Abstract
Collagen type XI is a constituent of the pericellular matrix of chondrocytes and plays a role in the regulation of fibrillogenesis. The amino-terminal domain of collagen type XI α1 chain is a noncollagenous structure that has been identified on the surface of cartilage collagen fibrils. The biochemical composition of the amino-terminal domain varies due to alternative splicing of the primary transcript. Recombinantly expressed α1(XI) amino-terminal domain isoforms were used in this study to investigate potential interactions. Purified products were analyzed for heparan sulfate binding properties. The results demonstrated that two additional binding sites exist within the α1(XI) amino-terminal domain, one within the amino propeptide and one within the variable region of the amino-terminal domain. Analysis of relative affinities indicated that the site located within the amino propeptide (site 1) was of similar affinity to sites that exist within the major triple helix of collagen type XI. Substitution of amino acid residues 147 to 152 within the amino propeptide by site-directed mutagenesis resulted in altered affinity for heparan sulfate. The binding site located within the variable region (site 2) demonstrated significantly higher affinity than other sites within the molecule. Displacement of collagen type XI within the pericellular matrix was observed in cell culture in the presence of excess heparan sulfate and by treatment with heparinase. These studies suggest two additional binding sites located within the noncollagenous amino-terminal domain that may play a role in the function of collagen type XI. The localization of collagen type XI within the pericellular matrix may be dependent upon interactions with heparan sulfate proteoglycans, and these are likely to take place in an isoform-specfic manner.
Collagens represent an extensive family of proteins that are found in the extracellular matrix. Collagens are assembled from α chains of which there have been reported at least 41 genetically distinct proteins comprising 28 different collagen types (1–3). All collagen family members are modular proteins characterized by regions of triple helix yet differ with respect to the combination and location of the nontriple helical domains present.
Collagen type XI, composed of α1, α2, and α3 chains, appears to be concentrated pericellularly and plays a role in the regulation of fibrillogenesis (4, 5). The presence of collagen type XI correlates to the location of thin collagen fibrils within the pericellular matrix (6). The α1 and α2 chains of collagen type XI each contain homologous noncollagenous amino-terminal structures that comprise two separate regions: an amino propeptide (Npp)2 and a variable region (VR). Together, the Npp and the variable region make up the amino-terminal domain (NTD) of the collagen α1(XI) chain. The α3 chain of collagen type XI is the gene product of Col2a1 and contains an amino-terminal domain that is distinct from the α1 and α2 chains (7).
Within the NTD portion of α1 and α2, the Npp domain demonstrates similarity to a domain found in laminin, neurexin, and sex hormone binding globulin (the LNS family members) and originally identified as the Thsp1 or TSPN domain of thrombospondin (8–12). Although the Npp domain shares features with other modular extracellular proteins, an understanding of the function of this domain is still incomplete.
The VR connects the Npp domain to the minor triple helix (13). The VR of the α1 and α2 chains possesses unique biochemical characteristics determined by alternative splicing of the mRNA (14). While the Col2a1 mRNA undergoes alternative splicing which results in two splice variants, the alternative splicing of three exons in Col11a1 is predicted to generate eight splice variants (7, 13). The individual splice variants are expressed during development and growth with unique tissue, temporal, and spatial specificities (16). Of the eight potential isoforms, three are most prevalent within cartilage tissue during fetal development and growth (17). In contrast, the splice variants of the α2 chain are present very early in differentiation but rapidly converge to a single splice variant that persists in developing cartilage (18). Isoform-specific functions may result from the differences in size and electrostatic characteristics of the individual splice variants during skeletal development, particularly in the case of the α1 chain that demonstrates sustained expression of several variants throughout development and growth.
After biosynthesis and secretion to the extracellular environment, the Npp of the α1 and α2 chains may be proteolytically processed. The Npp of the α2 chain is removed rapidly and completely after synthesis while the Npp of the α1 chain is processed more slowly (19). In vitro processing takes place at an isoform-specific rate and to different extents (20). The collagen α1(XI) Npp may still be detected on the surface of thin collagen fibrils of cartilage extracellular matrix by immuno-TEM (21) and on intact collagen molecules by immunoblot (17, 19).
