Abstract
To determine the mechanism for the increased osteoclastogenesis in the jaw of cherubism patients with SH3BP2 mutations we evaluated the effect of mutant compared to wild-type SH3BP2 on activation of osteoclast signaling pathways. Indeed mutant forms of SH3BP2 do induce greater osteoclastogenesis.
Heterozygous activating mutations in exon 9 of SH3BP2 have been found in most patients with cherubism, an unusual genetic syndrome characterized by excessive remodeling of the mandible and maxilla due to spontaneous and excessive osteoclastic bone resorption.
Here we have investigated the functional consequences of SH3BP2 mutations on sRANKL-induced osteoclastogenesis in RAW 264.7 pre-osteoclast cells. sRANKL-stimulated RAW 264.7 cells were transfected with wild-type or mutant SH3BP2 plasmids. NFAT-luciferase and tartrate resistant acid phosphatase (TRAP), a marker of osteoclastic differentiation, levels were evaluated. Western Immunoblots were also performed to determine phosphorylation of key proteins involved in the PI-PLC pathway leading to NFATc1 translocation.
Our results indicate that forced expression of mutant forms of SH3BP2 , found in cherubism patients, in RAW 264.7 cells induce greater NFAT activity and greater expression of TRAP than forced expression of wild-type SH3BP2. These findings indicate that missense SH3BP2 mutations cause a gain of protein function. Moreover, over expression of SH3BP2 in RAW 264.7 cells potentiates sRANKL-stimulated phosphorylation of PLCγ1 and PLCγ2.
Our studies demonstrate that cherubism is due to gain-of-function mutations in SH3BP2 that stimulate RANKL-induced activation of PLCγ; The consequent activation of calcineurin and NFAT proteins induces the excessive osteoclastic phenotype of cherubism.
Keywords: bone turnover, osteoclasts, bone and cartilage development, osteoporosis
Introduction
The canonical signal transduction pathway for osteoclastogenesis and osteoclast stimulation involves the binding of receptor activator of nuclear factor κB ligand (RANKL) to receptor activator of nuclear factor κB (RANK) on osteoclast precursor cells(1). This canonical or other non-canonical pathways activate signal transduction cascades that involve phosphorylation of PLCγ, Syk, and Vav3(2-5). Subsequently the obligate osteoclastogenic factor, NFATc1, is dephosphorylated, leading to its nuclear translocation and induction of the genetic signature for osteoclast differentiation(6, 7). The onset of cherubism is early, age 2-5 years, at which time patients develop characteristic giant cell lesions in the maxilla and mandible, substantial facial swelling and cervical lymphadenopathy(8-10). The localized nature of the bone lesions in patients with cherubism is unexpected as the disorder is associated with heterozygous germline mutations in the SH3BP2 gene (4p16.3), which is widely expressed throughout the osteoimmune system(8, 9, 11-14). SH3BP2 is a complex adaptor protein, with an N-terminal pleckstrin homology (PH) domain, a ten amino acid Src Homology 3 (SH3) binding domain, and a C-terminal Src Homology 2 (SH2) domain(15). Most SH3BP2 mutations described to date in patients with cherubism occur in exon 9 and cause amino acid substitutions within a restricted six-amino acid region of the SH3BP2 protein(8, 14, 16-20). The precise function of this region remains unclear, but recent work from our laboratory suggests that these missense mutations lead to a gain-of-function rather than a loss of activity in Jurkat TAg cells(16). A gain-of-function for the SH3BP2 mutations in cherubism is consistent with prior observations that deletions of 4p16.3 in patients with Wolf-Hirschhorn syndrome, which result in loss of one copy of SH3BP2, do not result in a bone resorptive phenotype(21-23).
Previous studies in B and T cells have shown that over expression of SH3BP2 leads to activation of a luciferase reporter gene that is under the control of the NFAT binding sequence from the interleukin 2 (IL-2) gene promoter(24-26). Because over expression of a constitutively active form of NFATc1 in the RAW 264.7 osteoclast precursor cell line is sufficient to induce osteoclast differentiation(27), we have hypothesized that mutant activated forms of SH3BP2 may increase NFAT activation and thereby induce the osteoclastic bone lesions of cherubism(1, 6, 8, 27). In the present study we evaluated the effect of forced expression of mutant (mutations found in cherubism patients) compared to wild-type SH3BP2 on activation of the NFAT and TRAP signaling pathways in RAW 264.7 preosteoclast cells. Here we report that mutant forms of SH3BP2, found in patients with cherubism, induce greater phosphorylation of PLCγ1 and PLCγ2, two downstream mediators of RANK signaling, and greater activation of osteoclastogenesis, than wild-type SH3BP2.
