Abstract
Apricot was introduced into the Mediterranean Basin from China and Asian mountains through the Middle-East and the Central Europe. Traditionally present in Tunisia, we were interested in accessing the origin of apricot species in the country, and in particular in the number and the location of its introductions. A set of 82 representative apricot accessions including 49 grafted cultivars and 33 seed propagated ‘Bargougs’ were genotyped using 24 microsatellite loci revealing a total of 135 alleles. The model-based Bayesian clustering analysis using both Structure and InStruct programs as well as the multivariate method revealed five distinct genetic clusters. The genetic differentiation among clusters showed that cluster 1, with only four cultivars, was the most differentiated from the four remaining genetic clusters, which constituted the largest part of the studied germplasm. According to their geographic origin, the five identified groups (north, centre, south, Gafsa oasis and other oases groups) enclosed a similar variation within group, with a low level of differentiation. Overall results highlighted the distinction of two apricot gene pools in Tunisia related to the different mode of propagation of the cultivars: grafted and seed propagated apricot, which enclosed a narrow genetic basis. Our findings support the assumption that grafting and seed propagated apricots shared the same origin.
Keywords: Prunus armeniaca L., Domestication, Rosaceae, SSR, Genetic structure
Introduction
Apricot (Prunus armeniaca L.) is a species of Prunus genus belonging to the family Rosaceae that is commercially grown world-wide. Apricot is diploid (2n = 16) and has a small genome size (5.9 Mbp/2n) (Arumuganathan and Earle 1991) compared to other fruit woody species as cherry (6.8 Mbp/2n) and apple (7.5 Mbp/2n) (Yuepeng and Korban 2007). Three centres of apricot origin were proposed by Vavilov (1992): north eastern, central and western China, central Asian mountains and near-eastern centre. Apricot was introduced into the Mediterranean Basin through two different ways. The first one was through the Middle-East allowing the identification of the Irano-Caucasian group (Kostina 1969) and the second was through the Central Europe (Faust et al. 1998). Four apricot cultivar groups named ‘Diversification’, ‘Geographically Adaptable’, ‘Continental Europe’ and ‘Mediterranean Basin’ were identified by Hagen et al. (2002). These groups displayed a gradient of decreasing genetic diversity of varieties from east to south-west.
Tunisia, one of the extreme dissemination zone of apricot in the Mediterranean Basin, enclosed a strongly distinct gene pool of local apricot cultivars with a low genetic diversity compared to the four previously defined groups (Khadari et al. 2006). Apricot in Tunisia included traditional cultivars propagated by grafting, cultivated from the north to the south of the country, and accessions propagated by seeds, specific to oasis agrosystems, locally called ‘Bargougs’ and characterized by their shadow contribution to the oasian ecosystem. Khadari et al. (2006) supported that seed propagation was more frequent than grafting propagation during apricot introduction in Tunisia. Thus, grafted cultivars could be the results of few introduced genotypes which have been firstly propagated by seeds. However, this hypothesis was not completely verified since the analysis was limited to only grafting propagated cultivars. Based on these results, a likely scenario could be proposed considering that grafting propagated cultivars shared a same gene pool with seed propagated accessions supporting a single apricot introduction in Tunisia. An alternative scenario based on historical events suggested two main apricot introductions (Valdeyron and Crossa-Raynaud 1950; Carraut and Crossa-raynaud 1974). The first one, for the grafting propagated cultivars, was located in the North of the country and originated from Andalusian germplasm, and the second one, for the seed propagated accessions, was situated in the South of Tunisia and originated from the Irano-Caucasian group.
Recently, several studies focused on characterization and genetic variability assessment of Tunisian apricot cultivars in order to preserve the local genetic resources and to understand the evolution of apricot in south Mediterranean areas. In fact, using microsatellites markers (or simple sequence repeats; SSRs), Krichen et al. (2006) established an identification key for the discrimination of 54 cultivars on the basis of only five loci. While, using amplified fragment length polymorphism markers (AFLPs), Khadari et al. (2006) compared the genetic diversity among 31 grafting propagated Tunisian apricot cultivars and accessions from Europe, North America, Turkey, Iran and China, in order to give insights into the origin and historical selection process of local germplasm. Results revealed that Tunisian apricot constituted a distinct group with close genetic relationships among cultivars. Moreover, using the same set of molecular markers, Krichen et al. (2008) studied the genetic relationships among 31 different apricot cultivars and revealed a clustering closely related to their geographic origin with the distinction of two major groups suggesting probably at least the introduction of two independent gene pools in Tunisia. However, these studies considered only grafted propagated cultivars and surveys were limited to some areas of apricot culture. In this paper, our investigations implied mapped microsatellites markers covering the eight linkage groups of Prunus genome to study a larger set of Tunisian apricot germplasm including both grafting propagated cultivars and seed propagated accessions.
We tested two working hypotheses related to the scenarios described above: (a) two independent introductions, a northern-central group including grafting propagated cultivars and a south-oasian group composed by seed propagated accessions; and (b) a single introduction followed by a local diversification. For that purpose, a set of 82 apricot accessions including 49 grafted cultivars and 33 seed propagated ‘Bargougs’ was analyzed using 24 single-locus SSR markers, polymorphic in apricot species and selected throughout the Prunus genome. Using both multivariate analysis and model-based Bayesian clustering methods, we argued for the assumption that grafting and seed propagated apricots shared the common origin.
