Abstract
The crystallographic phase problem is the primary bottleneck encountered when attempting to solve macromolecular structures for which no close crystallographic structural homologues are known. Typically, isomorphous “heavy-atom” replacement and/or anomalous dispersion methods must be used in such cases to obtain experimentally-determined phases. Even three-dimensional NMR structures of the same macromolecule are often not sufficient to solve the crystallographic phase problem. RNA crystal structures present additional challenges due to greater difficulty in obtaining suitable heavy-atom derivatives. We present a unique approach to solving the phase problem for novel RNA crystal structures that has enjoyed a reasonable degree of success. This approach involves modeling only those portions of the RNA sequence whose structure can be predicted readily, i.e., the individual A-form helical regions and well-known stem-loop sub-structures. We have found that no prior knowledge of how the helices and other structural elements are arranged with respect to one another in three-dimensional space, or in some cases, even the sequence, is required to obtain a useable solution to the phase problem, using simultaneous molecular replacement of a set of generic helical RNA fragments.
Keywords: Ribozyme, Crystallographic Phase Problem, Molecular Replacement, RNA Crystallography, RNA Structure Solution
Background
To solve a novel macromolecular crystallographic structure, the phase problem must be solved, typically using isomorphous replacement of heavy atoms or anomalous scattering [1]. If a homologous crystal structure exists, it may be possible to use that structure for molecular replacement [2], provided that the homologous search model has an r.m.s.d. less than 1.5 Å; even homologous NMR structures often are not sufficiently similar for molecular replacement to succeed, permitting crystallographers to speculate that NMR might really stand for “Not for Molecular Replacement.” [3] [4]
Novel RNA crystal structures are especially challenging, given the comparative lack of homologues (in contrast to the many thousands of protein structures available in the protein data base), as well as their limited ability to form useful heavy-atom derivatives [5].
We have found, counter-intuitively, that prior knowledge of how an RNA molecule folds in three dimensions is not required for successful solution of the crystallographic phase problem via molecular replacement, provided ideal models of a subset of helical fragments corresponding to the known secondary structure of a crystallized RNA can be generated and used as a set of search models for molecular replacement. This approach has been used to solve structures of the L1 ribozyme ligase [6], satellite tobacco ringspot virus hammerhead ribozyme [7], riboswitches [8,9], and other RNA structures; a detailed description of the solution of the L1 ligase ribozyme structure has appeared elsewhere [10]. Here we focus upon the generalized method for solving RNA structures using helical fragments, and several improvements that have been incorporated since the ligase ribozyme structure was solved.
Most structured RNAs consist mainly of a set of helical elements and connecting loops. Within these helical elements, most base-pairs are either standard Watson-Crick pairs, or are variants that do not grossly perturb the secondary structure. Because of this, it is possible to predict RNA secondary structure with a reasonably high degree of confidence using standard computational techniques such as those incorporated in freely available software such as MFOLD [11] and ViennaRNA [12], and these secondary structural predictions are readily testable using standard nucleic acid biochemical probing techniques. Hence, by the time one is crystallizing an RNA molecule, its secondary structure most likely has already been established. Many RNAs also incorporate other well-known structural elements, such as the GNRA tetraloop. Hence reasonably accurate models of individual secondary structural elements of a complex RNA molecule can be obtained simply from modeling standard A-form RNA helices. We have found the program COOT (Crystallographic Object-Oriented Toolkit) [13,14] to be the most efficient way to generate idealized model A-form RNA fragments, using the menu item “Calculate > Other Modelling Tools > Ideal DNA/RNA”. This instantly generates an ideal A-form RNA helix for any given sequence. If required, non-helical structures such as GNRA tetraloops may be obtained from the PDB and can be grafted onto a modeled helix. Typically, we start with up to four independent helical elements, in four separately named PDB files, even if the RNA represented by these fragments is less than 1/2 of the total RNA in the crystallographic asymmetric unit. In fact, this sort of “under-sampling” often improves the molecular replacement solution [6].