Collagen type XI, as well as the closely related collagen type V, have been implicated in the regulation of collagen fibril growth. This activity may be mechanistically linked to the NTD, primarily by steric hindrance, although this domain may also be the site of interaction with other constituents of the extracellular matrix. The location of the NTD of collagen α1(XI) on the surface of thin heterotypic cartilage collagen fibrils (21) makes this domain a candidate binding site for such interactions.
In vitro studies have established that glycosaminoglycans, particularly heparan sulfate, interact with the major triple helix of collagen type XI (22). The interaction of pepsinized collagen type XI with heparan sulfate and/or other glycosaminoglycans has been shown to involve more than one site within the major triple helix (23). Heparin binding activity has also been described for the amino-terminal domain of the closely related α3 and α4 chains of collagen type V (24, 25).
In the present work, we describe the production and purification of recombinant isoforms of the NTD of collagen α1(XI) and identification of two additional heparan sulfate binding sites within the noncollagenous region. We also describe the individual substitution of four lysine residues by site-directed mutagenesis and the resulting changes in binding affinity for heparan sulfate. Furthermore, isoform-specific interaction properties determined by the nature of the variable region are presented. The NTD of α1(XI) may mediate interactions with heparan sulfate proteoglycans of the extracellular matrix or the cell surface contributing to the regulation of extracellular matrix organization and collagen fibrillogenesis.
EXPERIMENTAL PROCEDURES
Construction and Expression of the NTD α1(XI) Collagen Expression Vector
The NTD α1(XI) collagen sequences from rat were amplified and cloned as previously described (20). The recombinant isoforms were expressed in BL21 (DE3) Escherichia coli cells (Novagen, Madison, WI) using the pET11a bacterial expression vector (Stratagene, La Jolla, CA). The cells were grown in either small batch of one liter or in large batches in a 100-liter fermentor (Biostat 100D, Sartorius BBI, Bethlehem, PA). Expression was induced with 0.4 mm isopropyl α-d-thiogalactopyranoside at an A600 of 0.05 to 1.0. Two hours after induction, the cells were collected in a Sorvall high speed centrifuge or, in the case of large scale batches, a continuous flow centrifuge (Sharples AS-14, Alfa Laval, Richmond, VA).
Site-directed Mutagenesis
Residues Lys-147, Lys-148, Lys-149, and Lys-152 were altered to alanine using QuikChange site-directed mutagenesis (Stratagene). The expression vector, pET11aVO, was mutagenized using two complimentary oligonucleotide primers that introduced the desired mutations by changing the codons for lysine to alanine (Table 1). PfuTurbo DNA polymerase was used to generate the mutated plasmid, followed by DpnI treatment to digest the parental template and select mutation-containing DNA. Following transformation of E. coli XL1-Blue supercompetent cells and clonal selection, clones were amplified, purified (Novagen Mobius1000), and sequenced in both directions to confirm the desired mutation and the absence of unwanted mutations. Sequencing reactions were performed at the Molecular Research Core Facility, Idaho State University (Pocatello, ID). The mutagenized plasmids were transformed into E. coli BL21(DE3) cells containing the T7 RNA polymerase gene under the control of the lac promoter/operator for expression. The recombinant proteins were collected after the induction of the promoter by 0.4 mmisopropyl β-d-thiogalactopyranoside. Nomenclature for the mutated recombinant plasmids and proteins follows standard guidelines, e.g. pET11aVOK147A and α1(XI)VOK147A.
TABLE 1. Mutagenic primer pairs.
