Summary
The oceans are the Earth's largest ecosystem, covering 70% of our planet and providing goods and services for the majority of the world's population. Understanding the complex abiotic and biotic processes on the micro‐ to macroscale is the key to protect and sustain the marine ecosystem. Marine microorganisms are the ‘gatekeepers’ of the biotic processes that control the global cycles of energy and organic matter. A multinational, multidisciplinary approach, bringing together research on oceanography, biodiversity and genomics, is now needed to understand and finally predict the complex responses of the marine ecosystem to ongoing global changes. Such an integrative approach will not only bring better understanding of the complex interplay of the organisms with their environment, but will reveal a wealth of new metabolic processes and functions, which have a high potential for biotechnological applications. This potential has already been recognized by the European commission which funded a series of workshops and projects on marine genomics in the sixth and seventh framework programme. Nevertheless, there remain many obstacles to achieving the goal – such as a lack of bioinformatics tailored for the marine field, consistent data acquisition and exchange, as well as continuous monitoring programmes and a lack of relevant marine bacterial models. Marine ecosystems research is complex and challenging, but it also harbours the opportunity to cross the borders between disciplines and countries to finally create a rewarding marine research era that is more than the sum of its parts.
Introduction
The planet Earth is a blue planet – with 70% of its surface covered by oceans and 40% of the World's population living within 50 km of a coastline, our heritage, economy and well‐being are inextricably linked with the marine environment (Bowler et al., 2009). Marine microbes are the ‘gatekeepers’ for the Earth System with an estimated contribution to global primary productivity of between 50% and 90% (Falkowski et al., 1998). Recent investigations have shown that beside the classic carbon and energy flow through photosynthetic Eukarya to herbivores and on to higher trophic levels, a rather effective microbial food web exists (DeLong and Karl, 2005). Autotrophic bacteria fix carbon and – in case of Cyanobacteria– release oxygen; heterotrophic bacteria use energy stored in the non‐living, detrital carbon pool, recycling nitrogen, sulfur and phosphorus and other elements. Cell‐associated ectoenzymes enable bacteria to use high‐molecular‐weight organic carbon in addition to the more traditional low‐molecular‐weight and gaseous carbon substances. Additionally, there is ample evidence that marine heterotrophic bacteria can use light as an additional energy source to survive under conditions of nutrient starvation (Beja et al., 2001; Fuchs et al., 2007; Moran and Miller, 2007) and that viruses are potential mediators of photosynthetic genes (Sharon et al., 2007; 2009).
Although about half of the annual primary production of the planet occurs in the ocean and bacterial metabolism is involved in the chemical transformation of most elements (Falkowski et al., 2008), very little is known of marine microbial diversity, how many species are present in the oceans, and what each individual species does – i.e. its ecological function and interactions. The vast majority (90–99%) of these organisms cannot be cultured under standard laboratory conditions and so are not amenable to study by the methods that have proved so successful with medically important microorganisms throughout the 20th century. It was only with the development of a molecular toolbox to sequence DNA from the natural environment that information about the exceptional prokaryotic diversity in the oceans began to accumulate. An example of these molecular tools is the powerful PCR‐based methods that have been established for direct amplification, cloning and analysis of ribosomal RNA (rRNA) genes from the environment. Beyond diversity, the design and application of specific rRNA‐targeted oligonucleotide probes allows insights into the structure of microbial communities in situ (Stahl and Amann, 1991; Glöckner et al., 1999). The impact of cultivation‐independent methods for biodiversity analysis becomes evident by noting that as of January 2010, around 1.4 million rRNA sequences are available in online databases like SILVA (Pruesse et al., 2007), with the vast majority originating from so far uncultured bacteria and, of which, 11% are of marine origin.
However, little is known about the function of these microbes in the ecosystem or of their metabolic activity, because no function can be assigned to a large proportion of their genes (DeLong, 2005). Although our current knowledge on marine bacteria is poor, and might appear a limitation for biotechnological exploitation, we believe that there are excellent opportunities for bioprospecting for novel enzymes of industrial interest and for metabolic products. The marine environment is very diversified, ranging from cold to hot, and with hypersaline and high‐pressure habitats. Organisms that have evolved to occupy this wide range of ecological niches must have diverse metabolisms and will therefore possess novel enzyme capability. That is, unusual habitats have unusual organisms with unusual metabolism (Bornscheuer, 2005). Many of the processes that these microbes can carry out are likely to be of interest to industry. The challenge is to access the organisms, by bringing them into laboratory culture, or to access the genetic information, through genomics and metagenomics.