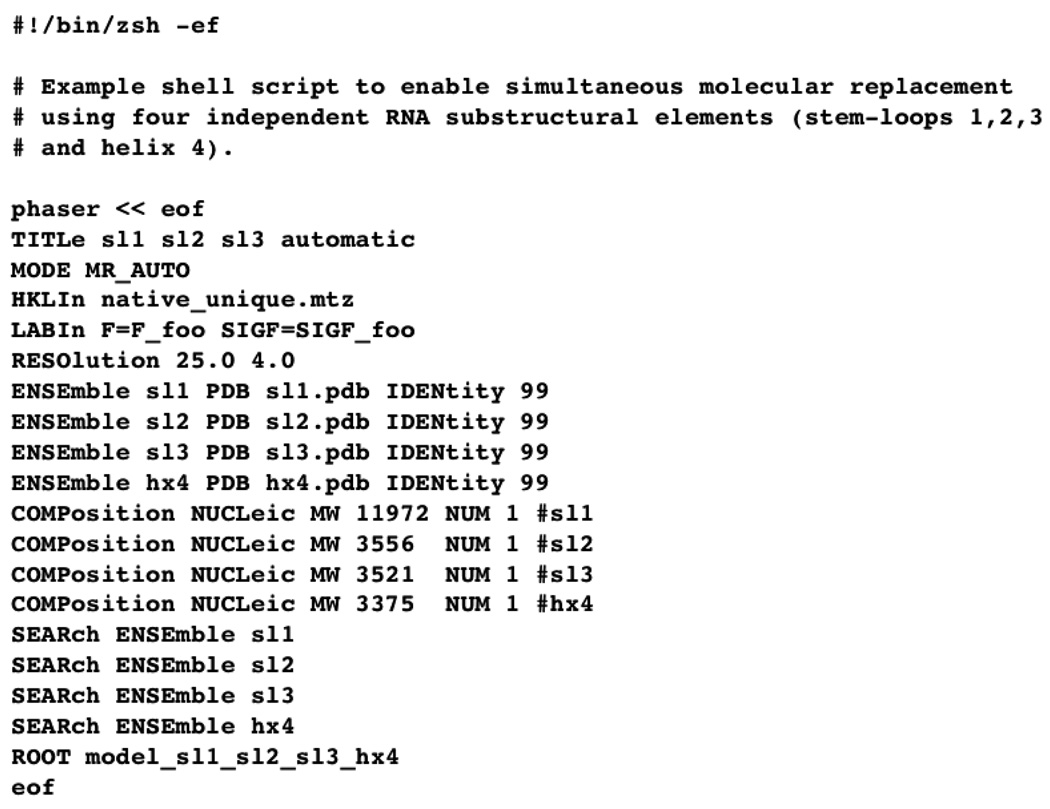
PHASER is an automated molecular replacement program [15] that is particularly well-suited to using several independent structural elements simultaneously. It automatically attempts to arrange the RNA fragments in three-dimensional space in a way that yields the best molecular replacement solution (and therefore best phase estimate). To make description of the process more concrete, Figure 1 depicts an example shell script in which four sub-structural elements, represented by four independent PDB files generated in COOT (called sl1.pdb, sl2.pdb, sl3.pdb and hx4.pdb), are used simultaneously for automatic molecular replacement (MODE MR_AUTO). A single “native” dataset is read by the program, and all the data between 25.0 and 4.0 Å resolution are employed. (Higher resolution data can be incorporated, if initial attempts fail, but it slows the calculation). Four “ENSEmble” entries are required for the four substructure pdb files, four “COMPosition NUCLeic” entries are required to designate these as nucleic acids, and to assign them molecular weights (based upon their sequences), and four “SEARch ENSEmble” entries are required to designate each as an independent, simultaneous search model.
Figure 1. Using Phaser with Four Independent Helical Fragments.
A shell script to use PHASER for the initial molecular replacement calculation is shown.
If all goes well, PHASER will output a pdb file and phase set corresponding to the most probable molecular replacement solution. The statistics for this solution will invariably be quite poor compared to what one might expect for standard molecular replacement, and within the present context are essentially meaningless. What is far more important is how the map appears, as this alone gives the most important indication of whether the procedure is beginning to work. Specifically, the PHASER-calculated sigma-A-weighted 2Fo-Fc map will show weak or broken up density where the model is incorrect, and more convincing density where the model is approximately correct. Typically, about 1/2 to 2/3 of the model will occupy reasonably strong density, and about 1/3 of the model will occupy weak or non-existent density. Again using COOT, the initial model can be edited manually. The most important form of manual intervention at this point of the procedure is to delete mercilessly any nucleotide that does not occupy reasonably strong electron density in a sigma-A-weighted 2Fo-Fc map contoured at 1. In addition, portions of any two pdb files that are involved in a steric clash (i.e., that try to occupy the same space in violation of van der Waals repulsion) should be deleted or adjusted to accommodate the electron density. When the editing process is complete, there should be few if any atoms that do not occupy electron density, and no steric clashes should remain. It is however most likely that there is no plausible physical connectivity between subsets of the RNA sequence. This is because the sequences themselves are not enough for the molecular replacement procedure to distinguish between one helix and another. It is in fact not necessary for the starting model to possess the correct sequence; all that is required is that each structural element represents an approximately correct secondary structure. (As an extreme test of this assertion, a solution of a 70 nucleotide RNA crystal structure was obtained using four randomly-generated five-base-paired helices, with no prior knowledge of the actual RNA sequence.)
