Abstract
Although the genetic basis of human sexual determination and differentiation has advanced considerably in recent years, the fact remains that in most subjects with disorders of sex development (DSD) the underlying genetic cause is unknown. Where pathogenic mutations have been identified, the phenotype can be highly variable, even within families, suggesting that other genetic variants are influencing the expression of the phenotype. This situation is likely to change, as more powerful and affordable tools become widely available for detailed genetic analyses. Here, we describe recent advances in comparative genomic hybridisation, sequencing by hybridisation and next generation sequencing, and we describe how these technologies will have an impact on our understanding of the genetic causes of DSD.
Key Words: Comparative genomic hybridization, Disorders of sex development, High-throughput sequencing, Next generation sequencing
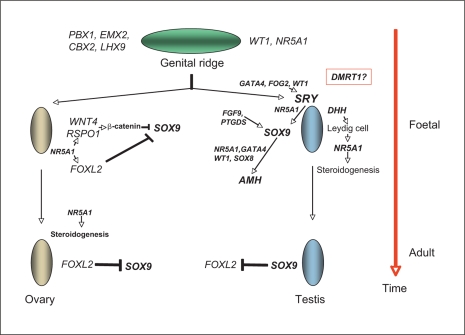
The term ‘disorders of sex development’ (DSD) has been defined as ‘congenital conditions in which the development of chromosomal, gonadal, or anatomical sex is atypical’ [Hughes et al., 2006]. DSD covers a wide spectrum of phenotypes. 46,XY DSD includes 46,XY complete or partial gonadal dysgenesis, or undervirilisation or undermasculinisation of an XY male due to defects in androgen synthesis or action. 46,XX DSD includes gonadal dysgenesis, or more commonly overvirilisation or masculinisation of an XX female due to androgen excess. Ovotesticular DSD refers to an individual with both ovarian and testicular material present in the same or different gonads, and 46,XX testicular DSD refers to an XX male with testes. Other forms of DSD include cloacal extrophy, severe hypospadias, vaginal atresia, and as part of other conditions such as Mayer-Rokitansky-Kuster-Hauser syndrome, Smith-Lemli-Opitz syndrome or genito-palato-cardiac syndrome [Porter, 2008; Sultan et al., 2009]. Although there have been considerable advances in our understanding of the genetic factors involved in gonadal differentiation in the last 20 years (fig. 1), it has been estimated that a molecular diagnosis is made in only around 20% of DSD, except in cases where the biochemical profile indicates a specific steroidogenic block [Hughes et al., 2006].
Fig. 1.
The molecular and genetic events in mammalian sex determination and differentiation. The bipotential genital ridge is established by several genes including NR5A1 [Wilhelm et al., 2007; Sekido and Lovell-Badge, 2009]. In the XY gonad the activation of SRY expression, possibly initiated by GATA4/FOG2/NR5A1/WT1, leads to the upregulation of SOX9 expression via a synergy with NR5A1 [Sekido and Lovell-Badge 2008, 2009]. In the XX gonad, the supporting cell precursors accumulate β-catenin in response to RSPO1/WNT4 signalling and repress SOX9 activity [Schlessinger et al., 2010]. Once SOX9 levels reach a critical threshold, several positive regulatory loops are initiated, including autoregulation of its own expression and formation of feed-forward loops via FGF9 or PGD2 signalling [Sekido and Lovell-Badge, 2009]. At later stages, FOXL2 may repress Sox9 expression [Uhlenhaut et al., 2009]. In the testis, SOX9 promotes the testis pathway, including Amh activation, and it also probably represses the ovarian genes Wnt4 and Foxl2 [Sekido and Lovell-Badge, 2009; Uhlenhaut et al., 2009; Schlessinger et al., 2010]. DMRT1 controls sex determination in some species of fish and may be the master sex-determining switch in birds, but its role in mammalian sexual development is unclear [Wilhelm et al., 2007; Smith et al., 2009]. Much of this data has been generated from studies in mice.
Identification of Novel Genetic Factors by Comparative Genomic Hybridisation
Comparative genomic hybridisation (CGH) was first described in 1992 as an approach to interrogate cancer genomic DNA using metaphase chromosomes as probes [Kallioniemi et al., 1992]. The strategy of the technique was to differentially label the DNA isolated from a test and reference cell population with different fluorochromes and to cohybridise the labelled samples to a metaphase spread from a reference cell. The relative hybridisation intensity of the test and reference signals at a given location is then proportional to the relative copy number of those sequences in the test and reference genomes. If the reference genome is normal, then increases and decreases in the intensity ratio directly indicate DNA copy number variation in the genome of the test cells. The ratio of the intensities of the 2 fluorochromes reflects the copy number differences between the cells of interest and the control cells.
The major technical challenge of array CGH is the generation of hybridisation signals that are sufficiently intense, specific and quantitative so that copy number changes can be accurately and reliably detected. In the late 1990s, array CGH was first developed by spotting DNA from large-insert clones such as BACs (bacterial artificial chromosomes) and cDNAs onto microarray slides [Solinas-Toldo et al., 1997; Pinkel et al., 1998]. Array elements made from genomic BAC clones (complexity 100–200 kb) typically provide more intense signals than do elements with shorter sequences such as oligonucleotides, so could be analysed by early detection systems. However, the advantage of smaller array elements, such as oligonucleotides is the much higher genomic resolution (often ×100) if measurement precision can be maintained. The most recent advancement in CGH microarrays has been the use of oligonucleotide sequences as probes [Lucito et al., 2003] (fig. 2) and the development of genotyping arrays from low-resolution SNP (single nucleotide polymorphism) arrays to high-resolution hybrid arrays that integrate both SNP and CNV (copy number variation) probes [Bignell et al., 2004]. Now many different oligoarray platforms are available for CGH analyses from various companies including Nimblegen (www.nimblegen.com) and Agilent (www.agilent.com) with ever increasing levels of resolution. In addition, several recent Next Generation Sequencing (NGS) approaches also provide information on structural variation (see below).