Marine microbial genomics
Marine ecosystems research has benefited greatly from the use of genomic approaches. DNA‐based sequencing, originally developed in the biomedical field, has been quickly incorporated in the marine sciences. The introduction of automated sequencing technologies has already led to a massive increase of sequence data. It should be noted that the amount of sequence data in the public data repositories has doubled every 18 months, and it is expected that this will significantly increase with the routine application of the next generation of sequencing technologies (Gupta, 2008).
Genomics can be defined as the study of the genetic complement of a single organism. Metagenomics refers to all of the genetic information of a natural assemblage – i.e. equivalent to the genomes of all of the organisms in the sample (Handelsman, 2004). This cultivation‐independent approach has resulted in an explosion of information on marine microbes. For example, the first part of the Global Ocean Sampling (GOS), which sampled the North Atlantic, Caribbean and a small part of the Pacific Ocean, added DNA sequence information that was equivalent to 50% of all protein‐encoding sequences that had previously been deposited in GenBank (Venter et al., 2004; Rusch et al., 2007). GOS confirmed that marine microbes are diverse, revealing how little is known about the genetic information of natural assemblages. But this study also highlighted the difficulties of making sense of metagenomic sequence data. A significant proportion of the open reading frames (ORFs, which are presumed to equate to genes) could not be characterized because there were no similar sequences in the databases (Yooseph et al., 2007).
This difficulty of interpreting the GOS sequence data is despite the number of marine microbes whose whole genome sequences are already in the databases. Largely as a consequence of funding from the Gordon and Betty Moore Foundation, 150 marine bacterial genomes have so far been released (Moore, 2009). So, although marine bacteria are well represented in the genomic databases, this information was still not sufficient to decipher the metagenomic information coming from the GOS.
The environmental context rules
The results demonstrate that even billions of nucleotides do not provide much information when it comes to understanding the function of organisms. This is a prerequisite for inferring an organism's ecology, as well as for discovering new catalytic mechanisms and enzymes. In every new marine genome or metagenome sequenced, approximately 40% of the potential protein‐encoding genes still lack any functional assignment. To be able to address the potential functions of these novel genes, a holistic approach is needed, based on contextual data (metadata) such as habitat parameters, the diversity and abundances of organisms, genomic information, as well as gene expression information on the transcriptome and proteome level (Doney et al., 2004; DeLong and Karl, 2005; Tringe and Rubin, 2005). To integrate these data, georeferencing has been shown to be extremely useful, especially for the open ocean, where any kind of genomic data can be easily linked with measured and remote sensing parameters based on location, time and depth (Field et al., 2008a; Kottmann et al., 2010).
Other important prerequisites to facilitate interoperability and exchange of data are standards for data formats, as well as quality management to provide confidence in prediction and interpolation. In 2005 the Genomic Standards Consortium, an open‐membership, international working body, was formed to provide mechanism that standardize the description of genomes and the exchange and integration of genomic data. Based on a collaborative effort, the first MIGS (Minimum Information about a Genome Sequence) and MIMS (Minimum Information about a Metagenome Sequence) checklists were published (Field et al., 2008b). Through specification of geographic location and sampling date, MIGS/MIMS intend to provide a better understanding of the origin of each genome and place genomes and metagenomes in their geospatial and temporal contexts. With the ongoing efforts of the Genomic Standards Consortium, an integrated view of the data will be possible (Green et al., 2008), this being a prerequisite to screen and filter the flood of data and to target sets of genes that will be primary targets for in‐depth functional analysis in the laboratory.
