Abstract
The development of high-throughput sequencing methods allows for the characterization of microbial communities in a wide range of environments on an unprecedented scale. However, insight into microbial community composition is limited by our ability to detect patterns in this flood of sequences. Here we compare the performance of 51 analysis techniques using real and simulated bacterial 16S rRNA pyrosequencing datasets containing either clustered samples or samples arrayed across environmental gradients. We find that many diversity patterns are evident with severely undersampled communities, and that methods vary widely in their ability to detect gradients and clusters. Chi-squared distances and Pearson correlation distances perform especially well for detecting gradients, while Gower and Canberra distances perform especially well for detecting clusters. These results also provide a basis for understanding tradeoffs between number of samples and depth of coverage, tradeoffs which are important to consider when designing studies to characterize microbial communities.
Keywords: beta-diversity, high-throughput, pyrosequencing, community analysis, ordination, clustering
Studies of complex microbial communities, including those found on and within humans (the human microbiome1) and those found in both natural and engineered environments, have been constrained by the enormous levels of diversity contained within these communities. The vast majority of this diversity cannot be observed using cultivation-based techniques2. However, recent advances in DNA sequencing technology such as pyrosequencing3 provide the opportunity to survey microbial diversity in unprecedented detail, through direct sequencing of the small ribosomal subunit rRNA gene. Hundreds of individual communities can now be analyzed simultaneously by coupling pyrosequencing with the use of error-correcting barcoded primers4, as has been demonstrated in a range of environments including rivers, the mammalian gut, multiple environments in the human body, soil, and the atmosphere1,4–6. Modern datasets from a single study may contain hundreds of thousands to millions of 16S rRNA sequences, drawn from hundreds of environmental samples. Such sequences are obtained without the biases inherent in culture-dependant methods, and typically include many sequences representing undescribed and uncharacterized species. The ability to obtain such extensive data relatively easily and cheaply has revealed important constraints in our ability to detect patterns in these increasingly large and complex datasets, and to relate such patterns to underlying biotic or abiotic variables.
The problems associated with assessing and explaining patterns in complex datasets are not unique to the field of microbiology. For example, plant and animal ecologists have developed a variety of strategies for the analysis of the relationships between individual biological communities17–21. The major goal of many of the techniques for the comparison of biological communities among samples is the identification of an environmental gradient (or gradients) instrumental in structuring community diversity, and/or the identification of factors that contribute to the clustering of compositionally similar communities. Several approaches exist for elucidating diversity relationships among samples, including cluster analyses (where samples are assigned to discrete groups), ordination methods (where samples are arranged in low-dimensional space), and explicit hypothesis testing methods (such as ANOVA and Mantel tests).
Humans in particular host a wide variety of microbial communities: microbial cells outnumber human cells by an order of magnitude7, and microbial communities inhabiting different body habitats such as the mouth and the skin differ more from one another than do microbial communities inhabiting non-host-associated environments such as soil and water8. Microbial community composition has been associated with the health of the host, and variations in a host’s microbiome are linked to myriad disorders including obesity, vaginosis, and inflammatory bowel disease (IBD)1.
The interplay between environmental or host factors and microbial communities can be subtle and complex. However, many ecological systems are driven by environmental gradients; for example, pH has a major and consistent influence on soil microbial communities, whether traditional fingerprinting methods such as denaturing gradient gel electrophoresis (DGGE), restriction fragment length polymorphism (RFLP). or pyrosequencing analyses are used9. Whether equivalent gradients are found in human-associated body habitats is less clear. Meta-analysis of large numbers of hand and gut samples suggests that they might, although larger numbers of subjects with more careful phenotypic characterization will be required to define the patterns10. Previous work on the efficacy of different methods for identifying gradients, although useful, has typically relied on simulated datasets that are far smaller in scale than those currently being collected by pyrosequencing11–13. Although environmental gradients in host-associated microbial communities have not been frequently described, datasets that demonstrate clusters or categorical differences between host-associated microbial communities are relatively common. For example, different samples collected along the distal gut in three humans cluster by subject14, mammalian fecal samples cluster by diet15, and fecal pellets of mice cluster by diet and physiological state16. Do the methods that generally work well for gradient analysis in ecological systems also work well for cluster detection?
