Abstract
Cancer cells manage to divide in the context of gross chromosomal abnormalities. These abnormalities can promote bypass of normal restraints on cell proliferation, but at a cost of mitotic vulnerabilities that can be attacked by chemotherapy. Determining how cancer cells balance these issues may permit chemotherapeutic sensitivity to be leveraged more efficiently. From a pan-genomic siRNA screen for modifiers of chemoresponsiveness, we identified the tumor antigen ACRBP/OY-TES-1 as a specifier of paclitaxel resistance. ACRBP expression is normally restricted to the testes but detected in a wide variety of cancers, including most ovarian cancers. We found that ACRBP is both necessary and sufficient for paclitaxel resistance in ovarian cancer cell lines and ovarian tumor explants. Moreover, high ACRBP expression correlated with reduced survival time and faster relapse among ovarian cancer patients. We identified the mitotic spindle protein NuMA as an ACRBP-interacting protein that could account for the effects of ACRBP on paclitaxel sensitivity. In cancer cells, ACRBP restricted a NuMA-dependent abrogation of mitotic spindle assembly that is otherwise pathologic. As a consequence, ACRBP depletion resulted in mitotic errors and reduced proliferative fitness that could be rescued by NuMA co-depletion. We propose that the co-dependent relationship of ACRBP and NuMA in cancer cells reflects their passage through a selection bottleneck during tumor evolution, one which requires the acquisition of traits which normalize mitotic perturbations that originally drove the plasticity of a pre-neoplastic genome. The molecular definition of such traits as defined by the ACRBP-NuMA complex may represent conceptually ideal intervention targets, based on the wide therapeutic windows they may offer.
Keywords: Tumor Antigen, RNAi Screen, Emergent Dependency
INTRODUCTION
Despite widespread use as a chemotherapeutic intervention for cancer treatment, the molecular underpinnings specifying tumor cell-selective sensitivity to microtubule poisons are enigmatic (1, 2). While clinically effective, patient responses are notoriously variable and associated with significant toxicity (3–7). From a pangenomic siRNA screen for cell autonomous specifiers of chemoresponsiveness, we isolated a cohort of gene products that, when depleted, significantly sensitized lung cancer cells to paclitaxel-induced cell death (8). An intriguing subclass of this cohort was previously recognized, through immunoreactive serum profiling, as encoding cancer testes tumor antigens with no known function (9). A member of this subclass, ACRBP (acrosin binding protein, a.k.a. OY-TES-1) (10–12), is normally expressed in developing and adult testis and anomalously expressed in a variety of tumor types, including lung, breast and liver (13). Significantly, gain of ACRBP/OY-TES-1 expression was reported in 69% of ovarian tumors (12).
Given the extensive expression correlation with cancer, together with the robust functional interaction with chemotherapeutic agents, we directly examined the contribution of ACRBP to the proliferative fitness of cancer cells. We found that anomalous ACRBP expression was both required and sufficient to support mitotic spindle assembly and function in the face of microtubule perturbagens. The mechanistic basis of this contribution to robust mitotic progression is at least in part through restriction of NuMA-dependent centrosomal fragmentation in cancer cells. The recapitulation of this functional relationship in ovarian tumor explants and the correlation of ACRBP expression with poor patient response to therapy suggests that ACRBP represents an acquired dependency within the pathological regulatory framework of cancer cell mitosis.
MATERIALS AND METHODS
Cell culture
H1155 and ES-2 cells were obtained from the ATCC (American Type Culture Collection). PEO1 and PEO4 cells were a gift from Hani Gabra (University of Edinburgh Cancer Research Center). H1155, PEO1 and PEO4 cell lines were validated by PowerplexTM genotyping prior to use.
Tumor Tissue and Primary Tumor Explants
Normal ovarian tissue RNA samples were obtained with consent from patients under the IRB 092004-010, “The University of Texas Southwestern Medical Center Tissue Resource (UTSTR). Patient ovarian tumor samples were isolated from ascites obtained during surgical removal of tumors (LCOV2 and LCOV17) or paracentesis (LCOV4) under appropriate HIPAA procedures (IRB 09-0412 at UNC-Chapel Hill “Identification of Linchpin Proteins Supporting Ovarian Tumorigenesis”). All numbers used to refer to samples are dummy identifiers that cannot be traceable back to individual patients. LCOV2 was derived from a patient with serous, stage IIIC ovarian cancer who had received 4 cycles of paclitaxel and carboplatin. LCOV4 was derived from a patient with primary peritoneal carcinosarcoma who had received 3 cycles of ifosfamide/carboplatin, single agent carboplatin, Taxotere®, GEMZAR®, followed by weekly paclitaxel. LCOV17 was from a newly diagnosed patient with Stage IIIC serous papillary ovarian cancer who had not received prior treatment. Ascites was plated in flasks and media changed weekly until cells were confluent. Cells were maintained in RPMI + 10% FBS (PEO1, PEO4 and ES-2) or RPMI + 5 % FBS (LCOV2, LCOV4, LCOV17).