Site-directed alteration of amino acid residues within putative binding site 1 147-KKKITK-152 is shown. Mutagenic primers are shown for each targeted change.
| Wild type | pET11aVO | 5′-GACAATGATTGTCGATTGTAAGAAGAAAATCACAAAGCCCCTCGATAGAAG-3′ |
| Mutagenic primer pair K147A | CMB226S | 5′-GACAATGATTGTCGATTGTAAGAAGAAAATCACAAAGCCCCTCGATAGAAG-3′ |
| CMB227A | 5′-CTTCTATCGAGGGGCTTTGTGATTTTCTTCGCACAATCGACAATCATTGTC-3′ | |
| Mutagenic primer pair K148A | CMB228S | 5′-GACAATGATTGTCGATTGTAAGGCGAAAATCACAAAGCCCCTCGATAGAAG-3′ |
| CMB229A | 5′-CTTCTATCGAGGGGCTTTGTGATTTTCGCCTTACAATCGACAATCATTGTC-3′ | |
| Mutagenic primer pair K149A | CMB230S | 5′-GACAATGATTGTCGATTGTAAGAAGGCAATCACAAAGCCCCTCGATAGAAG-3′ |
| CMB231A | 5′-CTTCTATCGAGGGGCTTTGTGATTGCCTTCTTACAATCGACAATCATTGTC-3′ | |
| Mutagenic primer pair K152A | CMB232S | 5′-GACAATGATTGTCGATTGTAAGAAGAAAATCACAGCGCCCCTCGATAGAAG-3′ |
| CMB233A | 5′-CTTCTATCGAGGGGCGCTGTGATTTTCTTCTTACAATCGACAATCATTGTC-3′ |
Purification and Refolding of NTD α1(XI) Collagen Recombinant Proteins
Cells were lysed by resuspension in a hypotonic detergent solution (B-PER, Pierce). Inclusion bodies were separated from soluble proteins by differential centrifugation and washed in 0.05 m Tris-HCl, pH 7.6. Inclusion body proteins were solubilized in 50 mm sodium phosphate, pH 7.6, containing 6 m guanidine hydrochloride, 1mm 2-mercaptoethanol and 80 mm NaCl. Solubilized proteins were clarified by centrifugation at 18,000 × g, 4 °C for 30 min and applied to a nickel-nitrilotriacetic acid-agarose (Qiagen Inc., Valencia, CA) column at a flow rate of 1 ml/min. Absorbance at 280 nm and conductivity were monitored throughout purification. Bound protein was eluted using 250 mm imidazole. Protein in each fraction was assessed by SDS-polyacrylamide gel electrophoresis and Coomassie Blue staining. Proteins were purified to 97% homogeneity. After refolding as previously described (26), proper structure of folded protein was confirmed by circular dichroism in the far-UV range (260–178 nm) and mass spectrometry to confirm proper disulfide bond formation.
Purification of α1(XI) Collagen Triple Helical Domain
Collagen type XI was extracted and purified from the hyaline cartilage harvested from the ends of long bones of fetal bovine hind and forelimbs (Gem Meat, Garden City, ID) as described previously (27). Briefly, collagen types II, IX, and XI were selectively precipitated from the homogenate by differential salt precipitation using 0.5 m CH3COOH, containing 0.9, 1.2, and 2.0 m NaCl, respectively. Only collagen type XI was used in this study.
Heparin Affinity Chromatography
Recombinant splice variants and pepsinized type XI collagen major triple helix were dialyzed against 0.05 m Tris-HCl, pH 7.5, containing 0.15 m NaCl. Samples were applied to a heparin-agarose (Sigma) column. Protein was eluted from the column during a linear concentration gradient of NaCl. Absorbance at 280 nm and conductivity were monitored continuously throughout the experiment. Fractions were collected and protein composition was monitored by SDS-polyacrylamide gel electrophoresis and Coomassie Blue staining to determine the concentration of NaCl required for displacement of the protein.
Circular Dichroism
Purified recombinant isoforms were dialyzed extensively against 0.02 m sodium phosphate, pH 7.5, with 0.1 m NaF. Each sample was analyzed in the far-UV range (260–178 nm) at 4 °C by CD spectroscopy on a Jasco J-810 spectropolarimeter. Protein samples were examined in a circular demountable 0.01-mm path length Supracil cell with a total volume of 18 µl (catalog number 124-QS 0.01 mm, Hellma, Plainview, NY). Three spectra and three base lines were collected at 1-nm intervals over the wavelength range from 270 to 178 nm at 4 °C. The high tension never exceeded 500 V.