From marine ecological genomics to blue biotechnology
In terms of the exploitation of molecules of high value, the exploration of the potential of marine biodiversity is still at an early stage – but the list of interesting marine biotech products is growing steadily. The inventory of marine high‐value‐added compounds, including a range of proteins, carbohydrates and lipids, stands at some 20 000 products (Blunt et al., 2007). Most of the marine compounds that have been successfully screened and structurally elucidated so far originate from isolated and culturable microorganisms, especially bacteria (Schweder et al., 2005). For example the presence of glycoside hydrolases in marine bacteria, which are specific for the degradation of sulfated polysaccharides, represents a unique source of enzymes for the production of sulfated (or unsulfated) oligosaccharides of algal origin. These compounds have proven to be of biotechnological importance because they are capable of stimulating defence reactions in plants. For instance, with the introduction of algal laminarin (Klarzynski et al., 2000), Goëmar Laboratories (France) have brought out Iodus 40, the first natural crop protection mechanism against winter wheat diseases. Moreover, simple bioengineering of these enzymes can potentially make them useful for the production of novel sugar compounds. An example of this is provided by the marine Planctomycete, Rhodopirellula baltica (Glöckner et al., 2003). This organism is able to desulfate complex materials and compounds and therefore is of great interest to the chemical industry (Wallner et al., 2005; Gadler and Faber, 2007). These enzymes are a viable alternative to chemical processes that often require arduous and environmentally unfriendly reaction conditions and produce toxic waste products (Uzawa et al., 2003). As other examples, the complete genome of the marine polysaccharide‐degrading Flavobacteria Zobellia galactanivorans reveals more than 100 carbohydrate‐active enzymes, many of which have undefined substrate specificities that await discovery (Michel et al., 2006). Thiocoraline, an antitumour drug from marine Actinomycetes, is under development by PharmaMar S.A. (Spain), and sulfated polysaccharides can act as antiviral drugs (Witvrouw et al., 1994). Metagenomic analysis has also led to the discovery of a crucial alternative to the nitrogen oxidation pathway (the anammox cycle) that opened up a whole new research field in water treatment (Strous et al., 1999). Also, an increased understanding of hydrocarbon‐degrading bacteria opens up perspectives for efficient oil degradation (Schneiker et al., 2006).
Status of marine research and biotechnology in Europe
Over the years, the European Commission has launched as set of programmes embedded in the framework programmes to support and stimulate research on marine diversity, and genomics in the environmental context. The two Networks of Excellence (NoEs), Marine Biodiversity and Ecosystems Functioning (MarBef) and Marine Genomics Europe (MGE), as well as the integrated research project HERMES (Hotspot Ecosystems Research on the Margins of European Seas) were able to bring together more than 150 institutions, over 1000 researchers in over 30 countries (within and outside Europe). Along the same lines the European Science Foundation (ESF) initiated the EUROCORES (European Collaborative Research) Scheme to promote collaborative research, networking and dissemination while targeting broad and complex topics of research across all scientific domains at the European level and in a global context. EuroDEEP has emerged as part of EUROCORES to describe, explain and predict variations of biodiversity within and between deep‐sea habitats, with respect to deep‐sea ecosystem functioning and the global biosphere.
All projects have contributed significantly to realizing the vision of a European Research Area (ERA) in the marine sector. This aims: (i) to improve dynamism and innovation into all sectors of industry and services, resulting in more and better jobs, (ii) to address important issues of European and even global dimension, such as health, energy supply and climate change, and (iii) to create a society in which knowledge is shared, taught and valued as an essential source of personal and collective development.
As pointed out in the Galway Declaration (Eurocean, 2004) marine science and technology plays an essential role in generating the knowledge needed to fuel the economy and welfare of humans on Earth. The Maritime Green paper (Greenpaper, 2006) highlights that marine research, and in particular marine biotechnological research, is a driver of economic activity. A reason for this is that it generates new knowledge in many scientific and technological disciplines. Therefore, the impact of the outputs extends much further than the marine field: it stimulates activity in areas such as health, food, energy, pharmaceuticals, environment and transport amongst others.
In the Bremen meeting on Marine Biotechnology Research (EC, 2007) experts proposed five action points that needed further support by the Commission: (i) to raise the awareness and visibility of marine biotechnology, (ii) to continue to support excellence in basic sciences, (iii) to provide access to and improve integrated research infrastructure, (iv) to streamline the management of intellectual property and (v) to establish cross‐cutting programmes to support commercialization.
In September 2008 the EC delivered a European Strategy for Marine and Maritime Research where marine biodiversity and biotechnology research was again prioritized and its potential to contribute new knowledge on which to base high‐value‐added products and processes through excellent scientific research was highly recognized (EC, 2008).
Recently the EC‐US Task Force on Biotechnology Research workshop in Monaco (EC‐US, 2008) emphasized that microbes play an important role in the marine and indeed global ecosystem and it is suspected that they are responsible for much biological activity. Great advances in ‘omics’ technologies to uncover new information on the bioactivity potential in cultivated and uncultivated marine microbes have been made with vast implications for basic scientific research as well as biotechnology research. While highlighting scientific research opportunities, the workshop also stressed the need for research to strengthen Europe's bioinformatics research capability.