Live-Cell Imaging and Quantitation
GFP-Histone H2B expressing lines were produced by transduction with pCLNCX-GFP-H2B retrovirus. Retrovirus was prepared by Fugene (Roche) transfection of 293gp cells with pCLNCX-GFP-H2B and VSVG according to the manufacturer’s instructions. 24 hours post-transfection the media was replaced and at 48 hours post-transfection supernatant was collected, centrifuged and filtered. H1155 cells were then transduced at 50% confluence in conjunction with 4 μg/mL polybrene. One day post-transduction, the cells were selected with 600 μg/mL Geneticin (Gibco) until a homogenously expressing population was obtained. For imaging of siRNA treated cells, the H1155-GFP-Histone H2B line was reverse transfected as described below and plated in a 96-well format followed by carrier or paclitaxel treatment 48 hours post transfection. At 72 hours post-transfection, imaging began on a BD Pathway 855 bio-imager using a 40X objective. Images were collected at 15-minute intervals for the next 48 hours. Image sequences were generated using ImageJ. Image sequences were then evaluated for indicated parameters manually.
siRNA Transfections
All transfections were performed with 50 nM final concentration of siGENOME SMART Pools and individual siRNA oligonucleotides obtained from ThermoFisher (Lafayette, CO). Reverse transfections were performed by mixing appropriate Dharmafect (ThermoFisher) with siRNA pools and incubating for 20 minutes followed by delivery of trypsin-derived single-cell suspensions. Cells were transfected for a minimum of 72 and maximum of 96 hours prior to analysis as indicated in figure legends. To optimize cell lines, cells were reverse transfected with control (DLNB14) or a toxic siGENOME pool (targeting ubiquitin B) using a panel of Dharmafect reagents and concentrations in 96 well format. 96 hours post transfection, cells were evaluated for viability using the Cell Titer Glo Assay (Promega). ACRBP off-target validation was performed by using 6 individual siRNAs representing two independent siRNA pools obtained from Dharmacon (siGENOME and On Target Plus (OTP)) in H1155 cells. Each individual siRNA from the original pool and 1 from the OTP pool effectively depleted ACRBP mRNA and resulted in paclitaxel-dependent mitotic delay. The target sequences for these siRNAs are : GGAGAATACCTGCATCATATT , TGGAAGAGCTCCTACAATCTT, CCAAGAGAGTCCTGTGTTCTT, GCCAAGGCCTGGAAGTACATT, TTTACGGGCTGGATTTGTATT. In subsequent assays, the siGENOME pool was used. An ACRBP depletion induced phenotype was also validated in PEO1 cells using a lentivirus-based system (TRC clone set, OpenBiosystems Catalog # RHS4533). Clones TRCN0000115843, TRCN0000115844 were effective.
cDNA Transient Transfections
H1155 or PE01 cells were transfected with cDNA expression vectors using Fugene according to the manufactures protocol for a 35 mm2 dish format. 24 hours post transfection, media was replaced with paclitaxel or vehicle containing media for an additional 24 hour incubation. Cells were then processed for immunofluorescence.
Quantitative real-time RT-PCR (qPCR)
Total RNA was isolated from tissue culture cells using the Sigma RNA isolation kit. The High-Capacity cDNA reverse transcription (RT) kit (Applied Biosystems) was used to synthesize cDNA from 3 μg of total RNA. Real-time RT-PCR was performed on diluted cDNA using the Applied Biosystems 7500 Fast real-time PCR system (standard program) and inventoried TaqMan gene expression assays. Human β-actin served as an endogenous control and “control transfected” was the calibrator for calculating differences in expression levels using the 2-ΔΔCT method. GAPDH was also used for normalization in many cases, and consistently returned the same biological observation. Final results represent the pooling of two test experiments and error bars represent average deviation from the mean.
Immunoblotting and Immunofluorescence
Monoclonal antibody raised in mouse against ACRBP was a gift from Kunle Odunsi and is previously described. Anti-NuMA rabbit antibody was obtained from Novus Biologicals. A monoclonal antibody against bromodeoxyuridine (BrdU) was obtained from Becton Dickinson.β-tubulin III, γ-tubulin, and Aurora-A antibodies were obtained from Sigma, Anti- Pericentrin was obtained from Abcam. Alexa 488 and Alexa 546(Invitrogen) were used as secondary antibodies for fluorescent labeling. Immunoblot assays were performed with proteins transferred to polyvinylidene difluoride membrane according the manufacturer's protocol for each antibody. For BrdU visualization, cells were treated with 30 uM BrdU for 14 hours and then fixed in 3.7 % HCHO. Cells were permeabilized with acetone for 10 min at −20°C. DNA was denatured with 1 mM HCl for 10 minutes and then blocked in phosphate-buffered saline-5% bovine serum albumin-1% Tween for a minimum of 15 minutes. Anti-BrdU was used at a dilution of 1:4. For β-tubulin/pericentrin staining, cells were fixed as described above, permeabilized for 10 minutes with 0.5 % Triton x-100 and blocked as above. Cells were stained with monoclonal β-tubulin III and pericentrin at a dilution of 1:100 or 1:200, respectively for 1 hour. Following extensive washes, an Alexa-488 or 546 secondary antibody was applied for 30 minutes at 37C. For γ-tubulin staining, cell were fixed as above and permeabilized for 10 minutes in methanol at −20C. For ACRBP staining, cells were fixed and permeabilized as for β-tubulin and UA199 antibody was used at a 1:100 dilution. Cytokeratin-16 staining was performed using a monoclonal cytokeratin-16 antibody obtained from Dako Laboratories. All cells were visualized with an Axiovert or Axioimager upright microscope (Zeiss) equipped with a Hamamatsu or Zeiss CCD camera.