Methods
Cell Culture
RAW 264.7 cells (American Type Culture Collection) were maintained at 37°C, 5% CO2 in growth media consisting of DMEM or RPMI (Invitrogen) supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin (Invitrogen). For osteoclastogenesis assays, cells were resuspended in fresh medium and cultured for 8 days in 96-well plates at a cell density of 1000 cells/well.
Plasmids and Transfection of RAW 264.7 cells
SH3BP2 wild-type and mutant inserts were in a pMT3 plasmid backbone and the mutations were created by overlap extension as previously described(16). The NFAT-luciferase assays were done as previously described and in triplicate(7, 16). Briefly, we obtained the mouse SH3BP2 cDNA in pMT3 plasmid from Professor Marcel Deckert (University of Nice, France) and NFAT luciferase reporter from Professor Gerald Crabtree. RAW 264.7 cells at 40-70% confluence were transiently transfected with SH3BP2 cDNA, a hybrid reporter gene in which luciferase expression is under the control of a 3X minimal IL-2 promoter containing the NFAT binding site (NFAT-luc), and a Renilla luciferase reporter as a control for transfection efficiency (Promega). Transfection was performed by electroporation using a Nucleofector (Amaxa). 24 hours after transfection, luminescence was evaluated with a luminometer (Sirius Berthold). To create stable cell lines, RAW264.7 cells were co-transfected with plasmids encoding the NFAT-luciferase reporter gene and the neomycin resistance gene and cultured in the presence of G418 (500 μg/ml).
Immunoblot Analysis
To analyze immunoactive SH3BP2 immunoblots were prepared as previously described(7, 16). To analyze phosphorylated proteins, cells were scraped in lysis buffer (20 mM tris-HCL, pH 7.5, 20 mM p-nitrophenyl phosphate, 1 mM EGTA, 50 mM sodium fluoride, 50 μM sodium orthovanadate, and proteinase inhibitor cocktail) and lysed by freezing and thawing 3 times. Samples were electrophoresed on a 4-12% bis-tris gel (Invitrogen), 200 volts for 40 minutes in MOPS buffer and transferred to PVDF membranes at 100 volts for 1 hour. Membranes were then blocked in 5% nonfat dried milk in tris-buffered saline with 0.1% Tween-20 for one hour and incubated overnight at 4°C with antibodies that are specific for phosphorylated Tyr1217-PLCγ2 (Cell Signaling), phosphorylated Tyr783 PLCγ1 (Cell Signaling), phosphorylated Tyr525/526 Syk, phosphorylated Tyr173 Vav3 (Abcam), SH3BP2 (custom-made) or β-actin (Sigma). These immunoblots were repeated three times with independent cell lysates.
Determination of Osteoclast Differentiation
Osteoclast differentiation was assessed by measuring enzymatic activity of TRAP, an intracellular marker of terminal osteoclast differentiation as previously described(7). All TRAP assays were performed as independent experiments three times. Osteoclast differentiation was also assessed by counting the number of TRAP staining cells with greater than or equal to 3 nuclei (TRAP staining multinucleated giant cells).
Statistics
Statistical evaluation of the data used the analysis of variance (ANOVA) and Fisher’s protected least significant difference multiple comparison test or an unpaired t-test to determine significance between groups.
Results
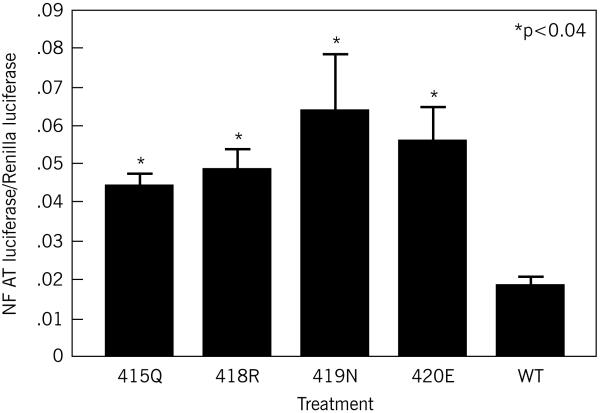
Each of the mutant SH3BP2 isoforms (found in patients with cherubism(8, 16) produced significant (p<0.04; 415Q p=0.036, 418R p=0.016, 419N p=0.0005, 420E p=0.003) and 2 to 3-fold greater increases in NFAT-luc activity compared to the wild-type SH3BP2 protein despite the fact that the total amount of SH3BP2 protein by immunoblot did not appear different between the mutants and the wild-type SH3BP2 (Figure 1a). There were no significant differences between any of the mutant forms in terms of NFAT luciferase activity (p>0.05).