Materials and methods
Plant material
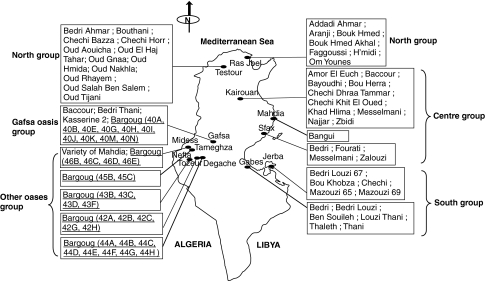
The plant material consisted of 82 Tunisian apricot accessions including 49 cultivars propagated by grafting and 33 spontaneous oasian ‘Bargougs’ propagated by seeds. Surveys were conducted in the northern (Ras Jbel and Testour), central (Kairouan, Mahdia and Sfax), and southern (Gabes and Jerba) areas of apricot culture in Tunisia and in six oasian zones (Gafsa, Midess, Tameghza, Nefta, Tozeur and Degache; Fig. 1). Surveyed areas belong to the sub-humid bioclimatic zone for northern areas to the Saharan superior bioclimatic one for some oasis. The studied accessions were classified into five geographic groups: north, centre, south, Gafsa oasis and other oases (Fig. 1).
Fig. 1.
Geographic origin of the 82 apricot accessions sampled in Tunisia. Surveys were conducted in the northern: Ras Jbel and Testour, central: Kairouan, Mahdia and Sfax and southern: Gabes and Jerba areas of apricot culture in Tunisia as well as in six oasis regions: Gafsa, Midess, Tameghza, Nefta, Tozeur and Degache. ‘Bargoug’ accessions located in oasian region and propagated by seeds were underlined (the number is related to the oasis origin and the letter represents accession of a same oasis). Studied accessions were classified into five geographic groups: north, centre, south, Gafsa oasis and other oases
DNA extraction and microsatellite amplification
Total genomic DNA was extracted from fresh young leaves according to Bernatzky and Tanksley (1986) protocol. Genotyping was conducted with a set of 24 SSR primers. These primers were mapped on the Prunus reference map: Texas × Earlygold (Joobeur et al. 1998) and distributed on the whole Prunus genome with the assumption of no linkage between pairs of loci (Table 1).
Table 1.
Genetic diversity parameters of the microsatellite loci used in the genetic apricot analysis
| Microsatellite primers | Linkage group position | Allele number | Size range (bp) | H o | H e | Fis | P-value |
|---|---|---|---|---|---|---|---|
| CPPCT034a | 1 | 5 | 197–207 | 0.512 | 0.534 | 0.040 | 0.442 |
| AMPA109b | 1 | 3 | 205–221 | 0.100 | 0.096 | −0.033 | 1.000 |
| BPPCT001c | 2 | 2 | 116–118 | 0.487 | 0.445 | −0.094 | 0.457 |
| BPPCT030c | 2 | 5 | 136–150 | 0.675 | 0.717 | 0.059 | 0.168 |
| BPPCT004c | 2 | 7 | 193–209 | 0.650 | 0.763 | 0.149 | 0.004 |
| AMPA116b | 2 | 6 | 115–141 | 0.612 | 0.722 | 0.152 | 0.055 |
| AMPA101b | 3 | 4 | 188–212 | 0.662 | 0.664 | 0.003 | 0.208 |
| AMPA119b | 3 | 5 | 098–112 | 0.087 | 0.085 | −0.023 | 1.000 |
| BPPCT040c | 4 | 3 | 128–144 | 0.387 | 0.365 | −0.059 | 0.881 |
| UDP97-402c | 4 | 5 | 120–144 | 0.500 | 0.457 | −0.094 | 0.798 |
| AMPA105b | 5 | 7 | 182–214 | 0.725 | 0.684 | −0.059 | 0.089 |
| BPPCT017c | 5 | 5 | 161–209 | 0.537 | 0.593 | 0.094 | 0.109 |
| BPPCT038c | 5 | 6 | 127–147 | 0.587 | 0.696 | 0.157 | 0.026 |
| AMPA100b | 6 | 6 | 196–214 | 0.537 | 0.712 | 0.247 | 0.000 |
| BPPCT008c | 6 | 7 | 107–129 | 0.687 | 0.767 | 0.105 | 0.510 |
| BPPCT025c | 6 | 3 | 147–159 | 0.450 | 0.55 | 0.183 | 0.187 |
| CPPCT030a | 6 | 10 | 147–185 | 0.787 | 0.781 | −0.007 | 0.209 |
| Ma014ae | 6 | 3 | 136–142 | 0.400 | 0.479 | 0.166 | 0.095 |
| Ma040ae | 6 | 6 | 207–227 | 0.537 | 0.62 | 0.133 | 0.418 |
| UDP98-412f | 6 | 5 | 083–115 | 0.487 | 0.632 | 0.230 | 0.008 |
| CPPCT022a | 7 | 7 | 240–272 | 0.762 | 0.709 | −0.075 | 0.106 |
| CPPCT033a | 7 | 6 | 137–161 | 0.312 | 0.332 | 0.061 | 0.197 |
| CPPCT006a | 8 | 8 | 175–201 | 0.662 | 0.777 | 0.148 | 0.001 |
| UDP98-409d | 8 | 11 | 122–164 | 0.750 | 0.854 | 0.122 | 0.000 |
| Mean | 5.62 | 0.515 | 0.561 | 0.067 | 0.290 |
Polymerase Chain Reactions (PCR) were carried out in a 20 μl reaction mix containing 20 ng of template DNA, 2 mM of MgCl2, 4 pmol of the reverse primer and 1 pmol of the forward primer, 0.2 mM of each deoxynucleotide triphosphate, and 1 unit of Taq polymerase (Sigma). Reverse primers were unlabeled while forward primers were labelled on their 5′ end using one of the following fluorescent compounds: FAM, NED or HEX, to enable analysis on automated sequencers. Amplifications were performed on a MasterCycler, ep gradient S (Eppendorf) with the following conditions: 5 min initial denaturation step at 94°C followed by 35 cycles of amplification at 94°C for 30 s, T° annealing (depending on the locus) for 1 min, and 72°C for 1 min. This was ended by a final extension step at 72°C for 10 min. PCR products were detected using capillary electrophoresis on an ABI prism 3130 × l automatic DNA sequencer (Applied Biosystems). Samples were prepared by mixing 3 μl of diluted PCR products with 7.85 μl formamide and 0.15 μl Rox-labelled GenSize 400HD. The GENEMAPPER V3.7 software (Applied Biosystems) was applied to size the peak patterns.