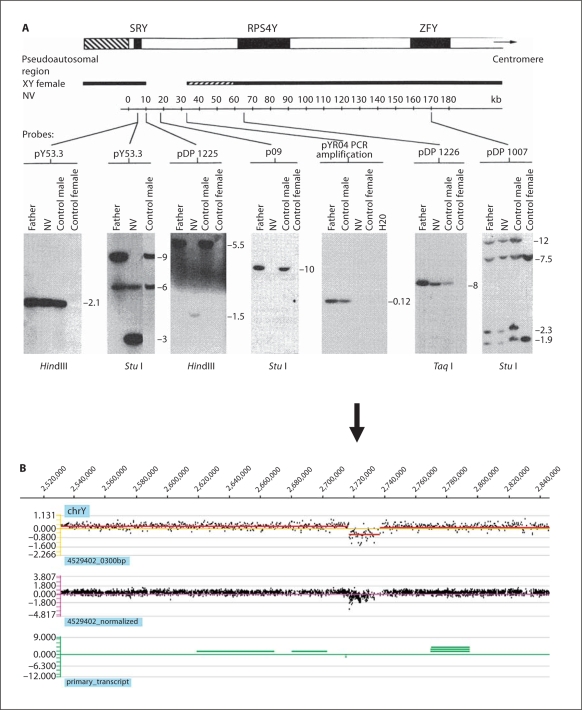
Fig. 2.
CGH analysis of a de novo Y chromosome deletion associated with 46,XY gonadal dysgenesis. A This case was first published almost 20 years ago and the deletion was detected and the extent of the deletion mapped by Southern blotting [McElreavy et al., 1992b]. B The same deletion identified by CGH analysis using a Y chromosome-specific platform (www.nimblegen.com). The genome profile of the patient versus normal male reference genomic DNA is shown for the short arm of the Y chromosome. The x-axis coordinates indicate the relative position of the oligo probes ordered by genomic map position on the Y chromosome. The y-axis shows the log2 ratio shift. The top panel shows window averaging of signals in 300 bp segments. The middle panel shows the normalized signal profile using qspline normalisation. The lower panel indicates the genomic position of transcripts on this region of chromosome Yp.
The recent impact of these technologies on medical genetics has been substantial [Zhang et al., 2009]. These advances include the detection of subtle changes in chromosome architecture associated with cancers [Beroukhim et al., 2010], severe developmental delay [Girirajan et al., 2010], epilepsy [de Kovel et al., 2010], and schizophrenia [Glessner and Hakonarson, 2009]. Like many developmental processes, human sex development is sensitive to gene dosage effects, and duplication and deletion events in the human genome are associated with DSD. For example, deletions of terminal 9p24 are associated with varying degrees of 46,XY gonadal dysgenesis in around 70% of XY individuals with this chromosomal change [Barbaro et al., 2009]. A number of reports have described deletions of terminal 10q associated with 46,XY gonadal dysgenesis together with somatic anomalies. The gene responsible for the gonadal phenotype remains unidentified although EMX2 has been suggested as a likely candidate [Ogata et al., 2000]. Deletions and duplications of chromosome 22q11.2 have been described in 3 cases of 46,XX SRY-negative testicular DSD [Aleck et al., 1999; Erickson et al., 2003; Seeherunvong et al., 2004]. A microduplication of chromosome 17q, including the SOX9 gene has been reported in a 46,XX man with ambiguous external genitalia and otherwise unremarkable phenotype [Huang et al., 1999]. Deletion of the dosage sensitive gene NR0B1, encoding DAX1 on chromosome Xp21.2 results in congenital adrenal hypoplasia (AHC), whereas an NR0B1 duplication in 46,XY individuals leads to gonadal dysgenesis and a female phenotype [Bardoni et al., 1994]. Duplications of NR0B1 gene have been described in a small number of other cases of 46,XY gonadal dysgenesis. Duplication of 1p has also been associated with 46,XY gonadal dysgenesis although the gene(s) responsible is not known [Wieacker and Volleth, 2007].
CGH analysis is beginning to reveal novel rearrangements associated with 46,XY and 46,XX DSD. This approach has been used to physically map translocation breakpoints in a case of 46,XX ovotesticular DSD associated with a 12;17 translocation upstream of the SOX9 gene [Refai et al., 2010]. An inherited 960 kb deletion, positioned 517 kb to 1.477 Mb upstream of the SOX9 gene, was detected by oligoarray CGH in a XY DSD child with acampomelic dysplasia and in her affected mother [Lecointre et al., 2009]. In both these examples, the rearrangements may have lead to the deregulation of SOX9 expression. A large 3 Mb deletion of 9q34 that includes the LMX1B and NR5A1 genes was identified using an array CGH approach in a 46,XY girl with clinical features of genitopatellar syndrome and ovotesticular DSD [Schlaubitz et al., 2007]. More recently a smaller 970 kb deletion that included the NR5A1 gene (encoding steroidogenic factor-1) was also detected in a 46,XY DSD girl without skeletal features [van Silfhout et al., 2009]. CGH analysis has also been used to define deletions on 11p13 and 9p24 associated with syndromic forms of 46,XY DSD [Le Caignec et al., 2007; Vinci et al., 2007].
CGH has successfully identified duplications of NR0B1 associated with 46,XY complete gonadal dysgenesis in the absence of somatic anomalies [Barbaro et al., 2007] and a unique 257 kb deletion located 250 kb upstream of NR0B1 in a 21-year-old girl with 46,XY complete gonadal dysgenesis [Smyk et al., 2007]. This deletion presumably results in deregulation of NR0B1 expression.
Aside from the detection of subtle rearrangements of genetic factors known to be involved in gonadal determination and differentiation, CGH analyses offer the possibility to detect new genes involved in these processes. CGH analyses revealed a novel recurrent 15q24 microdeletion syndrome (1.7–3.9 Mb in size) that is characterised by growth retardation, microcephaly, digital abnormalities, characteristic facial dysmorphia (high anterior hair line, broad medial eyebrows, hypertelorism, downslanted palpebral fissures, broad nasal base, long smooth philtrum and full lower lip), and genital anomalies in the male (micropenis/hypospadias) [Sharp et al., 2007; Andrieux et al., 2009].