The EC has taken action by funding the ASSEMBLE (Assemble, 2009) research infrastructure initiative and the EUROFLEETS (EuroFleets, 2009) infrastructure plus the integrated project MAMBA [Marine Metagenomics for new Biotechnological Applications (MAMBA, 2009)] in the seventh framework programme. EUROFLEETS and ASSEMBLE will provide the ‘hardware’ for marine research by offering transnational access to ships as well as marine ecosystems and facilities. ASSEMBLE will conduct research on ecological models, while the MAMBA project specifically sets out to explore the metagenomic approach for the discovery of new enzymes from the marine realm for use in biotechnology. Furthermore the new infrastructures EMBRC (European Marine Biological Resource Center) and ELIXIR [European Life‐Science Infrastructure for Biological Information (ELIXIR, 2009)] are on the horizon. They will provide access to marine model genomes and their integration with modern ‘omics’ technologies, as well as stable funding for Europe's most important publicly accessible databases of molecular biological information, combined with the development of a compute infrastructure that can cope with the biological data deluge.
In summary the marine environment and the services it provides have gained significantly more attention in Europe over the last years (Volckaert et al., 2008). Nevertheless, much work needs to be done to defragment the European Research Area based in the marine sector.
Obstacles
Bioinformatics
Although the giga‐base amounts of microbial DNA sequences and other high‐throughput approaches have made fundamental improvements to microbial biotechnology and the understanding of uncultivated marine microbes, bioinformatics is often the limiting factor in all kinds of ‘omics’ studies (Anonymous, 2009a). The major hurdles are: (i) the computational aspects of data archiving, analysis and visualization of thousands of millions of DNA sequences which are released to databases, (ii) the integration and interpretation of molecular data in their environmental context, and (iii) linking environmental studies with laboratory experiments so that hypotheses can be tested and unknown genes can be assigned a function. Novel computational techniques are required that allow numerical descriptions of the specific biological functions unique to specific niches and acting against particular elements.
Furthermore, knowledge databases, with quality management and expert curating in order to provide well‐documented and reliable reference datasets for academia and industry, either are not available, or lack funding for further development (Howe et al., 2008). User‐friendly visualization tools need to be developed to display fragment recruitment, genomic context, functional annotation, scaffold characteristics for binning, metabolic pathway overlays and sample comparisons. These tools will be crucial if biologists are to utilize the large and rapidly expanding datasets that potentially hold the key to understanding microbial function in the oceans.
In the USA a variety of annotation pipelines have been developed for data analysis. These include the CAMERA System (Seshadri et al., 2007) to meet the challenge of studying marine life and ecosystems for examining the genomic complexities of natural communities of microorganisms as they have evolved in their local environments. Examples for multipurpose annotation pipelines are IMG/M (Markowitz et al., 2008) and MG‐RAST (Aziz et al., 2008). Europe is lagging behind in this respect and a European Bioinformatics Infrastructure specific for the marine realm should be envisioned to generate further knowledge.
Data acquisition, exchange and monitoring
Another obstacle that needs to be overcome is the lack of awareness among biologists about consistent data acquisition and storage. Contextual data of sampling campaigns are still either incomplete or stored in hand‐written laboratory notebooks or on individual computers. Unfortunately, what is finally published represents only a small subset of the original data and is often only ‘human readable’ and unable to be assimilated into databases without major efforts in text recognition. These practices greatly hamper any kind of Europe‐wide electronic data exchange and integration. Although approaches exist to develop standardized exchange languages specific for genomics based on XML (Kottmann et al., 2008), and the new SIGS journal requests that a specified set of contextual data must be provided in an electronic format (Garrity et al., 2008), a change of mindset towards an open policy of sharing data is still needed (Anonymous, 2009b).
To fulfil the promise of a global understanding for the diversity and function of marine organisms, a much denser network of data is urgently needed. Accurate real‐time monitoring at all levels from oceanographic data to marine nutrient cycling and biomass production would be desirable (Deutsch et al., 2007; Bowler et al., 2009). One way to reach such a goal is the development of ocean observatories – permanent off‐shore stations and profiling buoys – for which biological and genetic systems (Jones et al., 2008) are being developed and backed up by remote sensing (Delaney, 2007). In the USA a primer has been recently launched with the Ocean Observatories Initiative (OOI) (Ramons, 2009) which is complemented in Europe by the EMODNET vision (Emodnet, 2009).
The increasing automation of data acquisition and genomics technologies will significantly impact our capacities to better understand the functioning of the marine ecosystem.