Genetic diversity and structure analyses
Based on SSR profiles defined among the 82 studied accessions, the total number of alleles and the observed (H o) and the expected (H e) heterozygosity values were calculated in GENETIX 4.05 program (Belkhir et al. 1996–2004). The allelic richness and private allelic richness per geographic group, as defined in Fig. 1, were computed by the generalized rarefaction approach ADZE (Szpiech et al. 2008), standardized at G = 16 individuals per group. Wright’s F-statistics were estimated according to the formula of Weir and Cockerham (1984) in GENEPOP 4.0 program (Raymond and Rousset 1995a). They represent a hierarchical series of fixation indices where Fis represents the deviation from Hardy–Weinberg expectation within groups and Fst measures the fixation of different alleles in different groups. The significance of pairwise Fst values was assessed by Fisher’s exact probability test (Raymond and Rousset 1995b). Pairwise standard genetic distances (Nei 1972) were calculated among geographic groups and an unrooted tree was constructed using the Neighbor-Joining algorithm with 10 000 bootstraps over SSR loci as implemented in PHYLIP 3.69 package (Felsenstein 1993). Factorial Correspondence Analysis (FCA) was performed with GENETIX to provide a synthetic representation of the genetic variability of studied accessions according to their genetic origin, as well as the position of nuclear microsatellite alleles. Finally, for analysis of molecular variance (AMOVA), we defined two levels of genetic partition: (a) a regional level identifying two main pools: grafted propagated apricots from north, centre and south of Tunisia; and seed propagated accessions from Gafsa oasis and other oases; (b) a population level distinguishing the five geographic groups. The partitioning of molecular variance within and among groups was calculated in GENALEX (Peakall and Smouse 2006).
Model-based Bayesian clustering
Two Bayesian Markov Chain Monte Carlo programs, STRUCTURE (Pritchard et al. 2000; Falush et al. 2003) and InStruct (Gao et al. 2007) were used to infer population structure of studied accessions. STRUCTURE program assumes Hardy–Weinberg equilibrium and linkage equilibrium within populations, while InStruct does not assume Hardy–Weinberg equilibrium and allows simultaneous inference of the selfing rate and the number and admixture of historical lineages. The STRUCTURE algorithm was run using the basic model with admixture and correlated allele frequencies, with the assumed number of genetic K clusters varying from 1 to 8, ten replicate runs per K value, a burnin period length of 100,000, and a post-burnin simulation length of 1,000,000. To identify the number of K clusters explaining the observed genetic structure, statistic parameters defined by Evanno et al. (2005) based on the rate of change in the log probability of data between successive K values were used. For InStruct program, we used the model 3, inferring population structure with admixture and selfing rates at the individual level. To infer the selfing rates at the individual level, we used the adaptative independence sampler method and the uniform distribution prior. We performed 20 independent chains for each K value ranging from 1 to 6. Each chain was iterated 200,000 times after burn-in with 100,000 iterations. To find optimal alignments of independent runs, the average pairwise similarity (H′) of run results for both STRUCTURE and InStruct was assessed by CLUMPP 1.1.2 program (Jakobsson and Rosenberg 2007) with greedy algorithm, 10,000 random input orders and 10,000 repeats. Graphical representation of clustering results was performed with the DISTRUCT software (Rosenberg 2004).