Immunoprecipitation
PEO4 cells grown to confluence on 100-mm2 dishes were lysed inbuffer containing 0.5% Triton X-100 and 150 mM NaCl and 0.5%deoxycholate. Soluble lysate was incubated overnight with anti-ACRBPmonoclonal antibody UA199 or mouse immunoglobulin G precoupled to cyanogen-bromide agarose beads. Beads were washed three times in lysis buffer with500 mM NaCl. Following washes, sample buffer was added and lysateswere boiled and separated by 10% sodium dodecyl sulfate-polyacrylamidegel electrophoresis (SDS-PAGE). HPLC-mediated tandem mass spectroscopy of ACRBP immunoprecipitates was performed by the UT Southwestern Protein Chemistry Core Research Facility in Mass Spectrometry. Multiple peptides corresponding to NuMA were identified in a 180 kDa silver stained band.
Gene expression association studies
Fresh tumor was collected from 29 advanced epithelial ovarian cancer patients between 2001 and 2006 as part of an ongoing Early Detection Research Network collaboration. All patients were treated with taxane/platinum-based chemotherapy. Specimens were immediately flash-frozen and stored at −80°C. The clinical course and outcome of the patients were prospectively determined as sample collection was ongoing. Genomic mRNA expression profiles were generated using the Affy Plus2 GeneChip. The RMA (Robust Multiarray Averaging) method was used to process microarray data and expression values were log2 transformed (14, 15). Patients were dichotomized to high ACRBP and low ACRBP groups based on a Gaussian mixture model (16). ACRBP and NuMA expression values were used to parse patient populations to high ACRBP-NuMA and low ACRBP-NuMA groups based on a hierarchical clustering analysis. Similar results are observed if median values were used to dichotomize expression values. Disease free survival time (DFS), defined as the time interval between chemotherapy conclusion and disease recurrence date or the last follow-up date, was used to measure patient’s response to chemotherapy. Survival curves were estimated using the product-limit method of Kaplan-Meier (17) and were compared using the log-rank test. Univariate Cox proportional-hazards was also performed, with survival as the dependent variable (18).
RESULTS
ACRBP copy number is amplified in tumors and confers paclitaxel resistance
The mechanisms driving the anomalous ACRBP expression in a variety of cancers are unknown (9). However, ACRBP genomic gain was recently detected in ovarian serous adenocarcinoma by the Cancer Genome Atlas consortium (TCGA, Figure 1A). Perhaps more clinically significant, gain of ACRBP/OY-TES-1 expression was reported in 69% of ovarian tumors (12). This relationship is iterated in ovarian tumor-derived cell lines and represents an attractive setting for analysis of ACRBP function in cancer. We examined the serous adenocarcinoma cell lines, PEO1 and PEO4 that were sequentially derived from a chemotherapy responsive tumor and the chemotherapy resistant relapse from the same patient (19). Of note, while ACRBP expression is activated in both lines relative to normal tissue, it is highest in the chemoresistant PEO4 cells (Figure 1B). Ectopic expression of ACRBP in PEO1, and in the original siRNA screen host NSCLC line H1155 (8), was sufficient to confer resistance to paclitaxel-induced mitotic arrest relative to controls (Figure 1C,D).
Figure 1. ACRBP gain-of-function in cancer.
A) Box plot of ACRBP copy number from the TCGA dataset. Oncomine (Compendia Bioscience, Ann Arbor, MI) was used for analysis and visualization. B) Quantitative rtPCR analysis of ACRBP expression is shown for indicated normal ovarian tissue samples and ovarian tumor cell lines. C) H1155 or D) PEO1 (right) cells were transfected with the indicated expression vectors. 24 hours post transfection cells were treated with indicated concentration of paclitaxel for 12 hours. Mitotic figures were scored microscopically following DAPI staining and are expressed as the % of total cells counted. Error bars represent average deviation from the mean of 2 independent experiments.