We consider only ordination analyses here, as they have been most useful for revealing patterns in large-scale surveys (Supplementary Table 1). In addition, we chose to address taxon-based (non-phylogenetic) methods in this paper because modeling phylogenetic approaches requires substantial additional decisions about the phylogenetic tree and the rate of environment switching, which make it more difficult to isolate the effects of ordination methods from the effects of model parameters. A discussion of such phylogenetic methods and their utility have been addressed previously10,19,22. We also consider only unconstrained ordination methods. Constrained methods (or direct gradient analysis methods) such as Canonical Correspondence Analysis (CCA) are useful when investigating the effect of measured environmental variables (sample pH, host health, or sample location) on microbial species present in a sample - in these methods the ordination axes are constrained to represent linear combinations of the measured environmental variables. However, here we assess techniques based on their ability to correctly reveal the diversity patterns inherent in microbial community sequence data, regardless of whether the researcher measured the underlying environmental variables responsible for shaping the communities. Finally, it is worth noting that although ordination methods allow simultaneous display of samples and species (biplots), we display only the samples here, as identification of the specific taxa responsible for differentiating samples does not affect a method’s usefulness at revealing sample clusters or gradients.
The optimal analysis approach depends on factors such as the size of the expected effect, the number of samples, the number of sequences per sample, the degree of replication, and the environmental data available for the sample set. The analysis techniques we compared were Principal Components Analysis (PCA) on raw abundance data as well as data subjected to chi-square, chord, hellinger, and species profile transforms, as well as both Principal Coordinates Analysis (PCoA) and Nonmetric Multidimensional Scaling (NMDS) techniques using each of the common dissimilarity metrics listed in Supplementary Table 2.
To assess the performance of these various analysis techniques, we used real and simulated pyrosequencing datasets modeling different microbial communities that we suspect are either shaped by a gradient in environmental conditions or partitioned by environmental factors into distinct groupings, or clusters of samples. We compared the performance of each analysis technique on real community data to the performance on simulated datasets where the inherent gradients and clusters of communities are known a priori. By using these simulated datasets we were able to distinguish between techniques that accurately reveal gradients and clusters inherent in the data versus those techniques that artificially generate patterns where they do not exist.
Results
Revealing Environmental Gradients
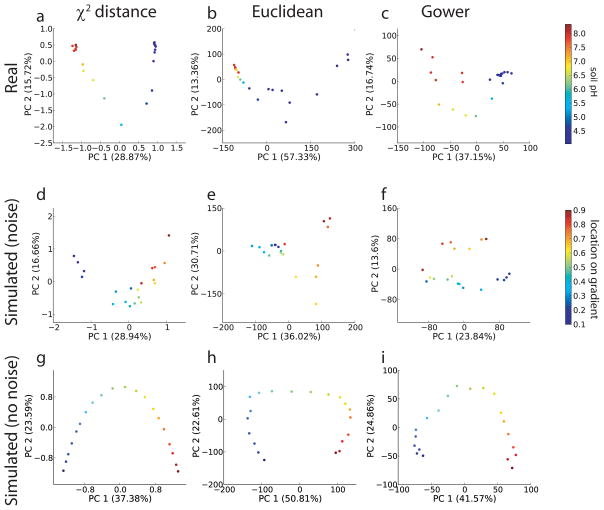
Our simulated gradient was fit from a soil microbial community dataset, where 16S rDNA sequences were acquired from samples of arable soil along an artificial pH gradient23,24 (Fig. 1a). We found that some techniques (notably those involving a χ2 distance: CA, or χ2 distance + PCoA or (NMDS) performed substantially better by our correlation quality metrics (see Methods) than other analysis techniques surveyed. These techniques also revealed a clear pH gradient when applied to the soil microbial community data described above (Fig. 2, and see Supplementary Table 3 for the full list of techniques surveyed). The close correspondence between the performance of these techniques with the soil pH data and the simulated data suggests that the simulations are relevant to analyses done on experimental data. The arch effect25 (where samples along a single environmental gradient are misleadingly placed in an arch configuration) is prominent in the simulated data, where we know there is only a single gradient. The presence of the same effect in the soil data suggests that the pH gradient in the soil is the single driving factor in these communities (compare top row and middle row of Fig. 2). Interestingly, the effects of noise differed substantially among methods: for example, the Gower distance + PCoA performs well in the absence of noise but is severely degraded in its presence (Fig. 2f and Fig. 2i), whereas the χ2 distance performed almost as well in the presence of noise as on the perfect dataset (Fig. 2d and Fig. 2g). Euclidean distance methods, as expected13, showed a strong arch effect. None of the methods we tested escaped the arch effect. See also Supplementary Tables 3 and 4 for more information on the performance of each method.