Results
SSR polymorphism
Eighty distinct genotypes were identified within the set of eighty-two apricot accessions analyzed using 24 SSR markers. All microsatellite loci were polymorphic with a number of alleles ranging from 2 (BPPCT001) to 11 (UDP98-409) (Table 1). A total of 135 alleles were observed with an average of 5.62 alleles per locus. Expected heterozygosity ranged from 0.085 (AMPA119) to 0.854 (UDP98-409) with an average across loci of 0.561. Observed heterozygosity ranged from 0.087 (AMPA119) to 0.787 (CPPCT030) with an average across loci of 0.515. A significant heterozygosity deficit was observed for five loci: AMPA100, CPPCT006, BPPCT004, UDP98-409 and UDP98-412 (Table 1).
Model-based Bayesian clustering
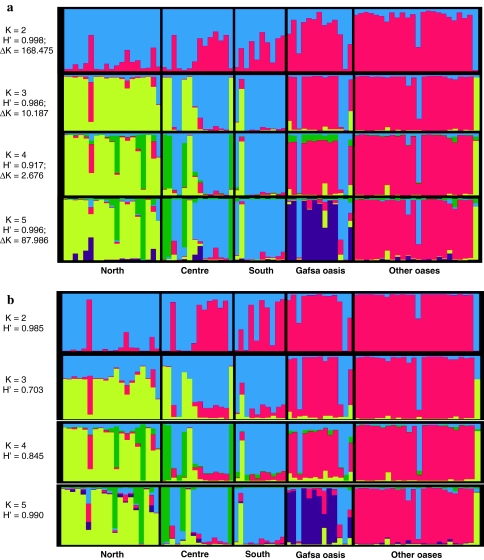
Based on the STRUCTURE program under admixture and correlated allele frequencies model, the estimated log probability of the data (ln Pr(X|K)), given the assumed number of ancestral populations K, was highest for K = 5 (data not shown). The ad hoc quantity based on the second order rate of change of the posterior probability of K (ΔK; Evanno et al. 2005) showed that the accurate representation of Tunisian apricot genetic structure, was observed for K = 2 (ΔK = 168.475) and K = 5 (ΔK = 87.986; Fig. 2a). Cluster visualisation for the permuted average pairwise similarity (H′) generated by CLUMPP among run results was presented in Fig. 2.
Fig. 2.
Estimated population structure. Each individual is represented by a thin vertical line, which is partitioned into K colored segments that represent the individual’s estimated membership fractions in K clusters. Black lines separate individuals of different populations on the basis of their geographic origin. Populations are labeled below the Figure. The Figure shown for a given K is based on the highest probability run at that K. a Results obtained using STRUCTURE program. For each K value, average pairwise similarity (H′) and rate of change of posterior probability of K (ΔK) were mentioned. b Results obtained using InStruct program based on the selfing rate model 3 using the adaptative independence sampler method and the uniform distribution prior. For each K value, average pairwise similarity (H′) was calculated
A first overview at K = 2 allowed to distinguish the seed propagated accessions (red) from the grafting propagated cultivars (blue). However, these cultivars include variable proportion of seed propagated gene pool especially in the centre and the south (Fig. 2a). The model under K = 5 classified the eighty apricot genotypes into five distinct genetic clusters. Taking into account the genotypes assigned to the five clusters, the first cluster (green) grouped the four traditional cultivars, mainly originated from Kairouan. The second cluster (yellow) was composed by 16 accessions originated from the north of Tunisia (Testour and Ras Jbel). The third cluster (blue) included 16 cultivars from the centre and the south, the fourth cluster (purple) was composed by 8 ‘Bargougs’ originated from Gafsa oasis. Finally the fifth cluster (red) included 20 ‘Bargoug’ accessions originated from the other prospected oases. In total, 64 accessions were clearly assigned with more than 80% of the assignation probability. The non assigned sixteen accessions were assumed as having a mixed ancestry. Four grafted propagated cultivars: ‘Baccour’, ‘Bedri Thani’, ‘Kasserine 2’ (Gafsa oasis) and ‘Variety of Mahdia’ (other oases group) were assigned to “Northern cultivars” and “Central and Southern cultivars” clusters, respectively (Fig. 2a). Based on the model 3 of the InStruct Program using the adaptative independence sampler method and the uniforme distribution prior, the posterior distribution of selfing rates varied from 0.294 to 0.459. Similar results were obtained with a higher probability of individual assignation to the five identified clusters (Fig. 2b).