ACRBP physically and functionally interacts with NuMA
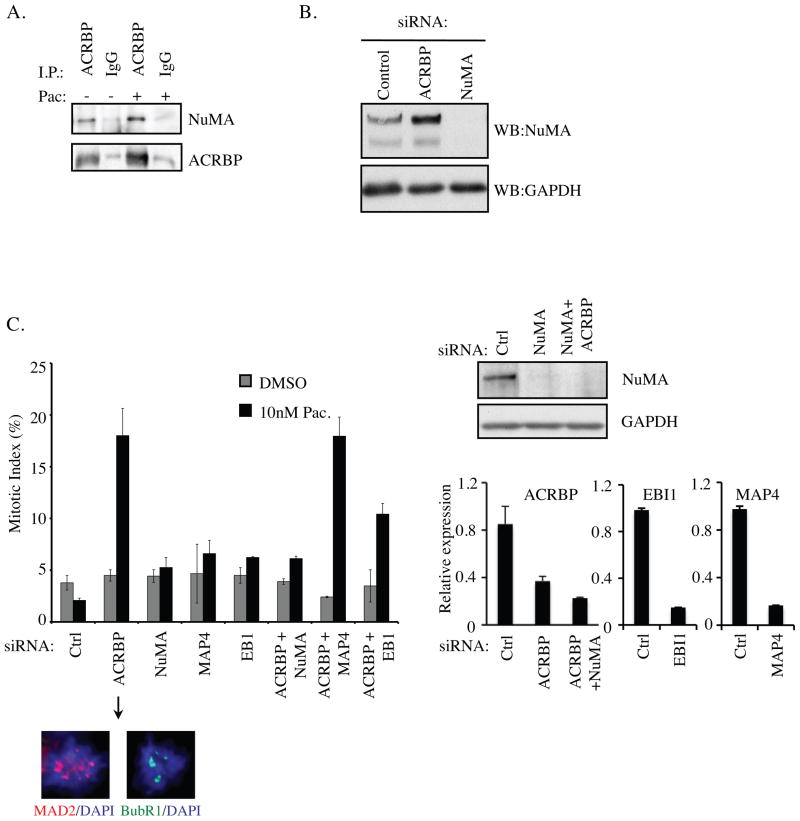
The gain-of-function activity of ACRBP indicates this molecule likely interacts with cellular components that otherwise promote paclitaxel responsiveness. To explore this, we examined ACRBP-associated proteins by nano-HPLC mass spectrometry analysis of endogenous ACRBP complexes. A number of candidates were returned including the nuclear and mitotic apparatus protein, NuMA, which was validated by immunoblotting anti-ACRBP immunoprecipitates from PEO4 cells with an anti-NuMA antibody (Figure 2A). In normal cells, NuMA is an essential component of the mitotic spindle, promoting spindle pole focusing and when overexpressed, microtubule bundling (20–28). The NuMA locus is in the 11q13 chromosomal band, which is frequently amplified in many tumors and includes tumor-promoting genes such as cyclin D1, EMSY and MEN1. The overexpression of NuMA in cancers disrupts centrosome integrity and increases genomic instability (21). The manifestation of spindle defects upon either loss or gain of NuMA strongly suggests that a balance of effective NuMA concentrations must be maintained for robust mitotic progression (20–22). Upon depletion of ACRBP, we found that NuMA protein levels were significantly elevated, suggesting that ACRBP expression restrains NuMA protein accumulation (Figure 2B). Remarkably, co-depletion of ACRBP and NuMA rescued the enhanced sensitivity to paclitaxel observed upon ACRBP depletion alone (Figure 2C). This effect was not observed upon codepletion of other modulators of microtubule dynamics, including the microtubule stabilizing protein MAP4 and the microtubule tip-binding protein EB1 (Figure 2C), indicating that ACRBP perturbation of paclitaxel sensitivity is NuMA-dependent (22, 23).
Figure 2. NuMA-dependent modulation of paclitaxel sensitivity by ACRBP.
A) Endogenous ACRBP was immunoprecipitated from PEO4 cell lysates as indicated and probed for coprecipitated NuMA. Normal mouse IgG was used as a negative control. Identical lysates were split for ACRBP and control IgG immunoprecipitations. Immunoprecipitations were performed in the presence and absence of 100 nM paclitaxel indicated as +/− pac. B) Whole cell extracts from PEO4 cells transfected with the indicated siRNAs were immunoblotted to detect accumulation of native NuMA as shown. C) H1155 cells were transfected with indicated siRNAs. 48 hours post-transfection cells were exposed to the indicated paclitaxel concentrations for 24 hours. Microtubules, genomic DNA and centrosomes were visualized by β-tubulin, DAPI and pericentrin immunostaining respectively. Mitotic figures were scored microscopically and are expressed as the % of total cells counted. MAD2 and BubR1 positive kinetochores in mitotic figures were used as an indication of failure to satisfy the spindle assembly checkpoint (representative panels are shown below the arrow). A minimum of 100 cells over at least 5 fields of view was scored for each group in each of 3 independent experiments. Error bars represent average error from the mean. NuMA protein depletion was evaluated by immunoblot (upper right) and ACRBP, EBI1 and MAP4 mRNA depletion were evaluated by quantitative rtPCR (lower right).