For these experiments, the concentration of each sample was determined in triplicate by amino acid analysis. Samples were gas-phase-hydrolyzed using 300 µl of 6 n HCl, 2% phenol at 110 °C for 22 h, and the hydrolysates were analyzed on a Beckman 6300 amino acid analyzer using sodium citrate buffers (AAA Services Laboratory, Boring, OR). As an alternative method, protein concentration was also determined by measuring the absorbance at 280 nm in 6 m guanidinium HCl with molar extinction coefficients calculated using ProtParam (28). Heparan sulfate was diluted to a 10 mg/ml stock in 18 mΩ water. The instrument was calibrated using d(+)-camphor sulfonic acid (Sigma) at 192 and 290 nm.
CD Spectral Analysis
CD spectra were processed using CDtool software (29). The spectra were averaged and base lines subtracted and smoothed with a Savitsky-Golay filter (30) with a smoothing window of seven. The calculated Δε values were determined using the mean residue weight value calculated from its sequence and calibrated to d(+)-camphor sulfonic acid. Reported error is the pooled standard deviation contributed by sample and baseline spectra.
Secondary structure analyses were performed using the DICHROWEB Web server (31–33) using the CONTINLL algorithm (34, 35). Reference data set 1 was used in fitting the spectra (36, 37). The normalized root-mean-square deviation parameter was used to evaluate the goodness of fit of the calculated data to the experimental data (38), defined as Σ[(θexp – θcalc)2/(θexp)2]1/2 over all wavelengths, where θexp and θcal are the experimental ellipticities and the back-calculated ellipticities, respectively, of the spectra for the predicted structure.
Heparan Sulfate Activation
Heparan sulfate (Sigma) was diluted to 0.5 mg/ml in 10 mm sodium acetate, pH 4.0. Sodium periodate was added to a final concentration of 3 mg/ml. The solution was allowed to react for 30 min at room temperate in the absence of light with gentle agitation.
Conjugation of Heparan Sulfate to Hydrazide-modified Surface
The activated heparan sulfate was diluted to 20µg/ml in 0.1msodium acetate, pH 5.5, and 100µl added to each well of a 96-well Carbo-BIND plate (Corning). The plate was incubated for 1 h at room temperature in the absence of light. Plates were rinsed three times with phosphate-buffered saline containing 0.05% (v/v) Tween 20 (PBST). Blocking and deactivation of any nonreacted hydrazide groups was accomplished with 1% (w/v) nonfat dry milk in 50 mm Tris, pH 8.2. Plates were rinsed once with PBS. Protein solution was diluted to 1 mg/ml in PBS, 10% horse serum. Serial dilutions of the protein of interest were performed to cover a range from 1 to 0.02 mg/ml. Aliquots of 100 µl per well were added and allowed to incubate for 30 min at 37 °C. Wells were rinsed three times with PBS prior to the addition of nickel-nitrilotriacetic acid-horseradish peroxidase conjugate in PBS with 10% horse serum to each well. After an overnight incubation at 4 °C, the wells were rinsed three times with PBS and incubated for 40 min with the horseradish peroxidase substrate 3,3′,5,5′-tetramethylbenzidine. To stop the reaction, 0.3 m sulfuric acid was added, and absorbance was measured at 450 nm. Data were analyzed using GraphPad Prism (GraphPad software, San Diego, CA) with a user-specifid equation. The Hill equation, Y = ((Bmax)*(Xn))/((Kd,n) + (Xn)), was used to fit the fractional binding saturation data, where n is a measure for the extent of cooperativity, with n > 1 indicative of positive cooperativity.