Figure 1a.
Activation of NFAT activity by over expression of SH3BP2. RAW 264.7 cells were transiently transfected with wild-type or mutant SH3BP2 and incubated with sRANKL for 48 hours. All mutant SH3BP2 proteins produced significantly greater increases in NFAT luciferase activity compared to wild-type SH3BP2 (versus wild-type, R415Q p=0.02, P418R p=0.007, D419N p=0.002, and G420E p<0.0001). This figure represents data pooled from three independent experiments.
Previous data and data of our own (supplemental Figure 1) showed that NFATc1 is increased in response to sRANKL(1, 6, 27, 28)(supplemental Figure 2).
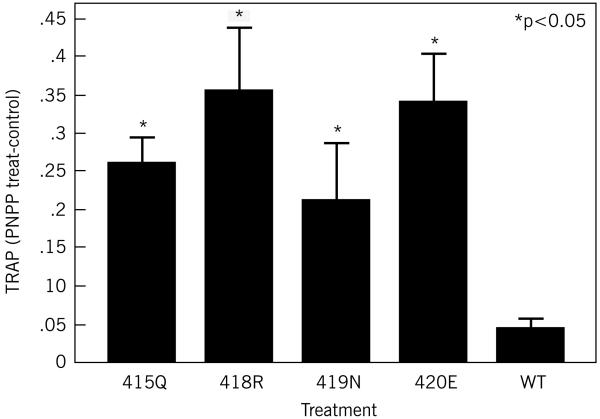
RAW 264.7 cells that had been incubated with 100 ng/ml sRANKL for one week showed a significant increase in TRAP activity (Figure 1b), with mutant forms of SH3BP2 (R415Q p=0.014, P418R p=0.009, D419N p=0.05, G420E p=0.004) producing significantly greater TRAP activity than cells that had been transfected with the wild-type SH3BP2 (Figure 1b) (p<or=to 0.05). There were no significant differences between any of the mutant forms of SH3BP2 in terms of TRAP activation (p>0.05).
Figure 1b.
TRAP activity is increased in the presence of SH3BP2. RAW 264.7 cells were transiently transfected with mutant (R415Q, G420E, P418R or D419N) or wild-type SH3BP2 and treated with sRANKL (100ng/ml). This figure demonstrates that in cells transiently transfected with mutant SH3BP2 cDNA there is significantly increased TRAP activity compared to the wild-type control (p<or=to 0.05). SH3BP2 levels, determined by immunoblot, show equivalent levels of the wild-type and mutant SH3BP2. This experiment was repeated three times with similar results.
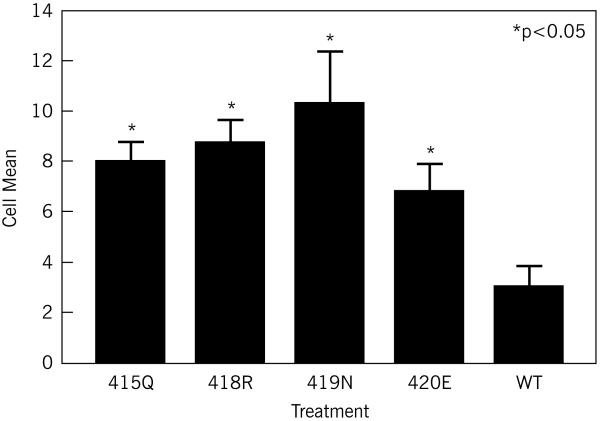
RAW 264.7 cells that had been incubated with 100 ng/ml sRANKL for one week also showed a significant increase in TRAP staining of multinucleated giant cells (Figure 1c), with mutant forms of SH3BP2 (R415Q p=0.01, P418R p=0.004, D419N p=0.0007, G420E p=0.045) producing significantly more TRAP staining multinucleated giant cells than cells that had been transfected with the wild-type SH3BP2 (Figure 1c) (p<or=to 0.05). There were no significant differences between any of the mutant forms of SH3BP2 in terms of TRAP staining of multinucleated giant cells (p>0.05).
Figure 1c.