Genetic differentiation
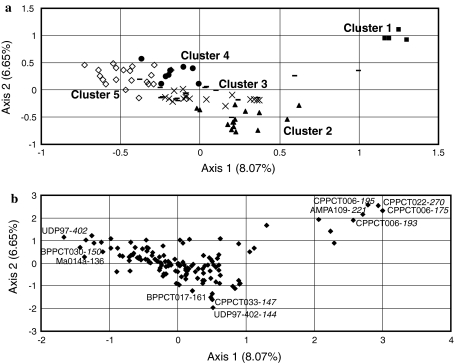
Based on the five clusters defined by the model-based Bayesian clustering algorithm using STRUCTURE program, we observed high significant genetic differentiation ranged from Fst = 0.121 (comparison between “Bargougs of Gafsa oasis” and “Bargougs from other oasis regions”) to 0.313 (between “Cultivars with rectangular-plate fruit shape and white flesh color” and “Bargougs from other oasis regions”) with a mean value of 0.183 (Table 2). The first cluster was the most differentiated from other clusters, with the Fst value ranging from 0.234 (compared to “Northern cultivars” cluster) to 0.313 (compared to “Bargougs from other oasis regions” cluster). Such differentiation could be biased due to the limited size of this group (only four cultivars). However, a similar differentiation was confirmed by multivariate analysis. The 2-dimensional scatter plot of Factorial Correspondence Analysis coordinates for the first and second axes, which explained 8.07 and 6.65% of variance, respectively, showed a clear separation of the first cluster including the four cultivars: ‘Bayoudhi’, ‘Chechi Khit El Oued’, ‘Chechi Dhraa Tammar’ and ‘Chechi Horr’ (Fig. 3a). This distinction evidenced mainly by five alleles at three SSR loci (CPPCT006-175, CPPCT006-193, CPPCT006-195, CPPCT022-270 and AMPA109-221) clustered as the four cultivar group (Fig. 3b). For the remaining clusters, except the genetic differentiation between “Central and Southern cultivars” and “Bargougs of Gafsa oasis” (Fst = 0.220), the pairwise Fst values do not exceed 0.194 (Table 2) indicating their narrow genetic basis. These results were confirmed by the FCA (Fig. 3).
Table 2.
Pairwise genetic differentiation index (Fst) among apricot clusters identified using model-based Bayesian clustering method (Global Fst = 0.183)
| Cluster 1 (4) | Cluster 2 (16) | Cluster 3 (16) | Cluster 4 (8) | Cluster 5 (20) | |
|---|---|---|---|---|---|
| Cluster 1 | – | ||||
| Cluster 2 | 0.234*** | – | |||
| Cluster 3 | 0.263*** | 0.136*** | – | ||
| Cluster 4 | 0.281*** | 0.194*** | 0.220*** | – | |
| Cluster 5 | 0.313*** | 0.180*** | 0.150*** | 0.121*** | – |
Cluster 1, cultivars with rectangular-plate fruit shape and white flesh color; cluster 2, northern cultivars; cluster 3, central and southern cultivars; cluster 4, Bargougs of Gafsa oasis and cluster 5 Bargougs from other oasis regions
*** P < 0.001
Fig. 3.
Plot based on the first and second axes of Factorial Correspondence Analysis (FCA) coordinate showing the genetic differentiation among the five defined genetic clusters. a Apricot genotype relationships with filled square cultivars with rectangular-plate fruit shape and white flesh color (cluster 1), filled triangle northern cultivars (cluster 2), times central and southern cultivars (cluster 3), filled circle Bargougs of Gafsa oasis (cluster 4) and diamond Bargougs from other oasis regions (cluster 5), dash non assigned cultivars. b Allele’s distribution explaining the apricot genotype relationships
Genetic variation
The total number of alleles detected within each geographic group varied from 77 (south group) to 101 (other oases group) with a mean of 91.4 alleles per group (Table 3). The allelic richness varied from 3.08 ± 0.21 (south) to 3.46 ± 0.25 (Gafsa oasis). Mean value of expected heterozygosity across loci ranged from 0.498 for other oases group to 0.551 for north one. No significant positive Fis values were observed suggesting the absence of heterozygosity deficit within groups. Based on the standardized maximum value G = 16 individual per group, the private allelic richness varied from 0.131 ± 0.058 (south) to 0.330 ± 0.096 (other oases). Seed propagated apricots from other oases group displayed a high level of allelic richness and private allelic richness but a low genetic diversity value; while grafted propagated apricots from the north group displayed both high level of allelic richness, private allelic richness and genetic diversity (Table 3).
Table 3.
Genetic diversity parameters within each group identified on the basis of their geographic origin (see Fig. 1)
| Number of accessions | H o | H e | Fis | Total allele number | Allelic richnessa | Private allelic richnessa | |
|---|---|---|---|---|---|---|---|
| North | 19 | 0.572 | 0.551 | −0.057 | 99 | 3.44 ± 0.25 | 0.306 ± 0.101 |
| Centre | 14 | 0.488 | 0.531 | 0.087 | 88 | 3.34 ± 0.28 | 0.224 ± 0.091 |
| South | 10 | 0.576 | 0.508 | −0.137 | 77 | 3.08 ± 0.21 | 0.131 ± 0.058 |
| Gafsa oasis | 13 | 0.480 | 0.549 | 0.134 | 92 | 3.46 ± 0.25 | 0.223 ± 0.083 |
| Other oases | 24 | 0.485 | 0.498 | 0.037 | 101 | 3.39 ± 0.27 | 0.330 ± 0.096 |
aStandardized at maximum value G = 16 individuals per group
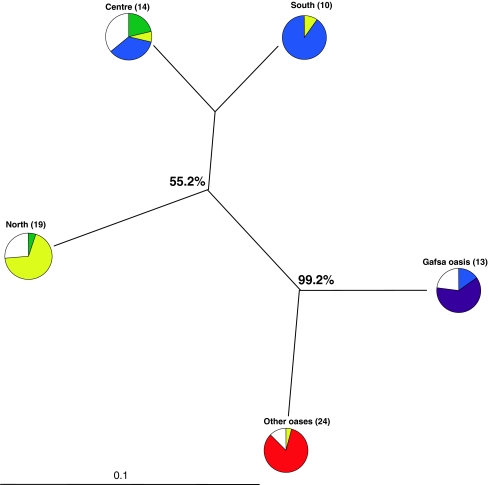
Genetic differentiation among the five geographic groups varied from Fst = 0.017 (between centre and south groups) to 0.122 (between north and other oases groups) with a global Fst value of 0.079 (Table 4). The lowest Nei’s genetic distance was obtained for the pair of groups centre-south (0.070), whereas the highest value was obtained for groups north-other oases (0.209; Table 4). The close relationships between centre and south groups were also illustrated in Fig. 4. AMOVA showed a significant genetic partition between and within grafted propagated cultivars and seed propagated accessions of about 5%, while it was 90% within geographic group (Table 5). These results were summarized in Fig. 4 revealing two main gene pools: grafted versus seed propagated apricots. This distinction showed a high bootstrap support with values higher than 50%.