Lack of relevant marine bacterial models
The wealth of sequence data from both marine microbes and diverse oceanic provinces presents considerable challenges. There are huge numbers of putative genes, the function of which is often unknown and at best only deduced from sequence comparisons. Because more is often known about the genetics and physiology of terrestrial organisms, the number of unknown/putative genes is overwhelming in marine samples, because there are so few experimental data on marine model organisms. For example, all phyla of marine algae synthesize sulfated polysaccharides that have no equivalent in land plants and most of these enzymes constitute completely new protein families: i‐ and l‐carrageenases (Michel et al., 2003), a‐agarases (Flament et al., 2007) or fucanases (Colin et al., 2006). It was only through the application of standard biochemical approaches that these enzymes could be identified – otherwise they would have been annotated as ‘conserved hypothetical proteins’.
Furthermore, there is an urgent need for more cultures of marine Bacteria, Archaea and viruses. Most culture collections are based on readily cultivated microbes. When these organisms were isolated, there were no techniques to establish if the isolate was abundant in the natural environment or even if it had any relevant function. Molecular biology has changed that, and the isolation of new cultivable microbes can now be based on their abundance and relevance in defined marine habitats. There are a number of novel and innovative approaches to the isolation of new potential‐model microorganisms (Giovannoni and Stingl, 2007). For example, Rappé and colleagues (2002) used a dilution approach to isolate SAR11 ‘Candidatus Pelagibacter ubique’– the bacterium that can be found in mostly all biodiversity or metagenomic studies. So, methods exist for isolating useful model microbes from the natural environment, but they remain labour‐intensive. Nevertheless, they are probably the only way in which relevant bacteria can be brought into culture since classical microbiology methods have not proved to be useful for difficult‐to‐cultivate microbes.
All together now
The wealth of new technologies ranging from improved laboratory systems and automation to next‐generation sequencing technologies have started to fuel our imagination to better understand fundamental questions like the impact of Man and global change on our oceans. Although long considered the perfect dump site, oceanic ecosystems are showing signs of environmental fatigue through nutrient enrichment (eutrophication), climate modification and the presence of novel chemicals (xenobiotics) in the food chain. A better understanding of the largest ecosystem on earth will not only help to develop sustainable management for the oceans, but lay the foundation for a wealth of new goods and services for all colours of biotechnology. To approach this challenge, it is essential to move on from piling up masses of data, to integrative approaches taking into account the organisms, their genetic repertoire and the environment surrounding them. This brave new world of life sciences is challenging because it is extremely complex. To best proceed we have to cross boundaries and involve different disciplines like chemistry, oceanography, ecology, genomics and information science. The reward will be the ability to make use of the Petabytes of data produced to generate an in silico model of the marine ecosystem. If this works out, a true ecosystems approach might be possible where, as for weather forecasting, we are able to predict the behaviour of our oceans (Fig. 1). What is most exciting in the field of marine genomics is that, as in physics or astronomy, it forces biology to become a multinational, multidisciplinary, integrative science.
Figure 1.
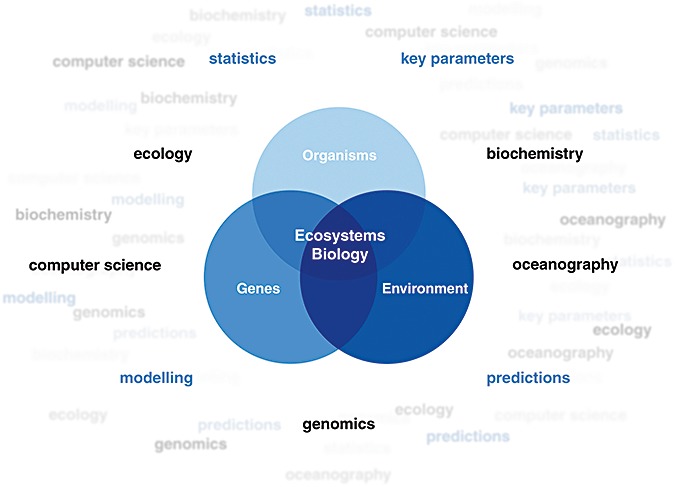
A multidisciplinary, integrative approach to ecosystems biology.
Acknowledgments
We would like to thank Garbiñe Guiu, Maurice Lex and John Claxton for fruitful discussions and comments on the paper. Thanks to the Max Planck Society and the Plymouth Marine Laboratory, a collaborative centre of NERC, for providing funding.
References
- Anonymous. Metagenomics versus Moore's law. Nat Methods. 2009a;6:623. [Google Scholar]
- Anonymous. Data's shameful neglect. Nature. 2009b;461:145. doi: 10.1038/461145a. [DOI] [PubMed] [Google Scholar]
- Assemble. 2009. ) Association of European Marine Biological Laboratories. [WWW document]. URL http://www.assemblemarine.org.
- Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beja O., Spudich E.N., Spudich J.L., Leclerc M., DeLong E.F. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- Blunt J.W., Copp B.R., Hu W.P., Munro M.H.G., Northcote P.T., Prinsep M.R. Marine natural products. Nat Prod Rep. 2007;24:31–86. doi: 10.1039/b603047p. [DOI] [PubMed] [Google Scholar]
- Bornscheuer U.T. Deep sea mining for unique biocatalysts. Chem Biol. 2005;12:859–860. doi: 10.1016/j.chembiol.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Bowler C., Karl D.M., Colwell R.R. Microbial oceanography in a sea of opportunity. Nature. 2009;459:180–184. doi: 10.1038/nature08056. [DOI] [PubMed] [Google Scholar]
- Colin S., Deniaud E., Jam M., Descamps V., Chevolot Y., Kervarec N. Cloning and biochemical characterization of the fucanase FcnA: definition of a novel glycoside hydrolase family specific for sulfated fucans. Glycobiology. 2006;16:1021–1032. doi: 10.1093/glycob/cwl029. et al. [DOI] [PubMed] [Google Scholar]
- Delaney J.R. 2007. pp. 1–2.
- DeLong E.E. Microbial community genomics in the ocean. Nat Rev Microbiol. 2005;3:459–469. doi: 10.1038/nrmicro1158. [DOI] [PubMed] [Google Scholar]
- DeLong E.F., Karl D.M. Genomic perspectives in microbial oceanography. Nature. 2005;437:336–342. doi: 10.1038/nature04157. [DOI] [PubMed] [Google Scholar]
- Deutsch C., Sarmiento J.L., Sigman D.M., Gruber N., Dunne J.P. Spatial coupling of nitrogen inputs and losses in the ocean. Nature. 2007;445:163–167. doi: 10.1038/nature05392. [DOI] [PubMed] [Google Scholar]
- Doney S.C., Abbott M.R., Cullen J.J., Karl D.M., Rothstein L. From genes to ecosystems: the ocean's new frontier. Front Ecol Environ. 2004;2:457–466. [Google Scholar]
- EC. 2007. ) Marine biotechnology research and the 7th research framework programme [WWW document]. URL http://ec.europa.eu/research/press/2007/maritime‐briefing/pdf/37‐bremen‐marine‐biotechnology‐research_en.pdf.
- EC. 2008. ) A European strategy for marine and maritime research [WWW document]. URL http://ec.europa.eu/research/press/2008/pdf/com_2008_534_en.pdf.
- EC‐US. 2008. ) Joint EC‐US CIESM workshop on marine genomics: at the interface of marine microbial ecology and biotechnological applications [WWW document]. URL http://ec.europa.eu/research/biotechnology/ec‐us/docs/monaco.pdf.
- ELIXIR. 2009. ) European Life Sciences Infrastructure for Biological Information. [WWW document]. URL http://www.elixir‐europe.org.
- Emodnet. 2009. ) The European Marine Observation and Date Network. [WWW document]. URL http://www.eurogoos.org/documents/eurogoos/downloads/mb_eurogoos_emodnet_vision_final.pdf.
- Eurocean. 2004. ) The Galway Declaration. [WWW document]. URL http://ec.europa.eu/research/press/2007/maritime‐briefing/pdf/24‐galway‐declaration_en.pdf.