Mutant SH3BP2 increases the number of multinucleated giant cells (at least 3 nuclei) per high power field compared to wild-type SH3BP2. RAW 264.7 cells were transiently transfected with mutant (R415Q, G420E, P418R or D419N) or wild-type SH3BP2 and treated with sRANKL (100ng/ml). This figure demonstrates that in cells transiently transfected with mutant SH3BP2 cDNA there is significantly increased TRAP staining of multinucleated giant cells (greater than or equal to 3 nuclei) compared to the wild-type control (p< 0.05).
To further characterize the effect of SH3BP2 on NFAT activation and osteoclastogenesis, we examined the effect of increasing the amount of SH3BP2 in RAW 264.7 cells that transiently expressed the NFAT-luc reporter gene. Transfection of cells with a low amount (0.5 μg plasmid DNA/well of a six well dish) of SH3BP2 cDNA showed that mutant SH3BP2 isoforms were significantly more active than wild-type SH3BP2 (Figure 1a). However, when a higher amount (2.0 μg plasmid DNA/well of a six well dish, data not shown) of cDNA was used there were similar increases in NFAT activity for mutant and wild-type SH3BP2 proteins. Immunoblot analysis showed that more SH3BP2 protein was expressed when RAW 264.7 cells were transfected with 2 μg of cDNA compared to 0.5 μg of cDNA, but for both experimental conditions the levels of mutant and wild-type SH3BP2 proteins were similar (supplemental figure 3).
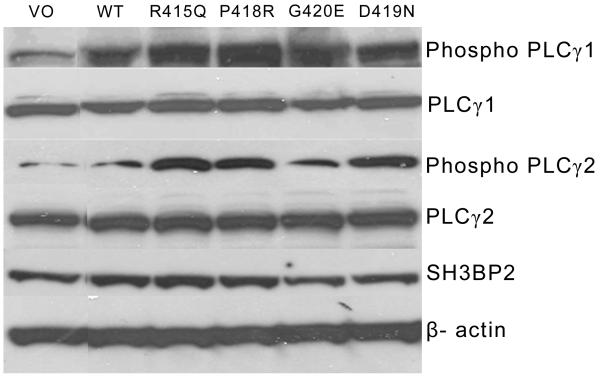
Because a previous study had shown that SH3BP2 can form complexes with PLCγ isoforms(26), we analyzed the effect of sRANKL and SH3BP2 on expression and phosphorylation of PLCγ1 and PLCγ2 in RAW 264.7 cells that had been transfected with SH3BP2. Levels of immunoreactive PLCγ2 and PLCγ1 were unchanged by expression of wild-type compared to mutant SH3BP2 proteins (Figure 2). On the other hand, protein levels of both phospho-Tyr1217PLCγ2 and phospho-Tyr783PLCγ1 were increased in cells that had been transfected with mutant SH3BP2 isoforms compared to wild-type SH3BP2 (2-4 fold) by ImageJ analysis (Figure 2).
Figure 2.
Mutant SH3BP2 increases PLCγ2 phosphorylation by immunoblot. Levels of PLCγ2 Tyr1217 phosphorylation and total PLCγ2 and levels of PLCγ1 Tyr783 phosphorylation and total PLCγ1 were examined in response to transient transfection of mutant or wild-type SH3BP2 in RAW 264.7 cells. β-actin is used as a control to ensure that equal protein was loaded in each lane. This experiment was repeated three times with the same results. WT represents cells transiently transfected with wild-type. VO represents cells transiently transfected with vector only plasmid with no SH3BP2 insert.
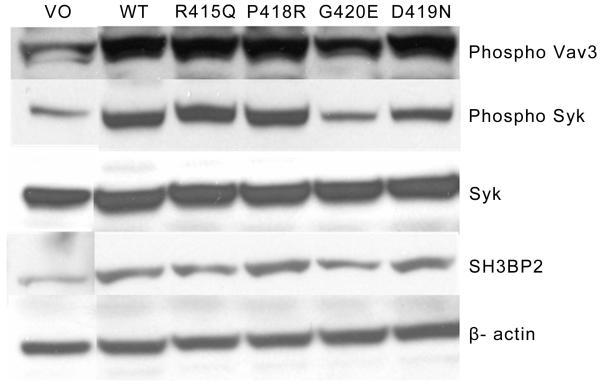
To determine whether the SH3BP2-enhanced phosphorylation was specific for PLCγ, we also analyzed phosphorylation of Syk and Vav3, two other proteins that potentially interact with SH3BP2(26). Both Syk and Vav3 showed increased phosphorylation in cells that were transfected with wild-type human or mutant SH3BP2, but there was no additional increase in phosphorylation in cells that expressed the mutant SH3BP2 proteins (Figure 3).
Figure 3.