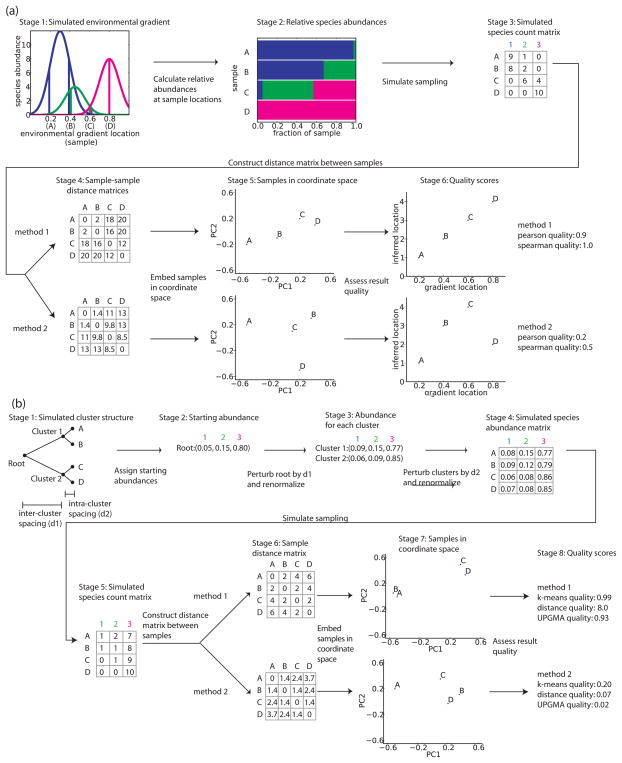
Figure 1.
Schematic of simulations and analysis of data. (a) 6 stages for the analysis of a simulated environmental gradient (b) Clustered samples. A hypothetical sample is formed at the root of a hierarchy which defines the relatedness of samples both inter- and intra-cluster (d1 and d2; stage 1). The species abundances at the root node (stage 2) are perturbed by an amount proportional to d1, and the results are renormalized to form the species abundances at each cluster (stage 3). The cluster nodes are then perturbed by d2 to produce species abundances at each sample (stage 4). Sample data is generated and analyzed similar to (a), and the analysis methods are then evaluated based on their ability to reveal the underlying cluster structure of the samples (stages 5–8).
Figure 2.
Comparison of different gradient methods on the soil dataset, a simulated gradient dataset with or without noise. Axes represent the first two principal coordinates maximizing the variance in the data, obtained via PCoA (the percentage of the total variance explained by each axis is shown in parentheses). Each data point is a microbial community sample, colored according to either a real gradient (soil pH) or a simulated gradient (arbitrary units). For simulated data, sequencing depth was 1,000 sequences per sample, and species rank-abundance distributions were fit from empirical data.
In addition, we generated simulated data with varying numbers of samples and depth of sequencing to determine how sequencing depth affects the performance of the ordination methods. We discovered that beyond approximately 100 sequences per sample, including more sequences was of rapidly diminishing utility for revealing the underlying gradient, provided one of the more effective ordination methods was used, and the gradient was sufficiently prominent. By resampling our empirical soil dataset, we saw that only below about 100 sequences per sample did analyses return substantially different results from analyses performed on the complete dataset (data not shown). These results are consistent with previous studies demonstrating that increasing the number of sequences per sample does not necessarily lead to an improvement in the ability to detect ecological patterns22,26. However, we did notice an improvement in the extent to which simulated subtle gradients were revealed at higher numbers of sequences per sample (Supplementary Fig. 1), suggesting that investigation of more subtle effects requires deeper sequencing.
Correlation-based distance methods (Pearson and Spearman) performed well at displaying the sample locations in a manner consistent with the underlying environmental gradient, as did chord distance methods, while the Gower distance performed notably poorly. Qualitative methods generally performed worse than quantitative (abundance-weighted) methods, and NMDS performed about as well as PCoA when the techniques were compared using the same distance measure (Supplementary Tables 3 and 4).