As increasing numbers of subjects with DSD are undergoing analysis using CGH platforms, more pathogenic rearrangements will be reported that may reveal unique insights into the mechanism of gonadal somatic cell fate determination and development.
Identification of Novel Genetic Factors Associated with DSD by Custom Array Sequencing
Diagnostic sequencing of genes known to be responsible for DSD using conventional Sanger sequencing is useful to identify novel mutations associated with gonad development and it is the mainstay of most diagnostic laboratories. However, this approach is both time-consuming and laborious. High-throughput, high-density sequencing using microarray technology offers the possibility of rapidly and accurately sequencing portions of the genome at reduced costs. Several next-generation sequencing technologies are emerging (see below) for whole genome analyses but at present the costs and considerable data handling that are involved means that these approaches will be limited to the research environment for the foreseeable future. In contrast, sequencing by hybridisation is suitable for situations in which a moderate amount of sequence is being analysed (10–300 kb, representing several to tens of genes) in a repetitive manner.
The principle of oligo-hybridisation sequencing is based on the differential hybridisation of target DNA to an array of oligonucleotide probes, thereby decoding its primary DNA sequence. This is most applicable to disorder-specific studies such as DSD where sequences can be compared with an established reference. CustomSeq arrays developed by Affymetrix rely on allele-specific hybridisation for determining the DNA sequence of interest and a single array can be used to sequence up to 300 kb of double-stranded unique DNA sequence. Every base to be sequenced is represented by probes that are 25 bp long and differ only at a single central position [Lipshutz et al., 1999]. The array uses 4 oligomers per base for each of the 2 DNA strands. The 4 oligomers differ at a single central position that could be A, C, G, or T, and they query complementary bases on the DNA strand. The remaining 24 positions are the same for all 4 oligomers and are complementary to the reference DNA sequence being queried.
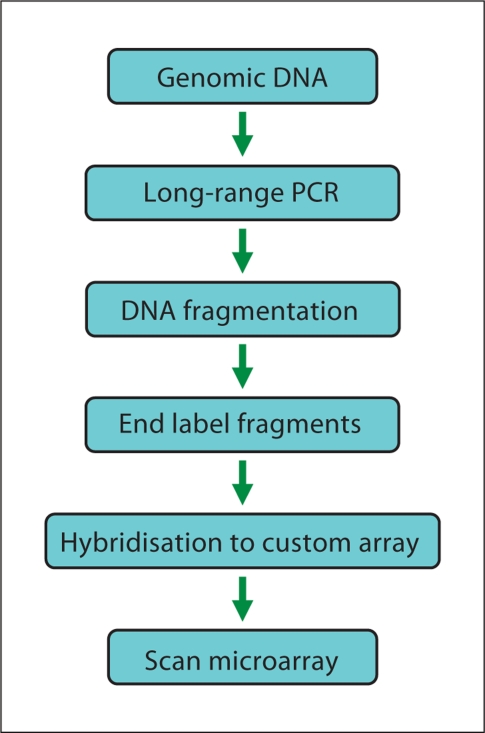
Sufficient DNA must be generated for hybridisation to the microarray. Current protocols for target-enriched patient material use locus-specific long-range PCR amplification of regions of interest. Long-range PCR minimises the number of reactions required, but genes at multiple loci, with highly dispersed exons, will require many PCR reactions. Long-range PCR products also usually require quantification and normalisation prior to hybridisation (fig. 3). The PCR products are then pooled and fragmented. The DNA fragments are labelled with biotin and subsequently hybridised with the microarray. Finally, the microarray is washed, stained and scanned to measure the fluorescence intensities (fig. 3). Software, such as the GeneChip Sequence Analysis Software (GSEQ; www.affymetrix.com) allows the user to perform sequence analysis of the data to produce the final sequence calls.
Fig. 3.
Schematic overview of procedure involved in sequencing by hybridisation.
Although relatively cheap, sequencing by hybridisation does have the major drawback of false positive calls. These can arise through various mechanisms such as PCR failure or SNP interference. The latter occurs when 2 variants are very close together on the same patient DNA fragment. If there is a homozygous variant, the performance of neighbouring probes, which assume a wild-type base at the variant position, can be poor. Therefore, clusters of SNP calls (i.e. SNPs within 9 bases of each other) should be interpreted with caution. Cross-hybridisation is a cause of false positives, so a single array cannot analyse homologous loci. Repetitive sequences can also generate false positive data, although algorithms have been developed to improve base call reading.
The elimination of false positive calls is essential to reduce the need to perform confirmatory capillary resequencing. Computational algorithms can be developed for specific arrays so that for the detection of rare mutations in clinical disease settings such as DSD, the use of an algorithm that compares a single patient data set to data from a group of controls is likely to improve call rates and false positive rates through a statistical comparison of signal strength for each probe. Algorithms may also help to solve another problem confronted by array sequencing technologies, the detection of novel insertion or deletion mutations. However, monoallelic deletions that are larger than a PCR amplicon could be missed due to the normalisation of PCR products prior to hybridisation.
Despite these disadvantages, custom sequencing arrays offer a high-throughput sequencing method that is efficient, fast, automatable, and cost-effective for repetitive resequencing of targeted regions of DNA. Relatively few examples of this approach have been reported in the literature to date. A microarray-based sequencing platform technology to detect sequence alterations in multiple autosomal recessive retinal disease genes was reported with a 99% accuracy and reproducibility [Mandal et al., 2005]. A 16 gene sequencing array has recently been described for studying genetic variation in genes involved in angiogenesis that had a 99.4% sequencing accuracy compared with Sanger capillary sequencing [Szoke et al., 2009]. Similarly, a comparison of 93 worldwide mitochondrial genomes sequenced using either the Affymetrix GeneChip Human Mitochondrial Resequencing Array 2.0 (MitoChip v.2.0) or Sanger sequencing an average call rate of 99.48% and an accuracy of 99.98% for the MitoChip [Hartmann et al., 2009]. The failure to detect some variants was largely due to the inherent difficulty of the array to detect insertions and deletions.