ACRBP supports high fidelity mitosis
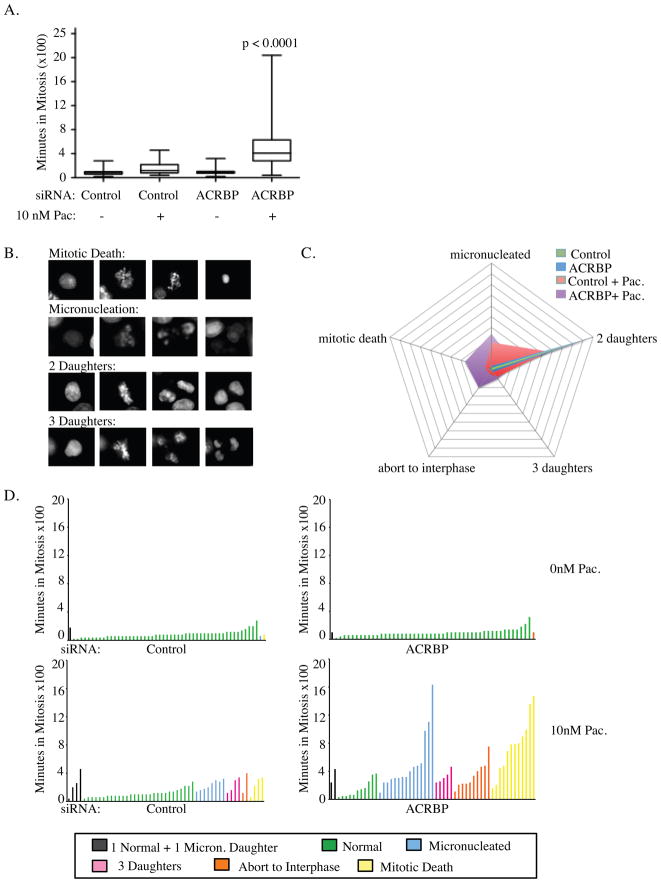
Given the role of NuMA in mitotic spindle assembly and function (20, 24–27), we directly assessed the consequence of ACRBP depletion on mitotic progression in real time at single-cell resolution using stable expression of GFP-Histone H2B. Consistent with previous observations in H1155 cells, depletion of ACRBP or exposure to 10nM paclitaxel had no measurable consequence on the timing and only slight consequences on the outcome of mitosis (Figure 3) (8). In contrast, depletion of ACRBP together with 10nM paclitaxel resulted in a significant mitotic delay as compared to controls (Figure 3A). The delay is consistent with spindle assembly checkpoint response as kinetochores in ACRBP depleted cells exposed to paclitaxel are MAD2 and BUBR1 positive (Figure 2C). Real-time single-cell lineage tracing indicated that this delay was coupled to aberrant resolution of mitosis that included apoptosis, micronucleation/nuclear fragmentation, premature exit to interphase, and tripolar mitosis (Figure 3B,C, D). Taken together, these observations indicate that ACRBP expressing cancer cells require this protein to support robust proper bipolar spindle formation.
Figure 3. ACRBP is required for high fidelity mitotic progression in tumor cells.
A) H1155 cells expressing GFP-Histone H2B were transfected with indicated siRNAs. 48 hours post-transfection, cells were exposed to paclitaxel or carrier and imaged live by time-lapse microscopy for 48 hours. Single-cell lineage tracing for each condition was performed to measure the length of mitosis as a function of the time from prometaphase to telophase in 50 cells over 2 independent experiments. Quartile ranges for condition are shown. P-values were calculated with the Mann-Whitney test. B) Representative examples of time-lapse sequences illustrating heterogeneity of fates exhibited by H1155 cells following ACRBP depletion and exposure to 10 nM paclitaxel. C) Radar plots indicating the population distributions of mitotic fates under the indicated conditions. D) Single-cell mitotic fate profiles and associated time in mitosis for H1155 cells transfected with indicated siRNAs and exposed to indicated paclitaxel concentrations. Each bar indicates the time in mitosis and the outcome for a single cell.
ACRBP restrains NuMA-driven mitotic pathology to enhance proliferative capacity
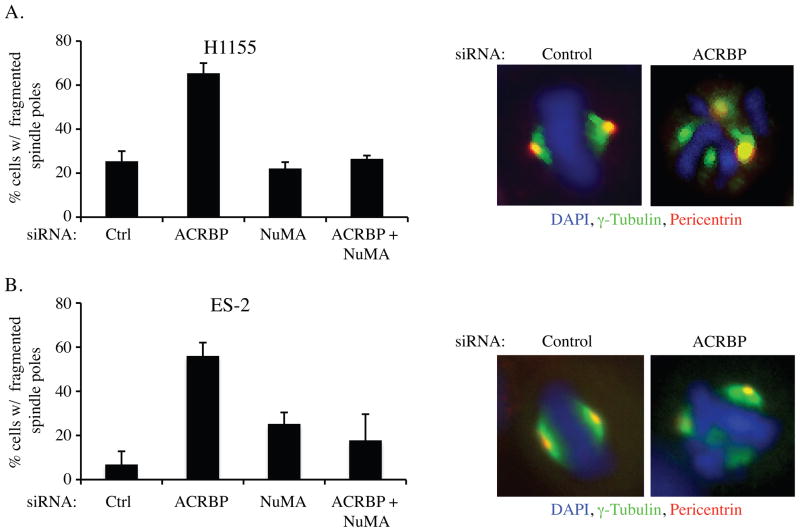
The overexpression of NuMA can promote multipolar spindle formation characterized by the presence of multiple gamma-tubulin positive spindle poles in mitotic cells. This observation has led to the hypothesis that aberrant accumulation of NuMA in cancers may promote genomic instability through inhibition of centrosomal coalescence (21, 28, 29). Close examination of mitotic spindle components, in lung and ovarian cancer cell lines, revealed that ACRBP depletion increased the frequency of spindle pole fragmentation upon exposure to low dose paclitaxel as indicated by non-colocalized supernumerary γ-tubulin and pericentrin foci (Figure 4A,B). This phenotype is reminiscent of pathological NuMA activity first described in esophageal cancer lines (21), and was reversed upon NuMA codepletion (Figure 4A,B). These observations suggest that ACRBP antagonizes NuMA in cancer cells to normalize defects that would otherwise lead to aberrant spindle assembly and consequent mitotic catastrophe.
Figure 4. ACRBP supports spindle-pole congression and proliferation in ovarian cancer cells.