Revealing Sample Clustering
We now focus on the analyses of microbial communities that are not structured by a continuous gradient in environmental conditions, but rather are partitioned into discrete clusters of communities. As in the generation of simulated gradient datasets, we varied the number of samples and depth of sequencing. For some of the analysis, we set the relatedness between clusters, and the relatedness between samples within the same cluster to values that produced simulated data with similar clustering behavior to a dataset of 16S rDNA sequences from microbial communities on keyboards and human fingertips27 (between-cluster distance of 1.0, within-cluster distance of 0.5; see Fig. 1b for an outline of the simulation methodology). However, in some simulations, we simulated a more subtle effect (between-cluster and within-cluster distances of 0.1). We applied various ordination techniques to each simulated dataset, and quantitatively assessed each technique’s effectiveness at revealing the inherent clustering of the samples.
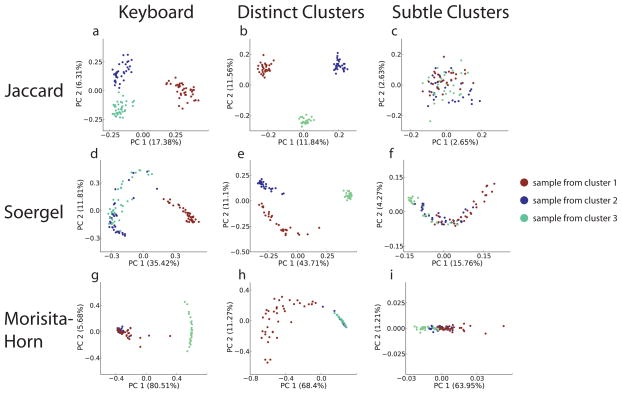
We found that the relative efficacy of different analyses was dependent on the relatedness of clusters, and that different analysis techniques applied to the same data were of substantially different effectiveness at revealing the underlying clusters (Fig. 3 and Supplementary Tables 3 and 5). The visual similarities between the results for the simulated prominent clusters and the results using actual keyboard data suggest that the model provides useful insight into the real dataset, and that the three-cluster structure is a good fit for the real data. Different methods behave differently. For example, the Jaccard distance + PCoA is able to recover the clusters well when they are prominent, although not when they are subtle, at a depth of coverage of 1,000 sequences per sample. In contrast, although they explain far more of the variance in the data, the Soergel and Morisita-Horn distance measures do not clearly recover the three-cluster pattern. Consequently, evaluating a method based on the percentage of the variance it explains rather than the biological insight it provides is likely to be a poor approach.
Figure 3.
Choice of analysis method reveals or obscures clusters. Keyboard data, simulated data resembling the keyboard data (distinct clusters), and simulated data representing less prominent sample clusters (subtle clusters) were analyzed by the indicated techniques All simulated data shown in this figure had 90 samples divided into 3 clusters, with 1,000 sequences per sample. Axes are labeled as in Figure 2.
Notably, the chi-squared distance measure, which performs superbly on gradient data, performs only moderately on cluster data (Supplementary Table 3). More generally, performance of methods on gradient and cluster data was weakly but negatively correlated (Spearman rank correlation r = −0.49), suggesting that high-performing methods from both classes should be applied to maximize the information extracted from a given dataset. For distance matrix based methods, the choice of distance measure typically had a more profound effect on the qualities of the analyses than did the choice of multivariate reduction technique (for example PCoA vs. NMDS, see Supplementary Tables 3 and 5).
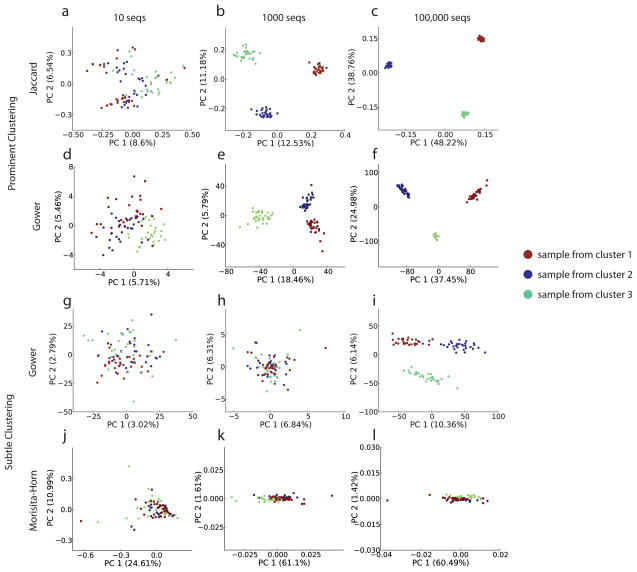
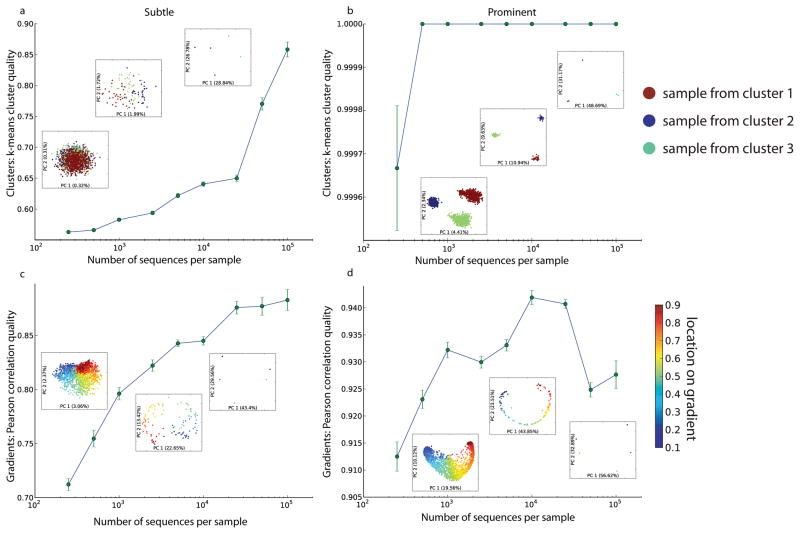