Identification of Novel Genetic Factors by Next Generation Sequencing
The advent of next generation sequencing (NGS) technologies has tremendously reduced the sequencing cost and experimental complexity. NGS technologies also offer improved template coverage, rendering sequencing-based genome analysis more readily available and useful to individual laboratories. These sequencing methods employ massively parallel approaches to sequence several hundred thousand to millions of reads simultaneously [Ansorge, 2009; Metzker, 2010].
The current approaches to NGS falls broadly into 2 types – polymerase based and ligase based [Ansorge, 2009; Metzker, 2010]. The recent use of ligases for massively parallel short-read DNA sequencing of human genomes offers several unique attributes compared to polymerases [Valouev et al., 2008]. In the context of mutation detection, the most important difference is the use of an error-correcting probe-labelling scheme (two-base encoding, or 2BE), which provides error correction concurrent with the color-called alignment of the data (i.e. without having to resequence the reads). There are currently 4 commercially available new-generation sequencing technologies. These are the polymerase based systems of Roche/454, Illumina/Solexa, Helicos HeliScope and the ligase-based Applied Biosystems SOLiD [Margulies et al., 2005; Bentley et al., 2008; Harris et al., 2008; Valouev et al., 2008]. All of these systems produce an abundance of short reads at a much higher throughput than is achievable with the state-of-the-art Sanger sequencer and each have their own unique properties (table 1; see also http://www.wellcome.ac.uk/Education-resources/Teaching-and-education/Animations/DNA/index.htm). Other NGS approaches are on the horizon. These include nanopore sequencing, where individual nucleic acid strands in solution are electrophoretically driven through nanopore-based devices. The narrow pore can ensure that the sequence of nucleic acids can be analysed in their native order without the need for prior amplification or labelling, therefore resulting in increased accuracy and reduced sequencing costs [Branton et al., 2008].
Table 1.
Next generation sequencing approaches
| Platform | Sequencing chemistry | Read length (bases) | Gb per run | Run time (fragment library) | System raw accuracy | Consensus base accuracy | Disadvantages |
|---|---|---|---|---|---|---|---|
| Roche/454 GS FLX titanium | Polymerase | 400 | 0.4-0.6 | 10 h | 99% to 400 bases | 99.99% at 15 × coverage | Prone to sequence error in homopolymer DNA sequences Cost per run expensive. |
| Illumina/Solexa GAII | Polymerase | 75-100 | 20a | 4 days | 98-99% | >99.999% at 20 × coverage | Low multiplexing capabilities |
| Life Technologies, ABI SOLiD 3+ | Ligase | 50 | 60b | 12 days | 99.94% | >99.999% at 15 × coverage | Duration of run |
| Helicos Biosciences, Heliscope | Polymerase | 32 | 21 | 8 days | 93-97% | 99.995% at 20 × coverage | Higher error rates |
Paired end sequences. The new Illumina/Solexa HiSeq 2000 generates up to 200 Gb per run.
The recent SOLiD™ 4 System generates over 100 gigabases.
In NGS, the sequencing is only a part of the entire process that includes template preparation, the sequencing reaction itself and the data handling and analyses [Medvedev et al., 2009; McPherson, 2009; Metzker, 2010]. The latter may in fact be more expensive than the costs involved in generating the billions of base pairs of sequence. Considerable challenges remain in terms of data transfer, storage and quality control. Specialised computational analysis and skill are required to align or assemble read data, along with laboratory information management systems for sample tracking and process management [McPherson, 2009].
DNA template for NGS is achieved either by solution-based hybridisation strategies or by solid phase amplification [Vogelstein, 2003; Fedurco et al., 2006]. In emulsion PCR (emPCR), a library of targets is created, and adaptors containing universal priming sites are ligated to the target ends, allowing complex genomes to be amplified with common PCR primers. After ligation, the DNA is separated into single strands and captured onto beads under conditions that favour one DNA molecule per bead. After the successful amplification and enrichment of emPCR beads, millions can be immobilised in a polyacrylamide gel on a standard microscope slide, chemically crosslinked to an aminocoated glass surface, or deposited into individual PicoTiterPlate (PTP) wells (Roche/454) in which the NGS chemistry can be performed [Leamon, 2003; Metzker, 2010].
Solid-phase amplification can also be used to produce randomly distributed, clonally amplified clusters from fragment or mate-pair templates on a glass slide. High-density forward and reverse primers are covalently attached to the slide, and the ratio of the primers to the template on the support defines the surface density of the amplified clusters. Solid-phase amplification can produce 100–200 million spatially separated template clusters (Illumina/Solexa), providing free ends to which a universal sequencing primer can be hybridised to initiate the NGS reaction. The Helicos BioSciences, HeliScope system is also based on polymerase sequencing-by-synthesis of single molecules in which labelled DNA bases are sequentially added to the nucleic acid templates captured on a flow cell (http://www.helicosbio.com). No prior amplification process is involved in this system resulting in a non-bias representation of templates for genome sequencing, although the error rate may be higher than other polymerase-based systems [Harris et al., 2008].
The Roche 454 system uses pyrosequencing technologies to enable the simultaneous sequencing of several hundred thousand DNA fragments, with a read of 400 bp (www.454.com). Pyrosequencing is a bioluminescence method that measures the release of inorganic pyrophosphate into a synthesised DNA strand by converting it into visible light by a series of enzymatic reactions [Ronaghi et al., 1996]. The order and intensity of the generated light is detected and recorded in the form of a peak signal, which reveals the underlying DNA sequence. The Illumina (Solexa) Genome Analyzer (GA) uses sequencing-by-synthesis to generate ∼200 million 75–100-bp reads (http://www.illumina.com/systems/genome_analyzer.ilmn). This is a cyclic method that comprises nucleotide incorporation, fluorescence imaging and cleavage. In the first step, a DNA polymerase, bound to the primed template, adds or incorporates just one fluorescently modified nucleotide, which represents the complement of the template base. The termination of DNA synthesis after the addition of a single nucleotide is an important feature of this approach. Following incorporation, the remaining unincorporated nucleotides are washed away. Imaging is then performed to determine the identity of the incorporated nucleotide. This is followed by a cleavage step, which removes the terminating/inhibiting group and the fluorescent dye. Additional washing is performed before starting the next incorporation.