Mutant SH3BP2 does not increase Syk or Vav3 phosphorylation relative to the wild-type. Levels of Syk phosphorylation and total Syk and levels of Vav3 phosphorylation were examined in response to transient transfection of mutant or wild-type SH3BP2 in RAW 264.7 cells. β-actin is used as a control to ensure that equal protein was loaded in each lane. The mutant SH3BP2s are not more effective at Syk Tyr525/526 or Vav3 phosphorylation compared to the wild-type. This experiment was repeated three times with the same results. WT represents cells transiently transfected with wild-type SH3BP2. VO represents cells transiently transfected with vector only plasmid with no SH3BP2 insert.
Discussion
We have recently shown that SH3BP2 mutations causing cherubism encode SH3BP2 proteins that potentiate sRANKL-induced activation of NFATc1 signaling(16). We have now demonstrated that transfection of RAW 264.7 cells with wild-type or mutant SH3BP2 induces not only nuclear translocation of NFAT but also osteoclastogenesis. This effect is dependent upon sRANKL, which induces expression of endogenous NFATc1(6) and stimulates translocation of NFATc1 into the nucleus(7). At the lower level of SH3BP2 expression (0.5 μg) each of the mutant SH3BP2s led to higher levels of NFAT activity compared to the wild-type SH3BP2(Figure 1a), however at the higher level of expression (2μg) NFAT activity reached a plateau, with similar activity with mutant and wild type SH3BP2. These results indicate that expression of high amounts of wild-type SH3BP2 protein can reproduce the effect of mutant SH3BP2 proteins, and suggest that some downstream component(s) of the SH3BP2 signaling pathway is or are rate-limiting under conditions of excessive SH3BP2 activity.
Because nuclear translocation of NFAT requires dephosphorylation by calcineurin, we further hypothesized that SH3PB2, which lacks catalytic activity, must interact with other proteins that then stimulate calcineurin activity. One candidate protein is PLCγ, which binds to SH3BP2 and is phosphorylated by sRANKL(3).
PLCγ, as well as other forms of PLC, cleaves the membrane phospholipid phosphatidyl inositol-4,5-biphosphate (PIP2) into the second messenger molecules inositol-1,4,5-triphosphate (IP3) and diacylglycerol (DAG)(29). IP3 directly increases intracellular calcium levels by inducing the release of endoplasmic reticulum calcium stores, which leads to activation of calcineurin. There are two forms of PLCγ (1 and 2)(3, 30-32); while PLCγ1 is widely distributed, expression of PLCγ2 is primarily limited to cells of hematopoetic lineage(30). Both PLCγ isoforms require phosphorylation on specific tyrosine residues for their catalytic activity(31). Targeted deletion of Plcγ2 but not Plcγ1 in mice results in an in vivo osteopetrotic phenotype(3), suggesting that PLCγ2 is the critical isoform for sRANKL-induced osteoclastogenesis. We found that forced expression of wild-type and mutant SH3BP2 in RAW 264.7 cells led to an increase in the relative amount of both phospho-PLCγ1 and phospho-PLCγ2, with no alteration in the total amount of either protein, and that mutant SH3BP2 was more active than the wild-type(1, 6, 33). We found that over expression of SH3BP2 augmented sRANKL-dependent phosphorylation of Syk, but there were no differences between wild-type and mutant SH3BP2 proteins in Syk phosphorylation. Similarly, both wild-type and mutant SH3BP2 produced comparable increases in sRANKL-induced activation of Vav3, which is phosphorylated by Syk. Thus, RANKL-induced phosphorylation of all four of these interacting proteins is enhanced by SH3BP2, but under the conditions that we used to replicate cherubism, mutant SH3BP2 proteins have a specific activating effect that appears to be limited to PLCγ1 and PLCγ2. The increase of PLCγ2 phosphorylation (and by inference activation) by the mutant forms of SH3BP2 compared to the wild-type are consistent with the recent finding that PLCγ2 activation can be dependent on Tec nonreceptor kinases rather than Syk(34). Thus the effect of mutant SH3BP2 on increased osteoclastogenesis would seem to be downstream of Syk activation (since Syk stimulation is not further increased but PLCγ is). No SH3BP2 mutant was consistently more active than the others in terms of phosphorylation of PLCγ2, and stimulation of NFAT and TRAP or TRAP staining of multinucleated cells.
