO. DNA methylation impacts on learning and memory in aging. Neurobiol Aging. 2009;30:549–560. doi: 10.1016/j.neurobiolaging.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic Regulation of bdnf Gene Transcription in the Consolidation of Fear Memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien SJ, McEwen BS, Gunnar MR, Heim C. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat Rev Neurosci. 2009;10:434–445. doi: 10.1038/nrn2639. [DOI] [PubMed] [Google Scholar]
- Macrì S, Laviola G, Leussis MP, Andersen SL. Abnormal behavioral and neurotrophic development in the younger sibling receiving less maternal care in a communal nursing paradigm in rats. Psychoneuroendocrinology. 2009;35:392–402. doi: 10.1016/j.psyneuen.2009.07.016. [DOI] [PubMed] [Google Scholar]
- Marmorstein R, Trievel RC. Histone modifying enzymes: Structures, mechanisms, and specificities. Biochim Biophys Acta. 2009;1789:58–68. doi: 10.1016/j.bbagrm.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez VE, Kelley JA, Agbaria R, Ben-Kasus T, Cheng JC, Yoo CB, Jones PA. Zebularine: A unique molecule for an epigenetically based strategy in cancer chemotherapy. Ann N Y Acad Sci. 2005;1058:246–254. doi: 10.1196/annals.1359.037. [DOI] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, Huang TCT, Unterberger A, Suderman M, Ernst C, Meaney MJ, Turecki G, Szyf M. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS ONE. 2008;3:e2085. doi: 10.1371/journal.pone.0002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Miranda TB, Jones PA. DNA methylation: The nuts and bolts of repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- Mirescu C, Peters JD, Gould E. Early life experience alters response to adult neurogenesis to stress. Nat Neurosci. 2004;7:841–846. doi: 10.1038/nn1290. [DOI] [PubMed] [Google Scholar]
- Moriceau S, Shionoya K, Jakubs K, Sullivan RM. Early-Life stress disrupts attachment learning: The role of amygdala corticosterone, locus ceruleus corticotropin releasing hormone, and olfactory bulb norepinephrine. J Neurosci. 2009;29:15745–15755. doi: 10.1523/JNEUROSCI.4106-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller BR, Bale TL. Sex-specific programming of offspring emotionality after stress early in pregnancy. J Neurosci. 2008;28:9055–9065. doi: 10.1523/JNEUROSCI.1424-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OFX, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Nair A, Vadodaria KC, Banerjee SB, Benekareddy M, Dias BG, Duman RS, Vaidya VA. Stressor-specific regulation of distinct brain-derived neurotrophic factor transcripts and cyclic AMP response element-binding protein expression in the postnatal and adult rat hippocampus. Neuropsychopharmacology. 2007;32:1504–1519. doi: 10.1038/sj.npp.1301276. [DOI] [PubMed] [Google Scholar]
- Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishchenko N, Nina KN, Sabri F, Castrén E, Ceccatelli S. Long-lasting depression-like behavior and epigenetic changes of BDNF gene expression induced by perinatal exposure to methylmercury. J Neurochem. 2008;106:1378–1387. doi: 10.1111/j.1471-4159.2008.05484.x. [DOI] [PubMed] [Google Scholar]
- Penner MR, Roth TL, Chawla MC, Hoang LT, Roth ED, Lubin FD, Sweatt JD, Worley PF, Barnes CA. Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2010.01.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry BD, Pollard R, Blakely T, Baker W, Vigilante D. Childhood trauma, the neurobiology of adaptation and ‘use-dependent’ development of the brain: How “states” become “traits”. Infant Mental Health J. 1995;16:271–291. [Google Scholar]
- Pryce CR, Feldon J. Long-term neurobehavioral impact of postnatal environment in rats: manipulations, effects and mediating mechanisms. Neurosci Biobehav Rev. 2003;27:57–71. doi: 10.1016/s0149-7634(03)00009-5. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Sullivan RM. Memory of early maltreatment: neonatal behavioral and neural correlates of maternal maltreatment within the context of classical conditioning. Biol Psychiatry. 2005;57:823–831. doi: 10.1016/j.biopsych.2005.01.032. [DOI] [PubMed] [Google Scholar]
- Sanchez MM. The impact of early adverse care on HPA axis development: Nonhuman primate models. Horm Behav. 2006;50:623–631. doi: 10.1016/j.yhbeh.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Schore AN. Dysregulation of the right brain: a fundamental mechanism of traumatic attachment and the psychopathogenesis of posttraumatic stress disorder. Aust N Z J Psychiatry. 2002;36:9–30. doi: 10.1046/j.1440-1614.2002.00996.x. [DOI] [PubMed] [Google Scholar]
- Teicher MH, Andersen SL, Polcari A, Anderson CM, Navalta CP, Kim DM. The neurobiological consequences of early stress and childhood maltreatment. Neurosci Biobehav Rev. 2003;27:33–44. doi: 10.1016/s0149-7634(03)00007-1. [DOI] [PubMed] [Google Scholar]
- Tian F, Hu XZ, Wu X, Jiang H, Pan H, Marini AM, Lipsky RH. Dynamic chromatin remodeling events in hippocampal neurons are associated with NMDA receptor-mediated activation of Bdnf gene promoter 1. J Neurochem. 2009;109:1375–1388. doi: 10.1111/j.1471-4159.2009.06058.x. [DOI] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- Tsankova N, Berton O, Renthal W, Kumar A, Neve R, Nestler E. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Weaver I, Champagne F, Brown S, Dymov S, Sharma S, Meaney MJ, Szyf M. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver I, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci U S A. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver ICG, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Westberry JM, Prewitt AK, Wilson ME. Epigenetic regulation of the estrogen receptor alpha promoter in the cerebral cortex following ischemia in male and female rats. Neuroscience. 2008;152:982–989. doi: 10.1016/j.neuroscience.2008.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PK, Kuroda MI. Noncoding RNAs and Intranuclear Positioning in Monoallelic Gene Expression. Cell. 2007;128:777–786. doi: 10.1016/j.cell.2007.01.032. [DOI] [PubMed] [Google Scholar]
- Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, Thatcher KN, Farnham PJ, LaSalle JM. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci U S A. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yossifoff M, Kisliouk T, Meiri N. Dynamic changes in DNA methylation during thermal control establishment affect CREB binding to the brain-derived neurotrophic factor promoter. Eur J Neurosci. 2008;28:2267–2277. doi: 10.1111/j.1460-9568.2008.06532.x. [DOI] [PubMed] [Google Scholar]
- Zimmerberg B, Foote HE, Van Kempen TA. Olfactory association learning and brain-derived neurotrophic factor in an animal model of early deprivation. Dev Psychobiol. 2009;51:333–344. doi: 10.1002/dev.20373. [DOI] [PubMed] [Google Scholar]