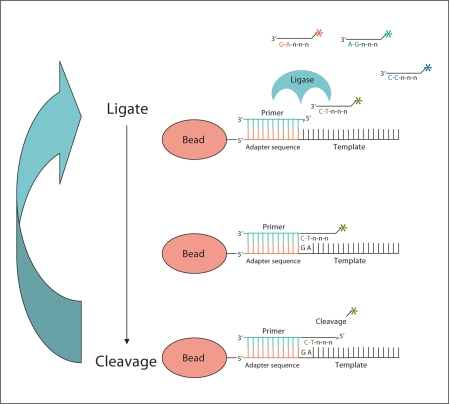
A novel massively parallel sequencing technology based on ligation of oligonucleotides as opposed to sequencing by synthesis has been developed by Applied Biosystems (http://solid.appliedbiosystems.com). In this approach, sequencing is carried out via sequential rounds of probe annealing and ligation with high fidelity and high read quality (fig. 4). Sequence errors are considerably reduced compared to polymerase-based systems because the ligation method is based on probe recognition rather than sequential nucleotide addition. In this system, there are 16 dinucleotide combinations with 4 fluorescent dyes, each dye corresponding to a probe pool of 4 dinucleotides per pool (fig. 4). Using this dinucleotide, 4-dye encoding scheme in conjunction with a sequencing assay that samples every base, each base is effectively probed by 2 different and independent reactions. The double interrogation of each base causes a true SNP polymorphism to result in a 2 consecutive colour change while a measurement error results in a single colour change. The SOLiD™ (Sequencing by Oligo Ligation and Detection) platform is capable now of producing 400 million 50-bp reads (combined total in 2 independent flow cells). This approach essentially eliminates the possibility of spurious insertions or deletions.
Fig. 4.
Schematic representation of SOLiD™ sequencing by ligation. Primers hybridise to the P1 adapter within the library template. A set of 4 fluorescence-labelled di-base probes competes for ligation to the sequencing primer. These probes have partly degenerated DNA sequence (indicated by n) and for simplicity only one probe is shown (labelling is denoted by asterisk). The specificity of the di-base probe is achieved by interrogating every 1st and 2nd base in each ligation reaction. Multiple cycles of ligation, detection and cleavage are performed with the number of cycles determining the eventual read length (modified from http://www3.appliedbiosystems.com).
Recently, the SOLiD system has been used to assay nucleotide variation in HapMap samples, in the framework of the 1000 Genomes Project [McKernan et al., 2009]. The 1000 Genomes Project is a collaboration among research groups in the US, UK, China, and Germany to produce an extensive catalogue of human genetic variation that will support future medical research studies (www.1000genomes.org). It will extend the data from the International HapMap Project and will allow genome-wide association studies to focus on almost all variants that exist in regions found to be associated with disease. The genomes of over 1,000 unidentified individuals from around the world will be sequenced using next generation sequencing technologies. Three HapMap samples were sequenced via the ligation-based approach utilised in the SOLiD sequencing system [McKernan et al., 2009]. In one sample with a sequence coverage estimated at 29×, 97% of heterozygotes SNPs and 99.96% of homozygotes SNPs were detected and false discovery rate was negligible [McKernan et al., 2009].
This approach is particularly useful for variation discovery in genetic association studies, where low sequence coverage (5×) and therefore lower cost, may detect a significant portion of the total sequence variation. To detect pathogenic mutations associated with disease a higher coverage is needed. What is the required accuracy for mutation discovery? A single base pair change may result in a DSD phenotype and any error in sequencing can give rise to false positives or false negatives, leading to time-consuming and challenging downstream studies. The raw sequencing accuracy is the accuracy of a single-pass sequencing read. This is an excellent measurement of each method's chemistry, fluorescence readout, process, and base-calling software (table 1). Ultimately, the more robust the raw reads, the fewer redundant reads are required, keeping the overall costs down. At the time of writing, the SOLiD system shows a high degree of sequence accuracy, even at low coverage, and this may be the system of choice for mutation detection.
Overall substantial cost reductions are associated with NGS technologies when compared with the Sanger method, but sequencing the whole human genome remains expensive for most laboratories. In the search for novel genetic markers of DSD, the medium term may see a more targeted approach. Specific regions of the genome may be enriched for sequencing: for example, sequencing all of the exons in the genome; sequencing specific gene families of interest; or focusing just on large chromosome regions or entire chromosomes that are implicated in disease. Exome capture approaches are available for targeting approximately 33 Mb or ∼180,000 coding exons across the human genome. Many platforms also offer capture of miRNAs and non-coding RNA sequences. Selective genomic enrichment of the human exome offers an attractive option for devising new experimental designs aimed at quick identification of potential disease-associated genetic variants. Human exome capture methods are currently based on either custom-designed oligonucleotide microarrays or solution-based hybridisation strategies. Roche/Nimblegen offers microarray capture platforms for solid phase hybridisation for the enrichment of exons [Albert et al., 2007]. The reported capture efficiency for these platforms ranges from 50–90% [Hodges et al., 2007]. Solution-based hybridisation methods for exon capture, such as molecular inversion probes (mIPs) [Porreca et al., 2007; Turner et al., 2009] and biotinylated RNA capture sequences [Gnirke et al., 2009; SureSelect Human All Exon Kit; Agilent Technologies] have also been developed that have capture efficiencies of 70–90%. Raindance technologies (www.raindancetechnologies.com) offers targeted sequence enrichment using a microfluidic device to create aqueous picolitre-volume droplets of forward- and reverse-targeting primers in an oil solution [Tewhey et al., 2009]. In a microfluidic chip, the primer pair droplets and template droplets, which contain fragmented genomic DNA and PCR reagents, are paired together in a 1:1 ratio. An electric field induces the 2 droplets to merge into a single PCR droplet. Around 1.5 million PCR droplets can be collected in a single PCR tube. This approach has been reported to have an 84% capture efficiency with 90% of the targeted bases showing uniform coverage when sequenced with either the Roche/454 or Illumina/Solexa platform [Tewhey et al., 2009]. From a medical perspective, this approach is interesting since it is particularly suitable for working with cells of limited availability, such as stem cells or primary cells from patients [Brouzes et al., 2009].