A previous study examined the role of the P416R SH3BP2 mutation in a knockin mouse model(4, 8). In this model there was a bone resorptive phenotype in the homozygous knockin mouse, which led the authors to conclude that there were two pathways by which SH3BP2 acted. In the second pathway hyperactive osteoclasts and Syk phosphorylation are involved. However we found phosphorylation of PLCγ2 at an earlier time point (24 hours) than they found Syk phosphorylation (48 hours) implicating SH3BP2 downstream of Syk in terms of PLCγ2 phosphorylation(4, 5). Aliprantis et al reported that mutant Sh3bp2 bone marrow cells from this same “cherubism” mouse displayed increased induction of the Nfatc1/A isoform in response to RANKL stimulation and that SH3BP2 is upstream of NFATc1 in RANKL-induced osteoclastogenesis.(35)
In conclusion, we show for the first time that RANKL-induced phosphorylation of proteins related to osteoclastogenesis is potentiated by SH3BP2. Moreover, mutant forms of SH3BP2 that occur in patients with cherubism have a specific activating effect that further potentiates RANKL-induced phosphorylation of PLCγ isoforms. The results of the current study suggest that SH3BP2, as well as PLCγ2, will be interesting targets for the development of novel therapies for disorders that are characterized by excessive osteoclastic development.
Supplementary Material
NFATc1 increases with RANKL treatment alone. Immunoblot showing an increase in NFATc1 over time with sRANKL treatment (100 ng/ml).
NFAT luciferase is increased with sRANKL in RAW 264.7 cells stably transfected with NFAT luciferase plasmid.
RAW 264.7 cells were incubated with sRANKL for 48 hours alone or transiently transfected with wild-type or mutant SH3BP2 and incubated with sRANKL for 48 hours. SH3BP2 levels, determined by immunoblot, show equivalent levels of the wild-type and mutant SH3BP2.
Acknowledgements
This work was supported by in part by US Public Health Service Research grants K08-AR47661 (SAL) and R01-DE018237 (SAL), DK34281 (MAL) and DK56178 (MAL), and General Clinical Research Center Grant RR0035. Dr. Lietman is the recipient of a Career Development Award from the Orthopaedic Research and Education Foundation.
Nonstandard abbreviations used
- SH3BP2
src homology 3 binding protein 2
- NFAT
nuclear factor of activated T cells
- PLCγ
phospholipase Cγ
- TRAP
tartrate resistant acid phosphatase
- sRANKL
soluble receptor activator of NFκB ligand
- SFK
src family kinase
- GFP
green fluorescent protein
- Jurkat T Ag
Jurkat T Antigen
- NFAT-luc
NFAT luciferase
- WT
wild-type
- SH2
src homology 2
- PH
pleckstrin homology
- OMIM
online mendelian inheritance in man
- M-CSF
macrophage-colony stimulating factor
- Tyr
tyrosine
- PKC
protein kinase C
Footnotes
All authors have no conflicts of interest.
References
- 1.Takayanagi H. Mechanistic insight into osteoclast differentiation in osteoimmunology. J Mol Med. 2005;83:170–179. doi: 10.1007/s00109-004-0612-6. [DOI] [PubMed] [Google Scholar]
- 2.Faccio R, Teitelbaum SL, Fujikawa K, Chappel J, Zallone A, Tybulewicz VL, Ross FP, Swat W. Vav3 regulates osteoclast function and bone mass. Nat Med. 2005;11:284–290. doi: 10.1038/nm1194. [DOI] [PubMed] [Google Scholar]
- 3.Mao D, Epple H, Uthgenannt B, Novack DV, Faccio R. PLCgamma2 regulates osteoclastogenesis via its interaction with ITAM proteins and GAB2. J Clin Invest. 2006;116:2869–2879. doi: 10.1172/JCI28775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ueki Y, Lin CY, Senoo M, Ebihara T, Agata N, Onji M, Saheki Y, Kawai T, Mukherjee PM, Reichenberger E, et al. Increased Myeloid Cell Responses to M-CSF and RANKL Cause Bone Loss and Inflammation in SH3BP2 “Cherubism” Mice. Cell. 2007;128:71–83. doi: 10.1016/j.cell.2006.10.047. [DOI] [PubMed] [Google Scholar]