When we investigated the effects of sequencing depth on the results of various analysis techniques, we again found that the recommended analysis methodology depends on the degree of separation between the underlying clusters and that even excessive sequencing does not provide reasonable resolution if the wrong analytical method is chosen. Resampling of our empirical keyboard dataset revealed that 100 sequences per sample is generally sufficient to obtain good analysis qualities, relative to the clustering observed when analyzing the complete dataset (data not shown). This result was confirmed in simulated data when the clusters were modeled as very prominent (cluster distance 1.0, sample distance 0.5), and additional sequencing beyond 1,000 sequences per sample did not substantially improve our ability to resolve the patterns relating the samples (Fig. 4a–c). However, when clusters were far less prominent (cluster/sample distance 0.1/0.1), we found that increasing sequencing depth beyond 10,000 sequences per sample was required to achieve any analysis of good quality (k-means quality above .85 and relative distance quality above 2.0, see methods). The Gower distance measure is effective on clustered data - it demonstrates that deep sequencing is required if and only if the sample clustering is subtle (Fig. 4d–i). The extent of variation explained by the axes is not a proxy for effectiveness of the technique, and many sequences per sample is insufficient to overcome an inappropriate technique, as the Morisita-Horn distance demonstrates (Fig. 4j–l).
Figure 4.
Deep sequencing is superfluous when clusters are prominent, but critical when clusters are subtle. Data representing either prominent or subtle clusters was generated (see methods) with varying sequencing depths. (a–c) Jaccard distance followed by PCoA was applied to prominent cluster data with 10, 1,000, or 100,000 sequences per sample. No substantial improvement in the effectiveness of the method was found above 1,000 sequences per sample. (d–f) Gower distance followed by PCoA was applied to the same data (g–i) Gower distance applied to more subtle clusters.. (j–l) Morisita-Horn distance followed by PCoA applied to the subtle clusters. Although substantially more of the variance is explained by this method, the clusters are not easily interpretable: this situation persists even with 10 million sequences per sample (data not shown).
Discussion
The difference in the performance of the various ordination methods is large, underscoring the importance of using an appropriate analysis strategy. For example, Morisita-Horn + PCoA frequently cannot reveal clusters in the data even at a depth of 10 million sequences per sample under conditions where methods based on other distance measures, such as the Canberra distance, are easily able to reveal the biological patterns with only 1,000 sequences per sample. Similarly, Spearman distance + PCoA was able to find the same quality of clustering with 1,000 sequences per sample that Euclidean distance + PCoA could only resolve with 10 million sequences per sample, showing that by using the appropriate analytical method, it is not necessary to gather as much sequence data per sample to detect the underlying patterns.
Several statistical artifacts remained resistant to analysis. Although several techniques were able to minimize the arch effect, none of the techniques considered here eliminated it. Certainly, the arch can be eliminated by detrending, using Detrended Correspondence Analysis28 (DCA). However, this technique rests on a poor theoretical foundation (or requires the a priori assumption that there is only one underlying environmental gradient) and has been found to be misleading in some cases, for example when there are multiple underlying environmental gradients11,29 Resolving the arch effect so that multiple gradients can be studied remains an important challenge for the field. In addition, the differences between NMDS and PCoA were usually minimal compared to the differences in which distance measure was used, and in general, qualitative methods performed well on cluster data but poorly on gradient data, while the reverse was true for quantitative methods. These results suggest that both types of methods should be applied to most datasets if it is unknown whether cluster or gradient structure is more likely.