A) H1155 and B) ES-2 cells were transfected with indicated siRNAs, incubated for 48 hours, then exposed to 10 nM paclitaxel for 24 hours. Following immunostaining with the indicated probes, mitotic figures were scored microscopically for fragmented centrosomes and are expressed as the % of total cells counted. Error bars represent average deviation from two independent experiments.
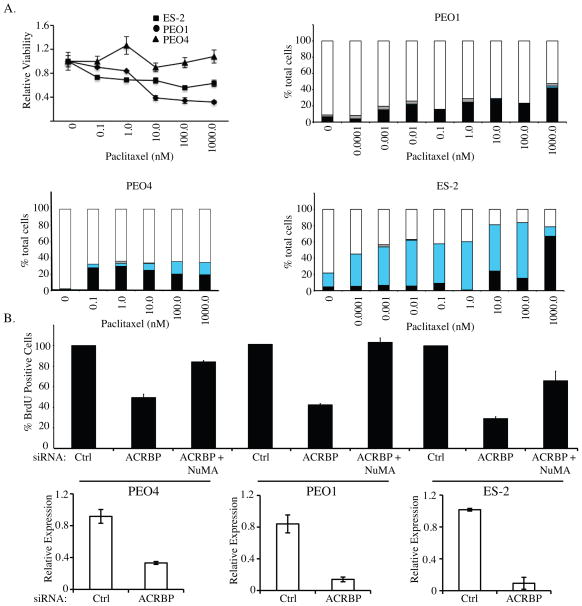
As mitotic spindle dysfunction can result in the production of daughter cells with incomplete genomes and thus reduced fecundity, we next examined the contribution of ACRBP to cell cycle progression in three different ovarian cancer cell lines. Each of these lines has a unique response to paclitaxel representing a diverse response to anti-mitotic agents frequently observed in culture and patient populations (Figure 5A). As shown in Figure 5B, ACRBP depletion resulted in a NuMA-dependent inhibition of BrdU incorporation in all lines tested. This occurred in the absence of paclitaxel challenge, indicating that gain of ACRBP expression confers proliferative fitness to ovarian cancer cells.
Figure 5. ACRBP depletion inhibits DNA Synthesis.
A) Line graph is paclitaxel dose-response curves generated for three ovarian cancer cell lines as indicated. Bar graphs are relative viability following a 48 hour exposure to the indicated paclitaxel concentrations in three different ovarian cell lines. Bars represent S.D. from triplicate analyses. Cells were immunostained for chromosomes, microtubules and centrosomes, and scored for the percentage of interphase cells (white bars), mitotic figures (black bars), micronucleated cells (Cyan bars), and apoptotic bodies (grey bars). Over 100 cells were counted for each condition, and means are shown from 2 independent experiments. B) The indicated ovarian cancer cell lines were transfected with the indicated siRNAs. 72 hours post-transfection, BrdU was added to the culture medium for 14 hours. BrdU incorporation was detected with an anti-BrdU antibody, nuclei were counterstained with DAPI and the percentage of BrdU-positive cells was quantified by microscopic observation. At least 100 cells were analyzed for each condition for each experiment across a minimum of 3 different fields of view. Values are normalized to the BrdU-incorporation frequency observed in control-transfected cells, which was set to 100%. Error bars indicate S.D. of 2 independent experiments each with 2 replicates. Quantitative rtPCR was performed on parallel samples to validate knockdown of ACRBP (bottom panels).
ACRBP is a marker of clinically aggressive disease
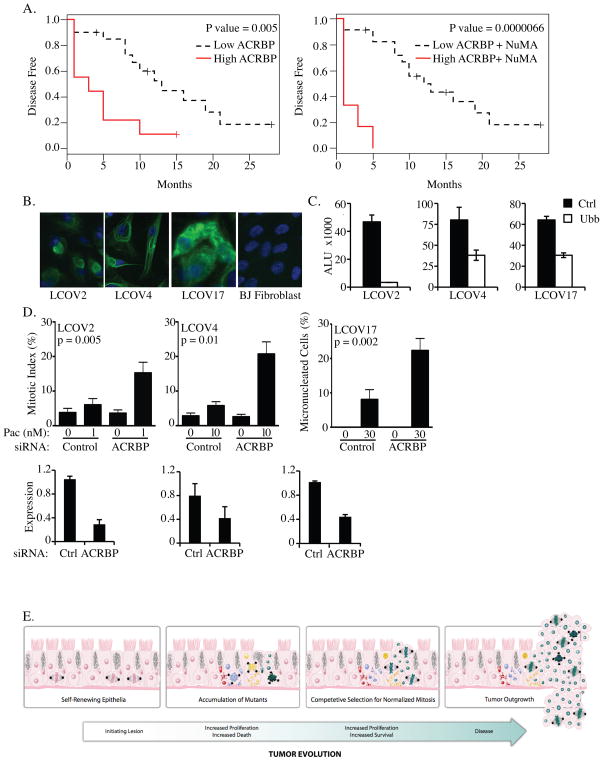
Ovarian cancer is typically treated by surgical debulking of the tumor mass, followed by taxane and platinum-based chemotherapeutic regimens (30). While the majority of patients respond, nearly all will relapse within 2–5 years, and the overall 5-year survival rate is less than 45% (30). To examine the correlation of ACRBP expression with patient outcome, we queried whole genome expression array data from tumors isolated from a cohort of 29 ovarian cancer patients with associated disease histories. We found high ACRBP expression is significantly correlated with poor prognosis, both with respect to overall survival (Hazard Ratio = 9.18 for high ACRBP group vs. low ACRBP group, p=0.0059 from Cox regression model) and time to disease recurrence (Hazard Ratio = 3.87, p=0.0064 from Cox regression model) (Figure 6A). Remarkably, the most aggressive disease was associated with both high ACRBP and high NuMA expression (Figure 6B). This correlation is consistent with the notion that ACRBP expression may be selected for to counter the consequence of aberrantly high NuMA activity.