Table 4.
Geographic group pairwise comparisons
| North (19) | Centre (14) | South (10) | Gafsa oasis (13) | Other oases (24) | |
|---|---|---|---|---|---|
| North | – | 0.105 | 0.155 | 0.162 | 0.209 |
| Centre | 0.047*** | – | 0.070 | 0.146 | 0.171 |
| South | 0.079*** | 0.017** | – | 0.159 | 0.151 |
| Gafsa oasis | 0.076*** | 0.065*** | 0.075*** | – | 0.097 |
| Other oases | 0.122*** | 0.100*** | 0.089*** | 0.048*** | – |
Above diagonal: Nei’s (1972) genetic distance. Below diagonal: Pairwise genetic differentiation index (Fst); Global Fst = 0.079
** P < 0.01; *** P < 0.001
Fig. 4.
Genetic relationships among the five apricot groups defined according to their geographic origin. The dendrogram is constructed based on Nei’s (1972) genetic distances and Neighbor-Joining algorithm. Numbers beside branches are bootstrap values. For each group, the genetic composition as estimated by model-based Bayesian clustering method is represented
Table 5.
Analysis of molecular variance (AMOVA) among the 80 apricots classified as two main pools: grafted and seed propagated apricots (at regional level). Grafted pool includes northern, central and southern geographic groups and seed propagated accessions includes Gafsa oasis and other oases groups. These five groups as defined as population level
| Source of variation | df | Sum of squares | Percentage of variation |
|---|---|---|---|
| Among regions | 1 | 46.721 | 5a |
| Within regions/populations | 3 | 51.069 | 5a |
| Within populations | 155 | 1019.729 | 90a |
| Total | 159 | 1117.519 |
aSignificant at alpha 0.001
Discussion
Our study deals with the genetic structure analysis of Tunisian local apricot germplasm in the south-west Mediterranean area. The obtained results showed that SSR markers can be successfully used in Tunisian apricot germplasm characterization. The 24 SSR loci used were carefully selected for their polymorphism among Prunus armeniaca L. species and covering the whole Prunus genome. All the microsatellite markers were polymorphic and revealed a total of 135 alleles allowing to distinguish 80 genotypes from the 82 studied apricot accessions. The expected heterozygosity value was 0.561, indicating that the Tunisian apricot accessions displayed a substantial genetic diversity. By comparison the expected heterozygosity value for apricot germplasm issued from Chinese, central Asian and Irano-Caucasian ecogeographical groups was highest (H e = 0.645) (Zhebentyayeva et al. 2003), and (H e = 0.741), when including North American material to this previous germplasm (Maghuly et al. (2005)). Even if these studies used different SSR markers, and different set of accessions, they can be compared to our study since SSR loci display a similar mutation rate. Compared to the previously defined ecogeographical groups, apricot in Tunisia displayed a substantial but lower genetic diversity. Our results confirmed the study of Khadari et al. (2006) based on AFLP markers and could be explained by the decreasing eastern-western gradient of genetic diversity at the Mediterranean Basin level related to historical introduction and diffusion of apricot in Tunisia.
In previous works, the phenetic approach allowed to classify genotypes into groups referred to geographic origins or to phenotypic characters as demonstrated by Sánchez-Pérez et al. (2005) who clustered apricot genotypes into seven principal groups according to both their pedigree and their geographic origin. However, the clustering method was limited in the identification of significantly distinct genetic groups. We adopted an alternative model-based Bayesian clustering analysis using both STRUCTURE (Pritchard et al. 2000) and InStruct (Gao et al. 2007) programs to infer genetic structure and assign individuals to groups. InStruct do not assume Hardy–Weinberg equilibrium and allows inferring genetic structure with individual selfing rate estimation, a relevant model in the case of apricot which displays a ratio variability of selfing versus outcrossing (Faust et al. 1998). Despite of the posterior distribution of selfing rates ranging from 0.294 to 0.459, similar results were obtained by both programs with the validation of two and five clusters indicating the low impact of selfing rate in the genetic structure of Tunisian apricot. The first cluster included four cultivars with rectangular-plate fruit shape and white flesh color, which were classified into north Mediterranean apricot (unpublished data), and was the most genetically differentiated from the other clusters as attested by the Fst values and the multivariate analysis. For the remaining germplasm (clusters 2–5), based on the genetic structure, the geographic origin and the propagation mode, we identified two main gene pools: grafted propagated cultivars from north, centre and south; and seed propagated accessions from oasis region. Comparing these two propagation modes, we noted that seed propagated apricots from other oases group and grafted propagated apricots from the north group shared similar allelic richness but were contrasted at the genetic diversity level. These findings suggested the impact of seedling events from probably a limited size of apricot genotypes as proposed by Khadari et al. (2006). Otherwise, the genetic diversity observed in grafting propagated cultivars is probably related to the varietal diversity maintained by clonal reproduction as the case of other Mediterranean fruit species like fig (Achtak et al. 2010).