Most methods that performed well for prominent clusters also performed well for subtle clusters, the exceptions being the qualitative methods which, as a class, performed much better on prominent than on subtle clusters. This suggests that effect size is important in choosing a method. Note that our simulations of prominent clusters were fit to the differences between the fingertips of three different subjects: these distances are small compared to, for example, the distances between different body sites or different free-living environments8. Furthermore, the required sequencing depth is inversely related to the size of the effects separating different samples (Fig. 5). However, the effect sizes for specific diseases, and hence the required depth of coverage, remains unknown, although differences between IBD (Inflammatory Bowel Disease) and non-IBD subjects have been reported at depth of coverage of only ~100 sequences per sample30. In contrast, lean and obese individuals do not cluster separately at depth of coverage of ~10,000 sequences per sample5, either because the clustering is subtle or because other genotypic or phenotypic characteristics cause more prominent clustering. The simulations presented here were performed by varying many of the simulation parameters, allowing one to generalize the conclusions we reached beyond simply which methods are ideal for the soil and keyboard data we used as reference. However, it is infeasible to simulate all effects found in the wide variety of microbial sequence data now being collected, and the reference empirical datasets used here were chosen for their relative simplicity and clarity. Clearly, additional work is needed to estimate the effect sizes in other environments, and simulations using more complex empirical data as references would be welcome.
Figure 5.
Tradeoff between number of samples and number of sequences per sample with prominent and subtle gradients and clusters. Panels show (a) subtle clusters, (b) prominent clusters, (c) subtle gradients, and (d) prominent gradients, with a survey budget of 500,000 sequences allocated to varying numbers of samples, and thus an inversely varying number of sequences per sample. Insets show examples of data at specific sampling depths. The inset panels show examples of the gradients and clusters at 5, 100, and 2,000 samples, corresponding to 100,000 5,000 and 250 sequences per sample respectively (arranged right to left in each panel). All comparisons use the Pearson distance + PCoA ordination method. Note that the fraction of the variance explained by the PCoA decreases as the number of samples increases, even when the patterns are clearer with more samples. Error bars represent ± s.e.m. of 12 simulations.
In general, our results are encouraging: on datasets with effect sizes comparable to the effects seen in real datasets, simple simulations are able to recapture the same trends, and powerful analysis methods are available to reveal the patterns in those datasets. The advantages of having large numbers of samples at shallow coverage (~1,000 sequences per sample) clearly outweigh having a small number of samples at greater coverage for many datasets, suggesting that the focus for future studies should be on broader sampling that can reveal association with key biological parameters rather than on deeper sequencing. However, if nothing is revealed by broad, shallow sampling it is possible that the community structuring effects are subtle, in which case deeper sequencing can be illuminating.
Methods
Ordination Methods
Most ordination methods considered here comprise two stages: first the abundance matrix is converted into a distance matrix that relates each sample to each other sample. That distance matrix is then used to produce a low dimensional representation of the samples via a multivariate reduction method such as Principal Coordinates Analysis (PCoA) or Non-metric Multi-Dimensional Scaling (NMDS). Some methods, such as Principal Component Analysis (PCA), skip the distance matrix step and proceed directly from the abundance matrix to the completed ordination.
The simulated and empirical data were subjected to the most widely used ordination analysis methods. We performed PCA on the abundance matrix data (the raw data, as well as transformed data as described in Legendre & Gallagher13), using the package ‘vegan’ in the R programming environment (specifically the function rda, using covariance: SCALE=FALSE). We computed the pairwise distance between samples using a variety of commonly used measures, such as the Bray-Curtis distance and the manhattan distance (Supplementary Table 2), using PyCogent. We performed Principal Coordinates Analysis (PCoA) on the distance data using PyCogent31, and 2 dimensional Nonmetric Multidimensional Scaling (NMDS) using the MASS package in the R programming environment (specifically the function isoMDS, seeded with results of a PCoA analysis, and limited to a maximum of 50 iterations or a convergence specified by a tolerance of 10−3, following the function’s default values). The NMDS results were rotated such the variance along the horizontal axis was maximized (this was for display purposes only, as the axes have no intrinsic meaning in NMDS). See Supplementary Table 2 for a list of all distance measures used, and Supplementary Table 3 for a list of all ordination methods considered here. In all cases, the result of the analysis was displayed in two dimensions.