Figure 6. ACRBP correlates with aggressive disease and paclitaxel resistance in tumor explants.
A) Kaplan-Meier plots of disease-free survival association with ACRBP expression (left) or ACRBP and NuMA expression (right). B) Primary tumor cultures derived from patient samples were immunostained with cytokeratin-16 (green) and DAPI (blue). C) siRNA transfection efficiency was monitored by measuring viability following transfection of siRNAs targeting an essential gene product, UBB. ALU: Arbitrary Light Units (right panels) D) Primary tumor cultures were transfected with indicated siRNAs for 48 hours then exposed to the indicated paclitaxel concentrations for 24 hours. For LCOV2 and LCOV4, paclitaxel doses were chosen as those which were at least 10-fold below that required to significantly increase the mitotic index of the parental cultures. For LCOV17, 30 nM was the lowest dose at which any defect, in control or ACRBP transfected cells, was detected. Following immunostaining with anti-β-tubulin and DAPI, mitotic figures were scored microscopically and are expressed as the % of total cells counted. A minimum of 200 cells were counted for each condition in each of 2 independent experiments. Error bars represent standard error from the mean from multiple, different fields of view. Quantitative rtPCR was performed to validate ACRBP mRNA depletion. E) Cartoon representation of epithelial tumor evolution indicating the selection bottleneck that can promote acquisition of anomalous components that normalize mitotic dysfunction and thus support clonal expansion.
Given the capacity of anomalous ACRBP expression to support robust mitotic progression in tumor-derived ovarian cancer cell lines, we next examined the consequence of ACRBP depletion on the mitotic progression of minimally cultured ascites-derived tumor explant cultures. Samples were obtained at the UNC Lineberger Cancer Hospital from the ascites of three patients: two of which had previously undergone multiple rounds of taxane based chemotherapeutic regimens (LCOV2, LCOV4) and one of which was chemo-naive (LCOV17)(see materials and methods). The derivation of epithelial tumor cells from these samples was validated by cytokeratin 16 staining (Figure 6C), and high efficiency siRNA transfection protocols were derived using cytotoxic siRNAs targeting ubiquitin B, UBB (Figure 6D). Upon ACRBP depletion, LCOV2 and LCOV4 displayed a significant increase in mitotic figures following exposure to doses of paclitaxel that had little to no effect on controls (Figure 6E). LCOV17s respond to paclitaxel by micronucleation as we have previously observed in ES-2 cells (Figure 4C). This response was also sensitized upon ACRBP depletion (Figure 6D). Thus, inhibition of ACRBP expression increased the sensitivity of all samples to paclitaxel induced mitotic defects.
DISCUSSION
Here we have uncovered a selective dependency of ovarian cancer cells on ACRBP for robust mitotic progression. Of note, the ACRBP locus is predominantly expressed in the spermatocyte lineage, and is not detected in normal lung or ovarian epithelia. As a consequence of expression in a sex-specific immune-privileged site, the gene product is not recognized as a “self” antigen, and can evoke a humoral response when displayed by cancer cells (13). The activation of ACRBP expression appears to counter what would otherwise be a NuMA-dependent pathological perturbation of microtubule dynamics and centrosome function. The detection of native ACRBP/NuMA complexes suggests this antagonistic relationship is relatively direct. Centrosome overduplication is a common characteristic of tumors that can be normalized by a centrosome coalescence process that occurs in metaphase. Aberrant accumulation of NuMA impairs centrosome coalescence and can therefore promote gross chromosomal instability. This type of spindle malfunction has been assigned as an early lesion in many tumor settings and is a source of ‘heritable’ variation that can both drive the emergence of adaptive traits as well as restrict population expansion through high frequency mitotic death (31–34). This intermediate state in tumor evolution likely selects for acquisition of traits, such as ACRBP expression, that restore mitotic fidelity and allow clonal expansion of variants with robust proliferative and survival capacity (35, 36). We suggest that the antagonistic relationship uncovered here is a consequence of the historic genomic instability that promotes stochastic acquisition of tumorigenic traits during evolution of disease (Figure 6E). Thus ACRBP is a member of a class of tumor-specific vulnerabilities that emerge as a consequence of tumor evolution, not as components that drive disrespect of proliferation or survival checkpoints, but rather as components that normalize the perturbed mitotic infrastructure responsible for disease-promoting genetic variation. Such “emergent dependencies”, if druggable, likely represent therapeutic intervention targets with large therapeutic windows.