NGS with exome capture has recently been applied to identify mutations associated with mendelian disorders [Ng et al., 2009, 2010]. Using Freeman-Sheldon syndrome (FSS) as a proof-of-concept, Ng and colleagues performed exome enrichment and sequencing using the Illumina system to identify the causal mutation for FSS. The same group has recently identified mutations in the DHODH gene in 4 individuals from 3 pedigrees with Miller syndrome (postaxial acrofacial dysostosis) using exome sequence capture and sequencing to a coverage of 40× with sufficient depth to call variants at approximately 97% of each targeted exome.
In addition, the recent NGS applications not only have the ability to generate huge amounts of sequence data but they also reveal structural variation, thereby bypassing the need for CGH analyses [McKernan et al., 2009].
These recent studies show the power of NGS approaches to detect pathogenic mutations causing disease. This approach also offers the opportunity to identify genetic modifiers. Familial cases of both 46,XY and 46,XX DSD often show considerable variation in the expression of the phenotype, including families where the underlying genetic mutation has been identified [Lourenco et al., 2009; Temel et al., 2007]. This phenotypic variability may be explained by variations in other genes that may interact with or influence the activity of the target gene. This has been shown in a pedigree with hypogonadotropic hypogonadism, where a compound heterozygous GNRHR and a heterozygous FGFR1 mutation were identified [Pitteloud et al., 2007].
When considering these NGS approaches that are currently costly and require considerable bioinformatics analyses, an important question to pose is what would be the cost benefits to the DSD field? The usefulness of this non-a priori approach to identify genetic modifiers has been mentioned above, but are there new genetic factors that when mutated could cause DSD? The reply to this question is yes. For example, almost all cases of non-syndromic ovotesticular DSD remain unexplained although there have been considerable genetic analyses of candidate genes [McElreavey et al., 1992a; Temel et al., 2007]. Despite advances in understanding the mechanisms of mammalian gonad formation, the underlying genetic cause of most cases of 46,XY gonadal dysgenesis remains unknown [McElreavey and Fellous, 1999]. Mutations in the androgen receptor (AR) gene are the most common genetic cause of 46,XY DSD. However, even in 46,XY underandrogenised subjects with testes who are suspected to have an AR defect, a pathogenic mutation is found only in less than half of the cases [Audi et al., 2010]. In cases of simple hypospadias or cryptorchidism a genetic cause is rarely detected despite epidemiological evidence suggesting a major genetic contribution to these phenotypes [Fukami et al., 2006; Schnack et al., 2008; Köhler et al., 2009]. Recently, a mutation in Map3k4 was associated with XY gonadal sex reversal in the mouse, suggesting that mutations in mitogen-activated protein kinase (MAPK) signalling pathway may be associated with DSD [McElreavey et al., unpublished data; Bogani et al., 2009].
It is clear that NGS will have a tremendous impact on medicine [Voelkerding et al., 2009]. Exciting developments are rapidly taking place in the field and these are leading to a deeper understanding of variation across individual genomes, genetic variation in tissues within an individual and the identification of disease causing mutations. Sequencing technology is continuing to evolve at an unprecedented pace. The generation of massive data sets of qualitative and quantitative information of both RNA and DNA sequences in a patient sample at a relatively limited cost will transform the field of reproductive disorders by offering novel insights into the genetics and physiology of DSD.
Acknowledgements
Supported by grants from the Agence Nationale de la Recherche-GIS Institut des Maladies Rares (to Dr. McElreavey); by a research grant (1-FY07-490) from the March of Dimes Foundation (to Dr. McElreavey); by a research grant from the EuroDSD in the European Community's Seventh Framework Programme FP7/2007-2013 under grant agreement No. 201444 (Drs Achermann, Wieacker, Ledig, McElreavey and Bashamboo); a Wellcome Trust Senior Research Fellowship in Clinical Science (079666, to Dr Achermann).