The edited molecular replacement solution is then refined, typically using REFMAC [16] within COOT [13,14], and used as a partial model for subsequent iterations of molecular replacement within PHASER [15]. At this point simply including one additional helical element is usually sufficient for further model improvement; each addition requires further manual editing as described in the previous paragraph [10]. When further addition of helical elements yields no further improvement in the electron density map, the initial structure is refined using REFMAC, and the resulting phase probability distributions are converted to Hendrickson-Lattmann coefficients using the CCP4 program HLTOFOM [17] or the corresponding module within CNS [18]. These phases, when combined with the experimentally measured amplitudes, may then be treated as if they were determined by isomorphous replacement, with accompanying phase error estimates. Specifically, improvement of the phases using solvent flattening (in solvent-flipping mode [19]) within CNS [18] will simultaneously reduce model bias and improve the electron density map. The initial model used to generate the phases at this point is discarded.
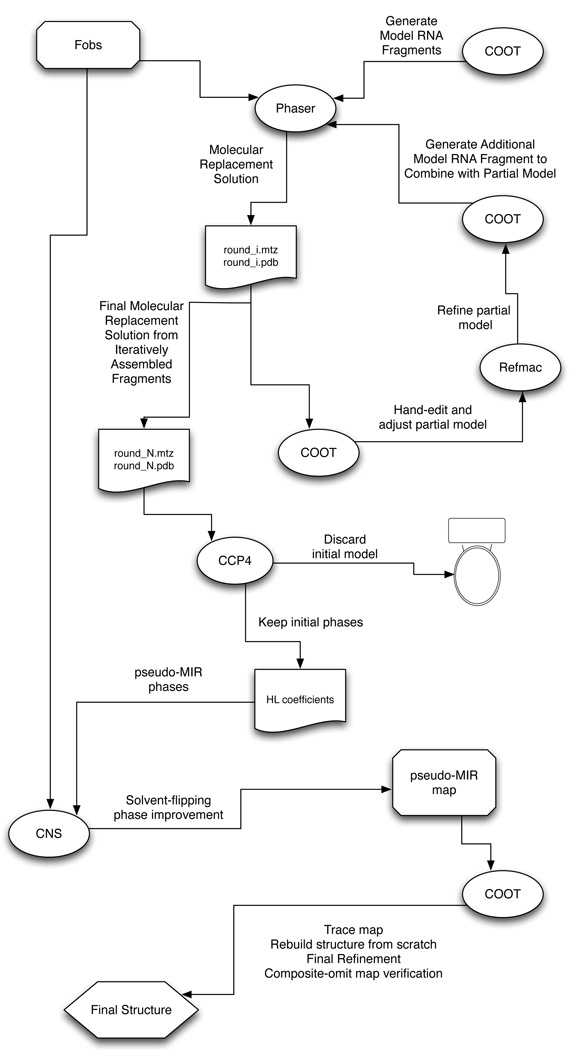
The newly solvent-flattened electron density map may now be treated as if it is an initial experimental map, and a poly-C nucleotide chain can be built into the density using COOT. Simulated annealing refinement of the initial poly-C structure within CNS, using all of the available amplitude data, will likely produce a significantly improved map, one that can be traced using the actual nucleotide sequence. The complete procedure is outlined schematically in Figure 2, and the initial and final, solvent-flattened, electron density maps are shown in Figure 3.
Figure 2. Schematic Representation of the Procedure.
An initial assembly of modeled RNA fragments (helices, loops) is used, in combination with a native data set (Fobs), to obtain a starting molecular replacement solution in PHASER [15] (round_1.mtz and round_1.pdb). This solution is examined in COOT [13,14], and all steric clashes are removed by manual editing, and any nucleotide that does not reside in strong electron density in the sigma-A-weighted 2Fo-Fc map is manually excised. The remaining model is then positionally refined using REFMAC [16] (which can be done within COOT) and any unoccupied density is modeled and refined in COOT. In addition, a new helical fragment (typically 5 base-pairs of A-from helix) for a subsequent iteration of molecular replacement is generated. Using the edited and refined model as a fixed partial model, the next (ith) round of molecular replacement in PHASER and subsequent editing and refinement is carried out with the new helical fragment. This cycle is repeated until no more helical fragments can be added. At this point, at the Nth cycle, the best molecular replacement solution (round_N.mtz and round_N.pdb) is used to generate a solvent-flattened electron density map in CNS [18]. The model (round_N.pdb) is discarded, and the calculated phase probability distributions are converted to Hendrickson-Lattmann coefficients within CCP4 [17] and are then imported into CNS, along with the native data set (Fobs), and treated as if they were experimentally determined MIR phases to be solvent-flattened. The resulting map is then used for building the final model from scratch (i.e., without reference to the discarded molecular replacement solution round_N.pdb) as if the map were derived from experimental MIR phases. The resulting structure is then checked against a composite-omit map generated in CNS in which 10% of the model is omitted from each element of the composite, and phases are regenerated from a standard simulated annealing procedure with a starting temperature of 4000K.