Acknowledgments
We thank Paola Gehrig, Division of Gynecological Oncology, for aid in obtaining live tumor explants, Kunle Odunsi for ACRBP monoclonal antibody, and Ben Neel for helpful discussions. This work was supported by the Robert Welch Foundation (I-1414) and the National Cancer Institute (CA71443 to MW and CA128926 to AW). Y.X. was supported by UL1 RR024982. K.C was supported by the National Cancer Institute training grant CA071341-14. AWW was supported in part by the American Cancer Society (PF-07-120-01-GMC).
References
- 1.Rieder CL, Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev Cell. 2004;7:637–51. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Gascoigne KE, Taylor SS. How do anti-mitotic drugs kill cancer cells? J Cell Sci. 2009;122:2579–85. doi: 10.1242/jcs.039719. [DOI] [PubMed] [Google Scholar]
- 3.Zalcberg J, Millward M, Bishop J, et al. Phase II study of docetaxel and cisplatin in advanced non-small-cell lung cancer. J Clin Oncol. 1998;16:1948–53. doi: 10.1200/JCO.1998.16.5.1948. [DOI] [PubMed] [Google Scholar]
- 4.Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–8. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 5.Ichiki M, Kawasaki M, Takayama K, et al. A multicenter phase II study of carboplatin and paclitaxel with a biweekly schedule in patients with advanced non-small-cell lung cancer: Kyushu thoracic oncology group trial. Cancer Chemother Pharmacol. 2006;58:368–73. doi: 10.1007/s00280-005-0168-3. [DOI] [PubMed] [Google Scholar]
- 6.Yumuk PF, Turhal NS, Gumus M, et al. Results of paclitaxel (day 1 and 8) and carboplatin given on every three weeks in advanced (stage III-IV) non-small cell lung cancer. BMC Cancer. 2005;5:10. doi: 10.1186/1471-2407-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramaswamy S. Rational design of cancer-drug combinations. N Engl J Med. 2007;357:299–300. doi: 10.1056/NEJMcibr072593. [DOI] [PubMed] [Google Scholar]
- 8.Whitehurst AW, Bodemann BO, Cardenas J, et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446:815–9. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- 9.Simpson A, Caballero OL, Jungbluth A, Chen Y-T, Old LJ. Cancer/Testis Antigens, Gametogenesis and Cancer. Nature Reviews Cancer. 2005;5:615–25. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 10.Okumura H, Noguchi Y, Uenaka A, et al. Identification of an HLA-A24-restricted OY-TES-1 epitope recognized by cytotoxic T-cells. Microbiol Immunol. 2005;49:1009–16. doi: 10.1111/j.1348-0421.2005.tb03688.x. [DOI] [PubMed] [Google Scholar]
- 11.Ono T, Kurashige T, Harada N, et al. Identification of proacrosin binding protein sp32 precursor as a human cancer/testis antigen. Proc Natl Acad Sci U S A. 2001;98:3282–7. doi: 10.1073/pnas.041625098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tammela J, Uenaka A, Ono T, et al. OY-TES-1 expression and serum immunoreactivity in epithelial ovarian cancer. Int J Oncol. 2006;29:903–10. [PubMed] [Google Scholar]
- 13.Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: review, standardization, and commentary. Cancer Immun. 2004;4:1. [PubMed] [Google Scholar]
- 14.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 16.Yeung KY, Fraley C, Murua A, Raftery AE, Ruzzo WL. Model-based clustering and data transformations for gene expression data. Bioinformatics. 2001;17:977–87. doi: 10.1093/bioinformatics/17.10.977. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan EL, Meier Nonparametric estimation from incomplete observations. Journal of the American Statistical Association. 1958;53:457–81. [Google Scholar]
- 18.Collett D. Modelling survival data in medical research. Chapman & Hall/CRC; 2003. [Google Scholar]
- 19.Langdon SP, Lawrie SS, Hay FG, et al. Characterization and properties of nine human ovarian adenocarcinoma cell lines. Cancer Res. 1988;48:6166–72. [PubMed] [Google Scholar]
- 20.Ban KH, Torres JZ, Miller JJ, et al. The END network couples spindle pole assembly to inhibition of the anaphase-promoting complex/cyclosome in early mitosis. Dev Cell. 2007;13:29–42. doi: 10.1016/j.devcel.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 21.Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–9. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 22.Kremer BE, Haystead T, Macara IG. Mammalian septins regulate microtubule stability through interaction with the microtubule-binding protein MAP4. Mol Biol Cell. 2005;16:4648–59. doi: 10.1091/mbc.E05-03-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tirnauer JS, Bierer BE. EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J Cell Biol. 2000;149:761–6. doi: 10.1083/jcb.149.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Compton DA, Luo C. Mutation of the predicted p34cdc2 phosphorylation sites in NuMA impair the assembly of the mitotic spindle and block mitosis. J Cell Sci. 1995;108 ( Pt 2):621–33. doi: 10.1242/jcs.108.2.621. [DOI] [PubMed] [Google Scholar]
- 25.Gaglio T, Saredi A, Compton DA. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J Cell Biol. 1995;131:693–708. doi: 10.1083/jcb.131.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol. 2000;149:851–62. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–58. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- 28.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bast RC, Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9:415–28. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weaver BA, Bonday ZQ, Putkey FR, Kops GJ, Silk AD, Cleveland DW. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162:551–63. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 33.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 34.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–86. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwon M, Godinho SA, Chandhok NS, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rancati G, Pavelka N, Fleharty B, et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879–93. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]