We asked for the origin of these two gene pools: grafted versus seed propagated accessions. Our first working hypothesis supports that the two apricot gene pools in Tunisia were originated from two independent introductions during the apricot dissemination. One was located in the north and center region of apricot culture allowing the presence of grafting propagated cultivars in the country (“Northern cultivars” and “Central and Southern cultivars” clusters), and the other was situated in the oasis region with the existence of seed propagated material (“Bargougs of Gafsa oasis” and “Bargougs from other oasis regions” clusters). Here, we proposed several results as genetic signature invalidating this hypothesis and supporting their common origin. First, compared to the global genetic differentiation, lower Fst values among the four clusters (clusters two to five) were observed. Second, the genetic partition within and between the two apricot gene pools was about 5%. These results, which were illustrated by the FCA, indicated the narrow genetic basis of these two gene pools. Finally, the genetic structure at K = 2 as identified by both STRUCTURE and InStruct programs allowed to distinguish the seed propagated accessions (in red color) from the grafting propagated cultivars which enclosed a variable proportion of seed propagated gene pool (in blue color). In conclusion, our findings argued for the assumption that these two gene pools: grafting propagated cultivars and seed propagated accessions come from a similar and single introduction of apricot in Tunisia followed by a local diversification. The results presented here will be useful to design strategies that improve the conservation and management programs of apricot genetic resources.
Acknowledgments
The authors would like to thank A. Weber for her participation to microsatellite genotyping, Dr. H. Achtak for his help in the data analysis, and Dr. M. Pitrat and N. Ranc for their kind remarks on the early version of the manuscript. This work has been conducted with a helpful assistance of the Tunisian “Ministère de l’Enseignement Supérieur et de la Recherche Scientifique” (Project Lab B02). It was supported by a bilateral Franco-Tunisian action under the frame of CMCU Project (n°05G0904).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Achtak H, Ater M, Oukabli A, Santoni S, Kjellberg F, Khadari B. Traditional agroecosystems as conservatories and incubators of cultivated plant varietal diversity: the case of fig (Ficus carica L.) in Morocco. BMC Plant Biol. 2010;10:28. doi: 10.1186/1471-2229-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranzana MJ, Garcia-Mas J, Carbo J, Arùs P. Development and variability analysis of microsatellite markers in peach. Plant Breed. 2002;121:87–92. doi: 10.1046/j.1439-0523.2002.00656.x. [DOI] [Google Scholar]
- Arumuganathan K, Earle E. Nuclear DNA content of some important plant species. Plant Mol Biol Report. 1991;9:208–218. doi: 10.1007/BF02672069. [DOI] [Google Scholar]
- Belkhir K, Goudet J, Chikhi L, Bonhomme F (1996–2004) Genetix 4.05, logiciel pour WindowsTM pour la génétique des populations. Laboratoire Génome et Populations, CNRS UPR 9060, Université de Montpellier II, Montpellier (France)
- Bernatzky R, Tanksley SD. Genetics of actin-related sequences in Tomato. Theor Appl Genet. 1986;72:314–321. doi: 10.1007/BF00288567. [DOI] [PubMed] [Google Scholar]
- Carraut A, Crossa-Raynaud P. L’amélioration des variétés d’abricotier en Tunisie. Ann Inst Natl Rech Agron Tunis. 1974;47(2):33. [Google Scholar]
- Cipriani G, Lot G, Huang WG, Marrazzo MT, Peterlunger E, Testolin R. AC/GT and AG/CT microsatellite repeats in peach (Prunus persica L. Batsch) isolation, characterization and cross-species amplification in Prunus. Theor Appl Genet. 1999;99(1–2):65–72. doi: 10.1007/s001220051209. [DOI] [Google Scholar]
- Dirlewanger E, Cosson P, Tavaud M, Aranzana MJ, Poizat C, Zanetto A, Arús P, Laigret F. Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.) Theor Appl Genet. 2002;105:127–138. doi: 10.1007/s00122-002-0867-7. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotypes data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust M, Suranyl D, Nyujto F. Origin and dissemination of apricot. Hortic Rev. 1998;22:225–266. [Google Scholar]
- Felsenstein J (1993) Phylogenetic Interference Package (PHYLIP): version 3.5c. Department of Genetics, University of Washington, Seattle, USA