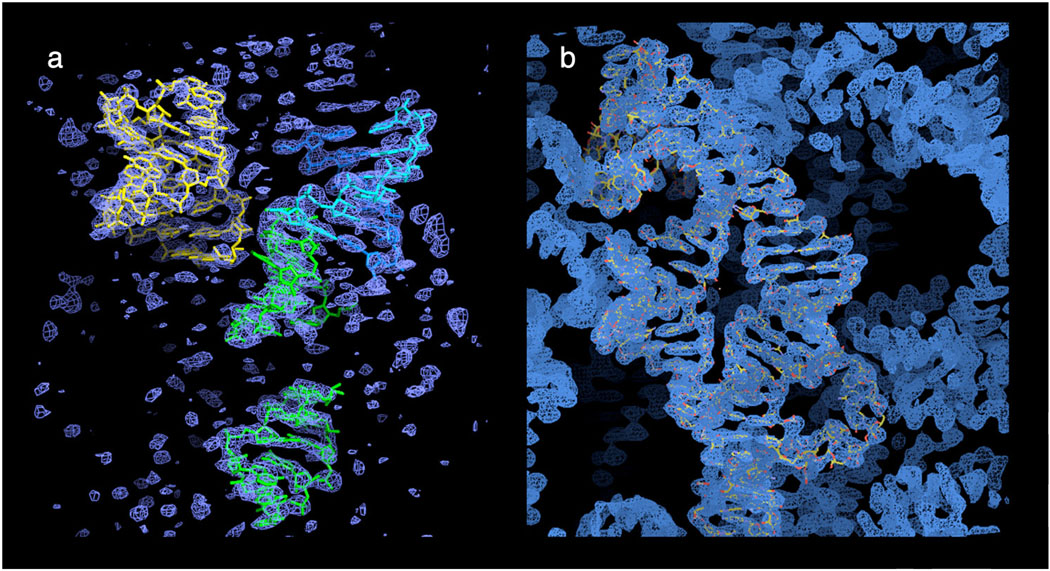
Figure 3. Electron Density Maps.
(a) Initial molecular replacement solution with four secondary structural fragments of the L1 ligase ribozyme. (b) The solvent-flattened electron density “pseudo-MIR” map created within CNS. A final refined model of the L1 ligase (2oiu) is superimposed on one asymmetric unit of electron density.
Software
The procedure outlined in the previous section makes use of a variety of readily available crystallographic software suites. These, along with their specific functionality and the purpose for their use, are described in Table 1, below.
Table 1.
Software of use for solving RNA Structures by piecewise molecular replacement
| Suggested Software | Functionality | Purpose |
|---|---|---|
| ViennaRNA | •RNAfold | •predict secondary structure of RNA |
| COOT | •Modelling Tools > Ideal DNA/RNA •Molecular Editing and Structural Manipulation |
•Create ideal A-form RNA helical fragments for initial molecular replacement. •Delete or adjust portions of initial molecular replacement solution within weak or nonexistent density. •Rebuilding of final model into pseudo-MIR map. |
| PHASER | •Automated Molecular Replacement |
•Position the various helical fragments correctly in 3D space to produce an approximate initial phase set |
| REFMAC | •Conventional Crystallographic Refinement |
•Optimize molecular geometry subsequent to editing |
| CCP4 | •Data Manipulation •Generation of Hendrickson- Lattmann Coefficients |
Create a pseudo-experimental phase set and convert to Hendrickson-Lattman coefficients for solvent-flattening and partial model refinement |
| CNS or PHENIX | •Solvent Flattening •Blurring Hendrickson-Lattmann Coefficients •Simulated Annealing Crystallographic Refinement •Composite-Omit map calculation |
•Improve initial phase estimate and reduce model bias using solvent flipping/flattening. •Further reduce model bias via “blurring” HL coefficients. •Calculation of “pseudo-MIR” electron density map. •Simulated annealing refinement of final model. •Composite-omit map calculation to check structural veracity. |
Troubleshooting
As with any molecular replacement procedure, the primary source of complications is model bias. Because the phases are not experimentally determined by model-free approaches, model bias is inherent. We have found the following approaches help to minimize the harmful effect of model bias [2].