- EuroFleets. 2009. ) Towards and Alliance of European Research Fleets. [WWW document]. URL http://www.eurofleets.eu/
- Falkowski P.G., Barber R.T., Smetacek V. Biogeochemical controls and feedbacks on ocean primary production. Science. 1998;281:200–206. doi: 10.1126/science.281.5374.200. [DOI] [PubMed] [Google Scholar]
- Falkowski P.G., Fenchel T., Delong E.F. The microbial engines that drive Earth's biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- Field D., Morrison N., Glöckner F.O., Kottmann R., Cochrane G., Vaughan R. Working together to put molecules on the map. Nature. 2008a;453:978. doi: 10.1038/453978b. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D., Garrity G., Gray T., Morrison N., Selengut J., Sterk P. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008b;26:541–547. doi: 10.1038/nbt1360. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flament D., Barbeyron T., Jam M., Potin P., Czjzek M., Kloareg B., Michel G. Alpha‐agarases define a new family of glycoside hydrolases, distinct from beta‐agarase families. Appl Environ Microbiol. 2007;73:4691–4694. doi: 10.1128/AEM.00496-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs B.M., Spring S., Teeling H., Quast C., Wulf J., Schattenhofer M. Characterization of a marine gammaproteobacterium capable of aerobic anoxygenic photosynthesis. Proc Natl Acad Sci USA. 2007;104:2891–2896. doi: 10.1073/pnas.0608046104. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadler P., Faber K. New enzymes for biotransformations: microbial alkyl sulfatases displaying stereo‐ and enantioselectivity. Trends Biotechnol. 2007;25:83–88. doi: 10.1016/j.tibtech.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Garrity G.M., Field D., Kyrpides N., Hirschman L., Sansone S.A., Angiuoli S. Toward a standards‐compliant genomic and metagenomic publication record. OMICS. 2008;12:157–160. doi: 10.1089/omi.2008.A2B2. et al. [DOI] [PubMed] [Google Scholar]
- Giovannoni S., Stingl U. The importance of culturing bacterioplankton in the ‘omics’ age. Nat Rev Microbiol. 2007;5:820–826. doi: 10.1038/nrmicro1752. [DOI] [PubMed] [Google Scholar]
- Glöckner F.O., Fuchs B.M., Amann R. Bacterioplankton compositions in lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckner F.O., Kube M., Bauer M., Teeling H., Lombardot T., Ludwig W. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc Natl Acad Sci USA. 2003;100:8298–8303. doi: 10.1073/pnas.1431443100. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J.L., Bohannan B.J.M., Whitaker R.J. Microbial biogeography: from taxonomy to traits. Science. 2008;320:1039–1043. doi: 10.1126/science.1153475. [DOI] [PubMed] [Google Scholar]
- Greenpaper. 2006. ) Green Paper on a Future Maritime Policy for the European Union. [WWW document]. URL http://ec.europa.eu/maritimeaffairs/policy_en.html.
- Gupta P.K. Single‐molecule DNA sequencing technologies for future genomics research. Trends Biotechnol. 2008;26:602–611. doi: 10.1016/j.tibtech.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe D., Costanzo M., Fey P., Gojobori T., Hannick L., Hide W. Big data: the future of biocuration. Nature. 2008;455:47–50. doi: 10.1038/455047a. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W.J., Preston C.M., Marin R., Scholin C.A., Vrijenhoek R.C. A robotic molecular method for in situ detection of marine invertebrate larvae. Mol Ecol Resour. 2008;8:540–550. doi: 10.1111/j.1471-8286.2007.02021.x. [DOI] [PubMed] [Google Scholar]
- Klarzynski O., Plesse B., Joubert J.M., Yvin J.C., Kopp M., Kloareg B., Fritig B. Linear beta‐1,3 glucans are elicitors of defense responses in tobacco. Plant Physiol. 2000;124:1027–1037. doi: 10.1104/pp.124.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottmann R., Gray T., Murphy S., Kagan L., Kravitz S., Lombardot T. A standard MIGS/MIMS compliant XML schema: toward the development of the Genomic Contextual Data Markup Language (GCDML) OMICS. 2008;12:115–121. doi: 10.1089/omi.2008.0A10. et al. [DOI] [PubMed] [Google Scholar]
- Kottmann R., Kostadinov I., Duhaime M.B., Buttigieg P.L., Yilmaz P., Hankeln W. Megx.net: integrated database resource for marine ecological genomics. Nucleic Acids Res. 2010;38:391–395. doi: 10.1093/nar/gkp918. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAMBA. 2009. ) Marine Metagenomics for New Biotechnological Applications. [WWW document]. URL http://mamba.bangor.ac.uk.
- Markowitz V.M., Ivanova N.N., Szeto E., Palaniappan K., Chu K., Dalevi D. IMG/M: a data management and analysis system for metagenomes. Nucleic Acid Res. 2008;36:534–538. doi: 10.1093/nar/gkm869. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel G., Helbert W., Kahn R., Dideberg O., Kloareg B. The structural bases of the processive degradation of iota‐carrageenan, a main cell wall polysaccharide of red algae. J Mol Biol. 2003;334:421–433. doi: 10.1016/j.jmb.2003.09.056. [DOI] [PubMed] [Google Scholar]
- Michel G., Nyval‐Collen P., Barbeyron T., Czjzek M., Helbert W. Bioconversion of red seaweed galactans: a focus on bacterial agarases and carrageenases. Appl Microbiol Biotechnol. 2006;71:23–33. doi: 10.1007/s00253-006-0377-7. [DOI] [PubMed] [Google Scholar]
- Moore. 2009. ) Microbial Genome Sequencing Project. [WWW document]. URL http://www.moore.org/microgenome/strain‐list.aspx.