- Gao H, Williamson S, Bustamante CD. A MCMC approach for joint inference of population structure and inbreeding rates from multi-locus genotype data. Genetics. 2007;176(3):1635–1651. doi: 10.1534/genetics.107.072371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen LS, Khadari B, Lambert P, Audergon JM. Genetic diversity in apricot revealed by AFLP markers: species and cultivars comparisons. Theor Appl Genet. 2002;105:298–305. doi: 10.1007/s00122-002-0910-8. [DOI] [PubMed] [Google Scholar]
- Hagen LS, Chaib J, Fady B, Decroocq V, Bouchet JP, Lambert P, Audergon JM. Genomic and cDNA microsatellites from apricot (Prunus armeniaca L.) Mol Ecol Notes. 2004;4:742–745. doi: 10.1111/j.1471-8286.2004.00802.x. [DOI] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23(14):1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Joobeur T, Viruel MA, de Vicente MC, Jauregui B, Ballester J, Dettori MT, Verde I, Truco MJ, Messeguer R, Batlle I, Quarta R, Dirlewanger E, Arùs P. Construction of a saturated linkage map for Prunus using an almond × peach F2 progeny. Theor Appl Genet. 1998;97:1034–1041. doi: 10.1007/s001220050988. [DOI] [Google Scholar]
- Khadari B, Krichen L, Lambert P, Marrakchi M, Audergon JM. Genetic structure in Tunisian apricot populations multiplicated by grafting: a signature of bottleneck effects and ancient propagation by seedlings. Genet Resour Crop Evol. 2006;53(4):811–819. doi: 10.1007/s10722-004-5721-0. [DOI] [Google Scholar]
- Kostina KF. The use of varietal resources of apricots for breeding. Trud Nikit Bot Sada. 1969;40:45–63. [Google Scholar]
- Krichen L, Mnejja M, Arús P, Marrakchi M, Trifi-Farah N. Use of microsatellite polymorphisms to develop an identification key for Tunisian apricots. Genet Resour Crop Evol. 2006;53:1699–1706. doi: 10.1007/s10722-005-1220-1. [DOI] [Google Scholar]
- Krichen L, Martins JMS, Lambert P, Daaloul A, Trifi-Farah N, Marrakchi M, Audergon JM. Using AFLP markers for the analysis of the genetic diversity of apricot cultivars in Tunisia. J Am Soc Hortic Sci. 2008;133(2):204–212. [Google Scholar]
- Maghuly F, Borroto Fernández E, Ruthner S, Pedryc A, Laimer M. Microsatellite variability in apricots (Prunus armeniaca L.) reflects their geographic origin and breeding history. Tree Genet Genomics. 2005;1:151–165. doi: 10.1007/s11295-005-0018-9. [DOI] [Google Scholar]
- Nei M. Genetic distance between populations. Am Nat. 1972;106:283–292. doi: 10.1086/282771. [DOI] [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49:1283–1286. doi: 10.2307/2410454. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. doi: 10.1046/j.1471-8286.2003.00566.x. [DOI] [Google Scholar]
- Sánchez-Pérez R, Ruiz D, Dicenta F, Egea J, Martinez-Gomez P. Application of simple sequence repeat (SSR) markers in apricot breeding: molecular characterization, protection, and genetic relationships. Sci Hortic. 2005;103:305–315. doi: 10.1016/j.scienta.2004.06.009. [DOI] [Google Scholar]
- Szpiech ZA, Jakobsson M, Rosenberg NA. ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics. 2008;24(21):2498–2504. doi: 10.1093/bioinformatics/btn478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testolin R, Marrazzo T, Cipriani G, Quarta R, Verde I, Dettori MT, Pancaldi M, Sansavini S. Microsatellite DNA in Peach (Prunus persica L. Batsch) and its use in fingerprinting and testing the genetic origin of cultivars. Genome. 2000;43:512–520. doi: 10.1139/gen-43-3-512. [DOI] [PubMed] [Google Scholar]
- Valdeyron G, Crossa-Raynaud P. Les fruits de Tunisie. Ann Inst Natl Rech Agron Tunis. 1950;23:65–82. [Google Scholar]
- Vavilov NI. Origin and geography of cultivated plants (tr. L. storr-best) Cambridge: CUP; 1992. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1158–1370. doi: 10.2307/2408641. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Mochida K, Imai T, Shi YZ, Ogiwara T, Hayashi T. Microsatellite markers in peach [Prunus persica (L.) Batsch] derived from an enriched genomic and cDNA libraries. Mol Ecol Notes. 2002;2:298–301. doi: 10.1046/j.1471-8286.2002.00242.x. [DOI] [Google Scholar]
- Yuepeng H, Korban SS. Spring: a novel family of miniature inverted-repeat transposable elements is associated with genes in apple. Genomics. 2007;90(2):195–200. doi: 10.1016/j.ygeno.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Zhebentyayeva TN, Reighard GL, Gorina VM, Abbott AG. Simple Sequence Repeat (SSR) for assessment of genetic variability in apricot germplasm. Theor Appl Genet. 2003;106(3):435–444. doi: 10.1007/s00122-002-1069-z. [DOI] [PubMed] [Google Scholar]