- 5.Shinohara M, Koga T, Okamoto K, Sakaguchi S, Arai K, Yasuda H, Takai T, Kodama T, Morio T, Geha RS, et al. Tyrosine kinases Btk and Tec regulate osteoclast differentiation by linking RANK and ITAM signals. Cell. 2008;132:794–806. doi: 10.1016/j.cell.2007.12.037. [DOI] [PubMed] [Google Scholar]
- 6.Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 7.Lietman SA, Yin L, Levine MA. SH3BP2 is an activator of NFAT activity and osteoclastogenesis. Biochem Biophys Res Commun. 2008 doi: 10.1016/j.bbrc.2008.04.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ueki Y, Tiziani V, Santanna C, Fukai N, Maulik C, Garfinkle J, Ninomiya C, doAmaral C, Peters H, Habal M, et al. Mutations in the gene encoding c-Abl-binding protein SH3BP2 cause cherubism. Nat Genet. 2001;28:125–126. doi: 10.1038/88832. [DOI] [PubMed] [Google Scholar]
- 9.Schultze-Mosgau S, Holbach LM, Wiltfang J. Cherubism: clinical evidence and therapy. J Craniofac Surg. 2003;14:201–206. doi: 10.1097/00001665-200303000-00012. discussion 207-208. [DOI] [PubMed] [Google Scholar]
- 10.Pulse CL, Moses MS, Greenman D, Rosenberg SN, Zegarelli DJ. Cherubism: case reports and literature review. Dent Today. 2001;20:100–103. [PubMed] [Google Scholar]
- 11.Petschler M, Stiller M, Hoffmeister B, Witkowski R, Opitz C, Bill JS, Peters H. Clinical and molecular genetic observations on families with cherubism over three generations. Mund Kiefer Gesichtschir. 2003;7:83–87. doi: 10.1007/s10006-002-0444-x. [DOI] [PubMed] [Google Scholar]
- 12.Lannon DA, Earley MJ. Cherubism and its charlatans. Br J Plast Surg. 2001;54:708–711. doi: 10.1054/bjps.2001.3701. [DOI] [PubMed] [Google Scholar]
- 13.Ladhani S, Sundaram P, Joshi JM. Sleep disordered breathing in an adult with cherubism. Thorax. 2003;58:552. doi: 10.1136/thorax.58.6.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo B, Faiyaz-Ul-Haque M, Kennedy S, Aviv R, Tsui LC, Teebi AS. Novel mutation in the gene encoding c-Abl-binding protein SH3BP2 causes cherubism. Am J Med Genet. 2003;121A:37–40. doi: 10.1002/ajmg.a.20226. [DOI] [PubMed] [Google Scholar]
- 15.Ren R, Mayer BJ, Cicchetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 16.Lietman SA, Kalinchinko N, Deng X, Kohanski R, Levine MA. Identification of a novel mutation of SH3BP2 in cherubism and demonstration that SH3BP2 mutations lead to increased NFAT activation. Hum Mutat. 2006;27:717–718. doi: 10.1002/humu.9433. [DOI] [PubMed] [Google Scholar]
- 17.Imai Y, Kanno K, Moriya T, Kayano S, Seino H, Matsubara Y, Yamada A. A missense mutation in the SH3BP2 gene on chromosome 4p16.3 found in a case of nonfamilial cherubism. Cleft Palate Craniofac J. 2003;40:632–638. doi: 10.1597/1545-1569_2003_040_0632_ammits_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- 18.Carvalho VM, Perdigao PF, Pimenta FJ, de Souza PE, Gomez RS, De Marco L. A novel mutation of the SH3BP2 gene in an aggressive case of cherubism. Oral Oncol. 2007 doi: 10.1016/j.oraloncology.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 19.de Lange J, van Maarle MC, van den Akker HP, Redeker EJ. A new mutation in the SH3BP2 gene showing reduced penetrance in a family affected with cherubism. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2007;103:378–381. doi: 10.1016/j.tripleo.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 20.Li CY, Yu SF. A novel mutation in the SH3BP2 gene causes cherubism: case report. BMC Med Genet. 2006;7:84. doi: 10.1186/1471-2350-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergemann AD, Cole F, Hirschhorn K. The etiology of Wolf-Hirschhorn syndrome. Trends Genet. 2005;21:188–195. doi: 10.1016/j.tig.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Battaglia A, Carey JC. Health supervision and anticipatory guidance of individuals with Wolf-Hirschhorn syndrome. Am J Med Genet. 1999;89:111–115. doi: 10.1002/(sici)1096-8628(19990625)89:2<111::aid-ajmg9>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 23.Battaglia A, Carey JC, Cederholm P, Viskochil DH, Brothman AR, Galasso C. Natural history of Wolf-Hirschhorn syndrome: experience with 15 cases. Pediatrics. 1999;103:830–836. doi: 10.1542/peds.103.4.830. [DOI] [PubMed] [Google Scholar]