Evaluation of Analysis Methods
To evaluate the efficacy of analysis methods applied to gradient data, we wanted to determine how faithfully the analysis revealed the underlying environmental gradient. An ideal technique would display all samples in the same order as they exist along the environmental gradient, with inter-sample distances proportional to the their separation along the gradient. To quantitatively assess this, we computed the Pearson correlation coefficient between the positions of the samples after analysis along the primary axis of variation (principal axis 1 for PCA and PCoA methods, and the axis of greatest variance for NMDS methods), with the position of those samples along the gradient from which they were drawn. Because we were also interested in whether samples were shown in their correct gradient order, we also evaluated the Spearman rank correlation coefficient of the samples’ displayed position and their order along the gradient. Also, because the direction of the gradient is not meaningful, we considered only the absolute value of the Pearson correlation and Spearman rank correlation coefficients when evaluating the quality of gradient analysis methods.
To address the efficacy of analysis methods applied to clustered data, we wanted to determine which analyses partitioned samples correctly, revealing the true clustering of the samples. We used three metrics to evaluate this. The first was the average displayed distance between two samples from separate clusters, divided by the average distance between two samples from the same cluster. The second was to apply k-means clustering to the results of the analyses (using the package ‘MASS’ in R), and computed the fraction of sample pairs whose k-means clustering matched the actual clustering of the data (samples from the same cluster found in the same k-means cluster, and those from different clusters found in different k-means clusters). The third was to perform UPGMA clustering on the pairwise sample distance as displayed in the ordination plots, and to test the extent to which the members of the clusters formed distinct groups in the tree. In general, the three quality assessment methods agreed well for assessing a given ordination analysis.
Empirical Data
We used two experimental datasets for comparison: a bacterial community survey of different fingertips and keyboards27, and a study examining the effects of soil pH change on bacterial communities23. These communities provide examples of relatively low- and high-diversity habitats (respectively), and span human and environmental datasets of practical importance to researchers. Both studies were pyrosequenced using error-correcting barcoding on the V2 region as previously described4,32.
Simulated Gradient Data
In a manner similar to Legendre & Gallagher13, we modeled each species as having peak abundance at a randomly chosen location along an artificial gradient, and a unimodal normal abundance curve (species response curve) centered at that gradient location. The relative abundances of the species were adjusted to match the species abundance distribution found in the combined samples from the soil dataset described in Methods (species-level phylotypes were defined as organisms with ≥ 97% 16SrRNA identity33). We did not assume any correlation between overall species abundance and location of peak abundance on gradient. To simulate the stochastic effects in species abundances, we then perturbed each species’ relative abundance by adding gaussian noise of a width proportional to that species’ relative abundance. Subtle gradients were those perturbed by noise drawn from a distribution of mean 0 and width equal to the species’ abundance, prominent gradients used a width of .5 times the species’ abundance. Each simulated environment was then sampled at either random or uniformly spaced positions along the gradient. Each sample consisted of a series of random selections, with replacement, from the species present, weighted by the relative abundances of the species at that gradient location. In other words, the abundance of each species was inferred using the probability distribution for each species, the total was renormalized to sum to a probability mass of 1, and individuals were sampled from the resulting distribution for that point. Sampling continued until a specified number of sequences were obtained. The number sequences varied from 10 per sample to over 10,000. The count of each species sampled at each sample location along the gradient formed the simulated dataset used to evaluate the ordination techniques.
Simulated Cluster Data
To generate simulated clustered data, we began with a species abundance distribution identical to that found in the keyboard dataset described in Methods (again, species-level phylotypes were defined as organisms with ≥ 97% identical 16S rRNA). We then perturbed each species’ relative abundance by multiplying the species abundance by a number drawn from a normal distribution of mean 1 and varying width (Fig. 1b). The resulting species abundance vectors were renormalized to sum to 1. These formed the basis for each cluster. These cluster level abundance vectors were again perturbed with gaussian noise of specified mean and standard deviation, and renormalized to form the sample abundance vectors. Each sample then consisted of a series of selections, with replacement, from these species abundance distributions.
Supplementary Material
Supplementary Figure 1. Required sequencing depth depends on prominence of gradient and analysis method used.