- Moran M.A., Miller W.L. Resourceful heterotrophs make the most of light in the coastal ocean. Nat Rev Microbiol. 2007;5:792–800. doi: 10.1038/nrmicro1746. [DOI] [PubMed] [Google Scholar]
- Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W.G., Peplies J., Glöckner F.O. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acid Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramons W. 2009. ) Ocean Observatories Initiative. [WWW document]. URL http://www.oceanleadership.org/2009/ocean‐observatories‐initiative‐receives‐award/
- Rappé M.S., Connon S.A., Vergin K.L., Giovannoni S.J. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418:630–633. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- Rusch D.B., Halpern A.L., Sutton G., Heidelberg K.B., Williamson S., Yooseph S. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiker S., Santos V., Bartels D., Bekel T., Brecht M., Buhrmester J. Genome sequence of the ubiquitous hydrocarbon‐degrading marine bacterium Alcanivorax borkumensis. Nat Biotechnol. 2006;24:997–1004. doi: 10.1038/nbt1232. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweder T., Lindequist U., Lalk M. Screening for new metabolites from marine microorganisms. In: Scheper T, editor. Springer; 2005. pp. 1–48. [DOI] [PubMed] [Google Scholar]
- Seshadri R., Kravitz S.A., Smarr L., Gilna P., Frazier M. CAMERA: a community resource for metagenomics. PLoS Biol. 2007;5:e75. doi: 10.1371/journal.pbio.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon I., Tzahor S., Williamson S., Shmoish M., Man‐Aharonovich D., Rusch D.B. Viral photosynthetic reaction center genes and transcripts in the marine environment. ISME J. 2007;1:492–501. doi: 10.1038/ismej.2007.67. et al. [DOI] [PubMed] [Google Scholar]
- Sharon I., Alperovitch A., Rohwer F., Haynes M., Glaser F., Atamna‐Ismaeel N. Photosystem I gene cassettes are present in marine virus genomes. Nature. 2009;461:258–262. doi: 10.1038/nature08284. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D.A., Amann R. Development and application of nucleic acid probes. In: Stackebrandt E., Goodfellow M., editors. John Wiley & Sons; 1991. pp. 205–248. [Google Scholar]
- Strous M., Fuerst J.A., Kramer E.H.M., Logemann S., Muyzer G., Van De Pas‐Schoonen K.T. Missing lithotroph identified as new Planctomycete. Nature. 1999;400:446–449. doi: 10.1038/22749. et al. [DOI] [PubMed] [Google Scholar]
- Tringe S.G., Rubin E.M. Metagenomics: DNA sequencing of environmental samples. Nat Rev Genet. 2005;6:805–814. doi: 10.1038/nrg1709. [DOI] [PubMed] [Google Scholar]
- Uzawa H., Nishida Y., Sasaki K., Minoura N., Kobayashi K. Synthetic potential of molluscan Sulfatases for the library synthesis of regioselectively O‐sulfonated d‐Galacto‐sugars. Chembiochem. 2003;4:640–647. doi: 10.1002/cbic.200300616. [DOI] [PubMed] [Google Scholar]
- Venter J.C., Remington K., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J.A. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. et al. [DOI] [PubMed] [Google Scholar]
- Volckaert F., Barbier M., Canario A., Olsen A., Wesnigk J., Clark M., Boyen C. Empowering marine science through genomics. Mar Genomics. 2008;1:33–35. doi: 10.1016/j.margen.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Wallner S.R., Bauer M., Würdemann C., Wecker P., Glöckner F.O., Faber K. Highly enantioselective sec‐Alkylsulfatase activity of the marine Planctomycete Rhodopirellula baltica shows retention of configuration. Angew Chem Int Ed. 2005;44:2–4. doi: 10.1002/anie.200501955. [DOI] [PubMed] [Google Scholar]
- Witvrouw M., Este J.A., Mateu M.Q., Reymen D., Andrei G., Snoeck R. Activity of a sulfated polysaccharide extracted from the red seawed Aghardhiella tenera against Human‐Immunodeficiency‐Virus and other enveloped viruses. Antivir Chem Chemother. 1994;5:297–303. et al. [Google Scholar]
- Yooseph S., Sutton G., Rusch D.B., Halpern A.L., Williamson S.J., Remington K. The Sorcerer II Global Ocean Sampling Expedition: Expanding the Universe of Protein Families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]