- 24.Deckert M, Tartare-Deckert S, Hernandez J, Rottapel R, Altman A. Adaptor function for the Syk kinases-interacting protein 3BP2 in IL-2 gene activation. Immunity. 1998;9:595–605. doi: 10.1016/s1074-7613(00)80657-3. [DOI] [PubMed] [Google Scholar]
- 25.Foucault I, Liu YC, Bernard A, Deckert M. The chaperone protein 14-3-3 interacts with 3BP2/SH3BP2 and regulates its adapter function. J Biol Chem. 2003;278:7146–7153. doi: 10.1074/jbc.M209509200. [DOI] [PubMed] [Google Scholar]
- 26.Foucault I, Le Bras S, Charvet C, Moon C, Altman A, Deckert M. The adaptor protein 3BP2 associates with VAV guanine nucleotide exchange factors to regulate NFAT activation by the B-cell antigen receptor. Blood. 2005;105:1106–1113. doi: 10.1182/blood-2003-08-2965. [DOI] [PubMed] [Google Scholar]
- 27.Hirotani H, Tuohy NA, Woo JT, Stern PH, Clipstone NA. The calcineurin/nuclear factor of activated T cells signaling pathway regulates osteoclastogenesis in RAW264.7 cells. J Biol Chem. 2004;279:13984–13992. doi: 10.1074/jbc.M213067200. [DOI] [PubMed] [Google Scholar]
- 28.Ishida N, Hayashi K, Hoshijima M, Ogawa T, Koga S, Miyatake Y, Kumegawa M, Kimura T, Takeya T. Large scale gene expression analysis of osteoclastogenesis in vitro and elucidation of NFAT2 as a key regulator. J Biol Chem. 2002;277:41147–41156. doi: 10.1074/jbc.M205063200. [DOI] [PubMed] [Google Scholar]
- 29.Patterson RL, van Rossum DB, Ford DL, Hurt KJ, Bae SS, Suh PG, Kurosaki T, Snyder SH, Gill DL. Phospholipase C-gamma is required for agonist-induced Ca2+ entry. Cell. 2002;111:529–541. doi: 10.1016/s0092-8674(02)01045-0. [DOI] [PubMed] [Google Scholar]
- 30.Wilde JI, Watson SP. Regulation of phospholipase C gamma isoforms in haematopoietic cells: why one, not the other? Cell Signal. 2001;13:691–701. doi: 10.1016/s0898-6568(01)00191-7. [DOI] [PubMed] [Google Scholar]
- 31.Rebecchi MJ, Pentyala SN. Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol Rev. 2000;80:1291–1335. doi: 10.1152/physrev.2000.80.4.1291. [DOI] [PubMed] [Google Scholar]
- 32.Katan M. Families of phosphoinositide-specific phospholipase C: structure and function. Biochim Biophys Acta. 1998;1436:5–17. doi: 10.1016/s0005-2760(98)00125-8. [DOI] [PubMed] [Google Scholar]
- 33.Hur EM, Park YS, Lee BD, Jang IH, Kim HS, Kim TD, Suh PG, Ryu SH, Kim KT. Sensitization of epidermal growth factor-induced signaling by bradykinin is mediated by c-Src. Implications for a role of lipid microdomains. J Biol Chem. 2004;279:5852–5860. doi: 10.1074/jbc.M311687200. [DOI] [PubMed] [Google Scholar]
- 34.Jongstra-Bilen J, Puig Cano A, Hasija M, Xiao H, Smith CI, Cybulsky MI. Dual functions of Bruton’s tyrosine kinase and Tec kinase during Fcgamma receptor-induced signaling and phagocytosis. J Immunol. 2008;181:288–298. doi: 10.4049/jimmunol.181.1.288. [DOI] [PubMed] [Google Scholar]
- 35.Aliprantis AO, Ueki Y, Sulyanto R, Park A, Sigrist KS, Sharma SM, Ostrowski MC, Olsen BR, Glimcher LH. NFATc1 in mice represses osteoprotegerin during osteoclastogenesis and dissociates systemic osteopenia from inflammation in cherubism. J Clin Invest. 2008;118:3775–3789. doi: 10.1172/JCI35711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NFATc1 increases with RANKL treatment alone. Immunoblot showing an increase in NFATc1 over time with sRANKL treatment (100 ng/ml).
NFAT luciferase is increased with sRANKL in RAW 264.7 cells stably transfected with NFAT luciferase plasmid.
RAW 264.7 cells were incubated with sRANKL for 48 hours alone or transiently transfected with wild-type or mutant SH3BP2 and incubated with sRANKL for 48 hours. SH3BP2 levels, determined by immunoblot, show equivalent levels of the wild-type and mutant SH3BP2.