References
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- Aleck KA, Argueso L, Stone J, Hackel JG, Erickson RP. True hermaphroditism with partial duplication of chromosome 22 and without SRY. Am J Med Genet. 1999;85:2–4. doi: 10.1002/(sici)1096-8628(19990702)85:1<2::aid-ajmg2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Andrieux J, Dubourg C, Rio M, Attie-Bitach T, Delaby E, et al. Genotype-phenotype correlation in four 15q24 deleted patients identified by array-CGH. Am J Med Genet A. 2009;149A:2813–2819. doi: 10.1002/ajmg.a.33097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansorge WJ. Next-generation DNA sequencing techniques. N Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Audi L, Fernández-Cancio M, Carrascosa A, Andaluz P, Torán N, et al. Novel (60%) and recurrent (40%) androgen receptor gene mutations in a series of 59 patients with a 46,XY disorder of sex development. J Clin Endocrinol Metab. 2010;95:1876–1888. doi: 10.1210/jc.2009-2146. [DOI] [PubMed] [Google Scholar]
- Barbaro M, Oscarson M, Schoumans J, Staaf J, Ivarsson SA, Wedell A. Isolated 46,XY gonadal dysgenesis in two sisters caused by a Xp21.2 interstitial duplication containing the DAX1 gene. J Clin Endocrinol Metab. 2007;92:3305–3313. doi: 10.1210/jc.2007-0505. [DOI] [PubMed] [Google Scholar]
- Barbaro M, Balsamo A, Anderlid BM, Myhre AG, Gennari M, et al. Characterization of deletions at 9p affecting the candidate regions for sex reversal and deletion 9p syndrome by MLPA. Eur J Hum Genet. 2009;17:1439–1447. doi: 10.1038/ejhg.2009.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardoni B, Zanaria E, Guioli S, Floridia G, Worley KC, et al. A dosage sensitive locus at chromosome Xp21 is involved in male to female sex reversal. Nat Genet. 1994;7:497–501. doi: 10.1038/ng0894-497. [DOI] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Huang J, Greshock J, Watt S, Butler A, et al. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res. 2004;14:287–295. doi: 10.1101/gr.2012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogani D, Siggers P, Brixey R, Warr N, Beddow S, et al. Loss of mitogen-activated protein kinase kinase kinase 4 (MAP3K4) reveals a requirement for MAPK signalling in mouse sex determination. PLoS Biol. 2009;7:e1000196. doi: 10.1371/journal.pbio.1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, et al. Droplet microfluidic technology for single-cell high-throughput screening. Proc Natl Acad Sci USA. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kovel CG, Trucks H, Helbig I, Mefford HC, Baker C, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133:23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson RP, Skinner S, Jacquet H, Campion D, Buckley PG, et al. Does chromosome 22 have anything to do with sex determination: further studies on a 46,XX,22q11.2 del male. Am J Med Genet A. 2003;123A:64–67. doi: 10.1002/ajmg.a.20489. [DOI] [PubMed] [Google Scholar]
- Fedurco M, Romieu A, Williams S, Lawrence I, Turcatti G. BTA, a novel reagent for DNA attachment on glass and efficient generation of solid-phase amplified glass and efficient generation of solid-phase amplified DNA colonies. Nucleic Acids Res. 2006;34:e22. doi: 10.1093/nar/gnj023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami M, Wada Y, Miyabayashi K, Nishino I, Hasegawa T, et al. CXorf6 is a causative gene for hypospadias. Nat Genet. 2006;38:1369–1371. doi: 10.1038/ng1900. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Hakonarson H. Common variants in polygenic schizophrenia. Genome Biol. 2009;10:236. doi: 10.1186/gb-2009-10-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, et al. Solution hybrid selection with ultralong oligonucleotides for massively parallel targeted sequencing. Nature Biotech. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- Hartmann A, Thieme M, Nanduri LK, Stempfl T, Moehle C, et al. Validation of microarray-based resequencing of 93 worldwide mitochondrial genomes. Hum Mutat. 2009;30:115–122. doi: 10.1002/humu.20816. [DOI] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, et al. Genome-wide in situ exon capture for selective resequencing. Nature Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- Huang B, Wang S, Ning Y, Lamb AN, Bartley J. Autosomal XX sex reversal caused by duplication of SOX9. Am J Med Genet. 1999;87:349–353. doi: 10.1002/(sici)1096-8628(19991203)87:4<349::aid-ajmg13>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Hughes IA, Houk C, Ahmed SF, Lee PA, LWPES Consensus Group, ESPE Consensus Group Consensus statement on management of intersex disorders. Arch Dis Child. 2006;91:554–563. doi: 10.1136/adc.2006.098319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Köhler B, Lin L, Mazen I, Cetindag C, Biebermann H, et al. The spectrum of phenotypes associated with mutations in steroidogenic factor 1 (SF-1, NR5A1, Ad4BP) includes severe penoscrotal hypospadias in 46,XY males without adrenal insufficiency. Eur J Endocrinol. 2009;161:237–242. doi: 10.1530/EJE-09-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamon JH. A massively parallel PicoTiterPlate based platform for discrete picoliter-scale polymerase chain reactions. Electrophoresis. 2003;24:3769–3777. doi: 10.1002/elps.200305646. [DOI] [PubMed] [Google Scholar]
- Le Caignec C, Delnatte C, Vermeesch JR, Boceno M, Joubert M, et al. Complete sex reversal in a WAGR syndrome patient. Am J Med Genet A. 2007;143A:2692–2695. doi: 10.1002/ajmg.a.31997. [DOI] [PubMed] [Google Scholar]
- Lecointre C, Pichon O, Hamel A, Heloury Y, Michel-Calemard L, et al. Familial acampomelic form of campomelic dysplasia caused by a 960 kb deletion upstream of SOX9. Am J Med Genet A. 2009;149A:1183–1189. doi: 10.1002/ajmg.a.32830. [DOI] [PubMed] [Google Scholar]
- Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;1(21 Suppl):20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- Lourenço D, Brauner R, Lin L, De Perdigo A, Weryha G, et al. Mutations in NR5A1 associated with ovarian insufficiency. N Engl J Med. 2009;360:1200–1210. doi: 10.1056/NEJMoa0806228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucito R, Healy J, Alexander J, Reiner A, Esposito D, et al. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal MN, Heckenlively JR, Burch T, Chen L, Vasireddy V, et al. Sequencing arrays for screening multiple genes associated with early-onset human retinal degenerations on a high-throughput platform. Invest Ophthalmol Vis Sci. 2005;46:3355–3362. doi: 10.1167/iovs.05-0007. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElreavey K, Rappaport R, Vilain E, Abbas N, Richaud F, et al. A minority of 46,XX true hermaphrodites are positive for the Y-DNA sequence including SRY. Hum Genet. 1992a;90:121–125. doi: 10.1007/BF00210754. [DOI] [PubMed] [Google Scholar]