Blurring of Hendrickson-Lattmann coefficients
Solvent-flattening and other density-modification procedures artificially inflate the figure of merit for estimated phases. Hence many partial models that have been crystallographically refined using modern approaches that attempt to model solvent effects will have phase probability estimates that are unrealistically high. This becomes problematic if the estimated phases are precise but rather inaccurate, as will be the case with the piecewise molecular replacement procedure we have described. The initial phases may be weighted too strongly and will thus not benefit from further attempts at improvement. The program CNS [18] is distributed with a module that permits the HL coefficients to be “blurred” by manually attenuating the temperature and scale factors (hlcoeff_blur.inp). Doing so inhibits a partial model from “taking over” density-modification procedures as a result of model bias, and can thus greatly improve the quality of the pseudo-experimental electron density map [20].
Phase Perturbation in EDEN [21]
The real-space density modification program EDEN uses a holographic procedure to minimize model bias, and further permits the user to randomly perturb initial phase estimates to test for reconvergence. An EDEN map generated from a partial model is often robust enough to enable construction of 2/3 of a missing asymmetric unit.
Composite-Omit Maps in CNS
Systematic elimination of 10% of a structure followed by simulated-annealing refinement of the remaining structure permits reconstruction of an electron density map corresponding to the omitted 10% of the structure. Ten such unique maps, when composited, will produce a a composite-omit map that has minimal model bias. This procedure is easily implemented within CNS [18] and should always be employed as a reality check.
Concluding Remarks
Contrary to initial assumptions, it is possible to use molecular replacement to solve crystal structures of RNAs having unique folds without prior knowledge of their tertiary structures. This approach appears to be possible due to the rather high degree of regularity of RNA secondary structures (such as A-form helices and tetraloops) and the comparative ease with which they may be predicted to form based upon known sequences. This in turn implies that, even in the absence of crystallographic phase information, there is sufficient information in a single crystal diffraction pattern to deduce the arrangement of secondary structural elements in three-dimensional space. In practice, this can be accomplished using simultaneous molecular replacement of several RNA helical fragments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Taylor G. The phase problem. Acta Crystallogr D Biol Crystallogr. 2003;59:1881–1890. doi: 10.1107/s0907444903017815. [DOI] [PubMed] [Google Scholar]
- 2.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 3.Chen YW, Dodson EJ, Kleywegt GJ. Does NMR mean "not for molecular replacement"? Using NMR-based search models to solve protein crystal structures. Structure. 2000;8:R213–R220. doi: 10.1016/s0969-2126(00)00524-4. [DOI] [PubMed] [Google Scholar]
- 4.Qian B, Raman S, Das R, Bradley P, McCoy AJ, Read RJ, Baker D. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450:259–264. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keel AY, Rambo RP, Batey RT, Kieft JS. A general strategy to solve the phase problem in RNA crystallography. Structure. 2007;15:761–772. doi: 10.1016/j.str.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robertson MP, Scott WG. The structural basis of ribozyme-catalyzed RNA assembly. Science. 2007;315:1549–1553. doi: 10.1126/science.1136231. [DOI] [PubMed] [Google Scholar]
- 7.Chi YI, Martick M, Lares M, Kim R, Scott WG, Kim SH. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol. 2008;6:e234. doi: 10.1371/journal.pbio.0060234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein DJ, Edwards TE, Ferré-D'Amaré AR. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulshina N, Baird NJ, Ferré-D'Amaré AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson MP, Scott WG. A general method for phasing novel complex RNA crystal structures without heavy-atom derivatives. Acta Crystallogr D Biol Crystallogr. 2008;64:738–744. doi: 10.1107/S0907444908011578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J Mol Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 13.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 14.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. logo. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 17.Winn MD. An overview of the CCP4 project in protein crystallography: an example of a collaborative project. J Synchrotron Radiat. 2003;10:23–25. doi: 10.1107/s0909049502017235. [DOI] [PubMed] [Google Scholar]
- 18.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 19.Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 20.Jones TA, Zou JY, Cowan SW. Kjeldgaard, Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 21.Szoke A. Diffraction of partially coherent X-rays and the crystallographic phase problem. Acta Crystallogr A. 2001;57:586–603. doi: 10.1107/s0108767301007322. [DOI] [PubMed] [Google Scholar]