Supplementary Table 1. Different choices in the analysis of gradient or cluster data.
Supplementary Table 2. Formulas of dissimilarity measures.
Supplementary Table 3. Summary results of ordination methods on simulated datasets.
Supplementary Table 4. Performance of each ordination method tested on gradient datasets.
Supplementary Table 5. Performance of each ordination method tested on cluster datasets.
Acknowledgments
This work was supported by the NIH (DK78669, HG4872, HG4866) the Crohns and Colitis Foundation of America, the Bill and Melinda Gates Foundation, and the Howard Hughes Medical Institute. We thank E. Costello, J. Zaneveld, and J. G. Caporaso for helpful comments on drafts of the manuscript.
Footnotes
Author Contributions
J.K. and R.K. wrote the manuscript; J.K., R.K., and Z.L. designed the research; C.L., D.M., J.K., R.K., and Z.L. contributed simulation and analysis code; all analyzed the data.
References
- 1.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 3.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costello EK, et al. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 8.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-Based Assessment of Soil pH as a Predictor of Soil Bacterial Community Structure at the Continental Scale. Appl Environ Microb. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2009 doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minchin PR. An Evaluation of the Relative Robustness of Techniques for Ecological Ordination. Vegetatio. 1987;69:89–107. [Google Scholar]
- 12.Faith DP, Minchin PR, Belbin L. Compositional Dissimilarity as a Robust Measure of Ecological Distance. Vegetatio. 1987;69:57–68. [Google Scholar]
- 13.Legendre P, Gallagher ED. Ecologically meaningful transformations for ordinations of species data. Oecologia. 2001;129:271–280. doi: 10.1007/s004420100716. [DOI] [PubMed] [Google Scholar]
- 14.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawford PA, et al. Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc Natl Acad Sci U S A. 2009;106:11276–11281. doi: 10.1073/pnas.0902366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jongman RH, ter Braak CJF, Van Tongeren OFR. Data analysis in community and landscape ecology. Cambridge University Press; Cambridge; New York: 1995. [Google Scholar]
- 18.Magurran AE. Measuring Biological Diversity. Blackwell; Oxford: 2004. [Google Scholar]
- 19.Lozupone CA, Knight R. Species divergence and the measurement of microbial diversity. FEMS Microbiol Rev. 2008;32:557–578. doi: 10.1111/j.1574-6976.2008.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legendre P, Legendre L. Numerical ecology. 2. Elsevier; Amsterdam; New York: 1998. [Google Scholar]
- 21.Ramette A. Multivariate analyses in microbial ecology. Fems Microbiol Ecol. 2007;62:142–160. doi: 10.1111/j.1574-6941.2007.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rousk J, et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- 24.Rousk J, Brookes PC, Baath E. Investigating the mechanisms for the opposing pH relationships of fungal and bacterial growth in soil. Soil Biology and Biochemistry. 42:926–934. [Google Scholar]
- 25.Gauch HG. Multivariate analysis in community ecology. Cambridge University Press; Cambridge Cambridgeshire; New York: 1982. [Google Scholar]
- 26.Kuczynski J, et al. Direct sequencing of the human microbiome readily reveals community differences. Genome Biol. 2010;11:210. doi: 10.1186/gb-2010-11-5-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fierer N, et al. Forensic identification using skin bacterial communities. P Natl Acad Sci USA. 2010;107:6477–6481. doi: 10.1073/pnas.1000162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill MO, Gauch HG. Detrended Correspondence-Analysis - an Improved Ordination Technique. Vegetatio. 1980;42:47–58. [Google Scholar]
- 29.Pielou EC. The interpretation of ecological data: a primer on classification and ordination. Wiley; New York: 1984. [Google Scholar]
- 30.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knight R, et al. PyCogent: a toolkit for making sense from sequence. Genome Biol. 2007;8:R171. doi: 10.1186/gb-2007-8-8-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stackebrandt E, Goebel BM. A Place for DNA-DNA Reassociation and 16s Ribosomal-Rna Sequence-Analysis in the Present Species Definition in Bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Required sequencing depth depends on prominence of gradient and analysis method used.
Supplementary Table 1. Different choices in the analysis of gradient or cluster data.
Supplementary Table 2. Formulas of dissimilarity measures.
Supplementary Table 3. Summary results of ordination methods on simulated datasets.
Supplementary Table 4. Performance of each ordination method tested on gradient datasets.
Supplementary Table 5. Performance of each ordination method tested on cluster datasets.