- McElreavey K, Vilain E, Abbas N, Costa JM, Souleyreau N, et al. XY sex reversal associated with a deletion 5′ to the SRY ‘HMG box’ in the testis-determining region. Proc Natl Acad Sci USA. 1992b;89:11016–11020. doi: 10.1073/pnas.89.22.11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElreavey K, Fellous M. Sex determination and the Y chromosome. Am J Med Genet. 1999;89:176–185. doi: 10.1002/(sici)1096-8628(19991229)89:4<176::aid-ajmg2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson JD. Next-generation gap. Nat Methods. 2009;6(suppl 11):S2–S5. doi: 10.1038/nmeth.f.268. [DOI] [PubMed] [Google Scholar]
- Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat Methods. 2009;6(suppl 11):S13–S20. doi: 10.1038/nmeth.1374. [DOI] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies – the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata T, Muroya K, Sasagawa I, Kosho T, Wakui K, et al. Genetic evidence for a novel gene(s) involved in urogenital development on 10q26. Kidney Int. 2000;58:2281–2290. doi: 10.1046/j.1523-1755.2000.00412.x. [DOI] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, et al. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007;117:457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porreca GJ, Zhang K, Li JB, Xie B, Austin D, et al. Multiplex amplification of large sets of human exons. Nature Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- Porter FD. Smith-Lemli-Opitz syndrome: pathogenesis, diagnosis and management. Eur J Hum Genet. 2008;16:535–541. doi: 10.1038/ejhg.2008.10. [DOI] [PubMed] [Google Scholar]
- Refai O, Friedman A, Terry L, Jewett T, Pearlman A, et al. De novo 12;17 translocation upstream of SOX9 resulting in 46,XX testicular disorder of sex development. Am J Med Genet A. 2010;152A:422–426. doi: 10.1002/ajmg.a.33201. [DOI] [PubMed] [Google Scholar]
- Ronaghi M, Karamohamed S, Pettersson B, Uhlén M, Nyrén P. Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- Schlaubitz S, Yatsenko SA, Smith LD, Keller KL, Vissers LE, et al. Ovotestes and XY sex reversal in a female with an interstitial 9q33.3–q34.1 deletion encompassing NR5A1 and LMX1B causing features of Genitopatellar syndrome. Am J Med Genet A. 2007;143A:1071–1081. doi: 10.1002/ajmg.a.31685. [DOI] [PubMed] [Google Scholar]
- Schlessinger D, Garcia-Ortiz JE, Forabosco A, Uda M, Crisponi L, Pelosi E. Determination and stability of gonadal sex. J Androl. 2010;31:16–25. doi: 10.2164/jandrol.109.008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnack TH, Zdravkovic S, Myrup C, Westergaard T, Christensen K, et al. Familial aggregation of hypospadias: a cohort study. Am J Epidemiol. 2008;167:251–256. doi: 10.1093/aje/kwm317. [DOI] [PubMed] [Google Scholar]
- Seeherunvong T, Perera EM, Bao Y, Benke PJ, Benigno A, et al. 46,XX sex reversal with partial duplication of chromosome arm 22q. Am J Med Genet A. 2004;127A:149–151. doi: 10.1002/ajmg.a.20630. [DOI] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–934. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R. Sex determination and SRY: down to a wink and a nudge? Trends Genet. 2009;25:19–29. doi: 10.1016/j.tig.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Selzer RR, Veltman JA, Gimelli S, Gimelli G, et al. Characterization of a recurrent 15q24 microdeletion syndrome. Hum Mol Genet. 2007;16:567–752. doi: 10.1093/hmg/ddm016. [DOI] [PubMed] [Google Scholar]
- Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- Smyk M, Berg JS, Pursley A, Curtis FK, Fernandez BA, et al. Male-to-female sex reversal associated with an approximately 250 kb deletion upstream of NR0B1 (DAX1) Hum Genet. 2007;122:63–70. doi: 10.1007/s00439-007-0373-8. [DOI] [PubMed] [Google Scholar]
- Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, et al. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. [PubMed] [Google Scholar]
- Sultan C, Biason-Lauber A, Philibert P. Mayer-Rokitansky-Kuster-Hauser syndrome: recent clinical and genetic findings. Gynecol Endocrinol. 2009;25:8–11. doi: 10.1080/09513590802288291. [DOI] [PubMed] [Google Scholar]
- Szoke D, Molnar B, Solymosi N, Racz K, Gergics P, et al. Polymorphisms of the ApoE, HSD3B1, IL-1beta and p53 genes are associated with the development of early uremic complications in diabetic patients: results of a DNA resequencing array study. Int J Mol Med. 2009;23:217–227. [PubMed] [Google Scholar]
- Temel SG, Gulten T, Yakut T, Saglam H, Kilic N, et al. Extended pedigree with multiple cases of XX sex reversal in the absence of SRY and of a mutation at the SOX9 locus. Sex Dev. 2007;1:24–34. doi: 10.1159/000096236. [DOI] [PubMed] [Google Scholar]
- Tewhey R, Warner JB, Nakano M, Libby B, Medkova M, et al. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat Biotechnol. 2009;27:1025–1031. doi: 10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner EH, Lee C, Ng SB, Nickerson DA, Shendure J. Massively parallel exon capture and library-free resequencing across 16 genomes. Nature Methods. 2009;6:315–316. doi: 10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenhaut NH, Jakob S, Anlag K, Eisenberger T, Sekido R, et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell. 2009;139:1130–1142. doi: 10.1016/j.cell.2009.11.021. [DOI] [PubMed] [Google Scholar]
- Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Silfhout A, Boot AM, Dijkhuizen T, Hoek A, Nijman R, et al. A unique 970 kb microdeletion in 9q33.3, including the NR5A1 gene in a 46,XY female. Eur J Med Genet. 2009;52:157–160. doi: 10.1016/j.ejmg.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Vinci G, Chantot-Bastaraud S, El Houate B, Lortat-Jacob S, Brauner R, et al. Association of deletion of 9p, 46,XY gonadal dysgenesis and autistic spectrum disorder. Mol Hum Reprod. 2007;13:685–689. doi: 10.1093/molehr/gam045. [DOI] [PubMed] [Google Scholar]
- Voelkerding KV, Dames SA, Durtschi JD. Next-generation sequencing: from basic research to diagnostics. Clin Chem. 2009;55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci USA. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieacker P, Volleth M. WNT4 and RSPO1 are not involved in a case of male-to-female sex reversal with partial duplication of 1p. Sex Dev. 2007;1:111–113. doi: 10.1159/000100032. [DOI] [PubMed] [Google Scholar]
- Wilhelm D, Palmer S, Koopman P. Sex determination and gonadal development in mammals. Physiol Rev. 2007;87:1–28. doi: 10.1152/physrev.00009.2006. [DOI] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]