Cell Culture Conditions
A cell line was derived in our laboratory from the Swarm rat chondrosarcoma (39). To allow the observation of newly synthesized pericellular matrix, cells were treated with 0.02% (w/v) collagenase 1A (Sigma) for 8 min followed by 0.25% trypsin/EDTA for 5 min immediately before plating. Cells were plated at 3.5 × 104 cells/cm2 onto poly-d-lysine/laminin-coated chamber slides (BD Biosciences) and incubated in DMEM supplemented with 10% fetal bovine serum, 50 µg/ml ascorbate 2-phosphate, 50 units/ml penicillin, and 50 µg/ml streptomycin. Accumulated pericellular matrix was analyzed 48 h after plating by indirect immunofluorescence. Cells were grown in the presence of 0.1, 0.2, 0.5, and 1 unit/ml heparinase III to determine optimum concentration by monitoring the depletion of heparan sulfate by indirect immunofluorescence using a heparan sulfate-specific monoclonal antibody (Upstate Biotechnology, Charlottesville, VA). Experiments were carried out with 1 unit/ml heparinase III or 100 µg/ml heparan sulfate as indicated in the figure legends of Figs. 7 and 8.
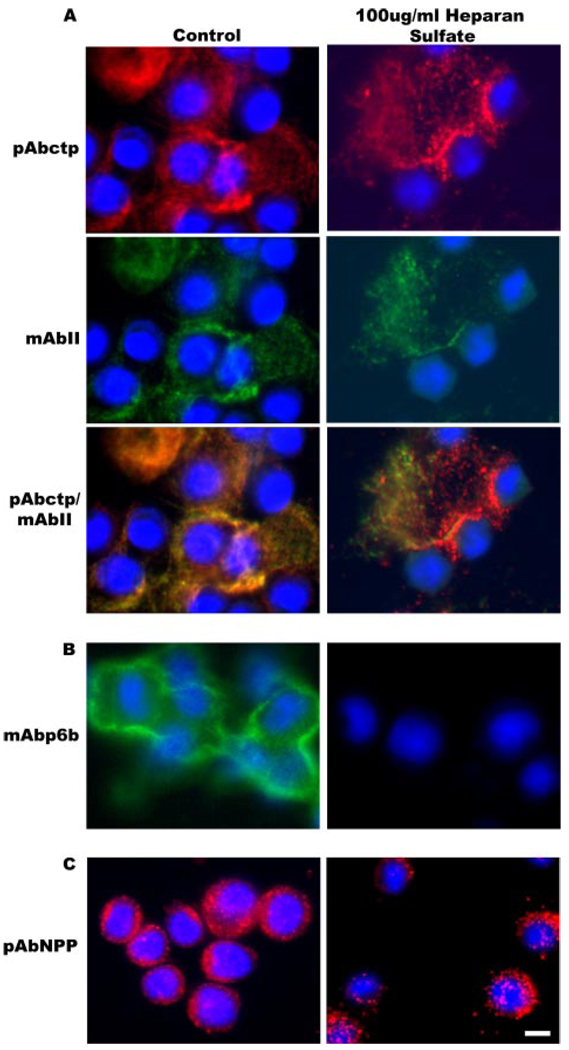
FIGURE 7. Specific localization of Npp[p6b7] within the pericellular matrix.
Cells were maintained in culture for 48 h during which the pericellular matrix was allowed to assemble in the absence or presence of heparan sulfate. A, double label with polyclonal antibody to the carboxyl telopeptide (ctp) of collagen α1(XI), which remains associated with the major triple helix after proteolytic removal of the carboxyl propeptide, and a monoclonal antibody to collagen type II. Localization of collagen type XI and collagen type II are not significantly affected by the presence of heparan sulfate in the growth medium. B, immunolabel with monoclonal antibody to p6b (containing heparan sulfate binding site 2). Note diminished labeling with monoclonal antibody to p6b in the presence of heparan sulfate. C, immunolabel with polyclonal antibody to collagen α1(XI)Npp (containing heparan sulfate binding site 1). Note punctate distribution of the Npp epitope with relatively unaltered localization in the presence of heparan sulfate. Nuclei were stained with DAPI (blue). Scale = 10 µm.
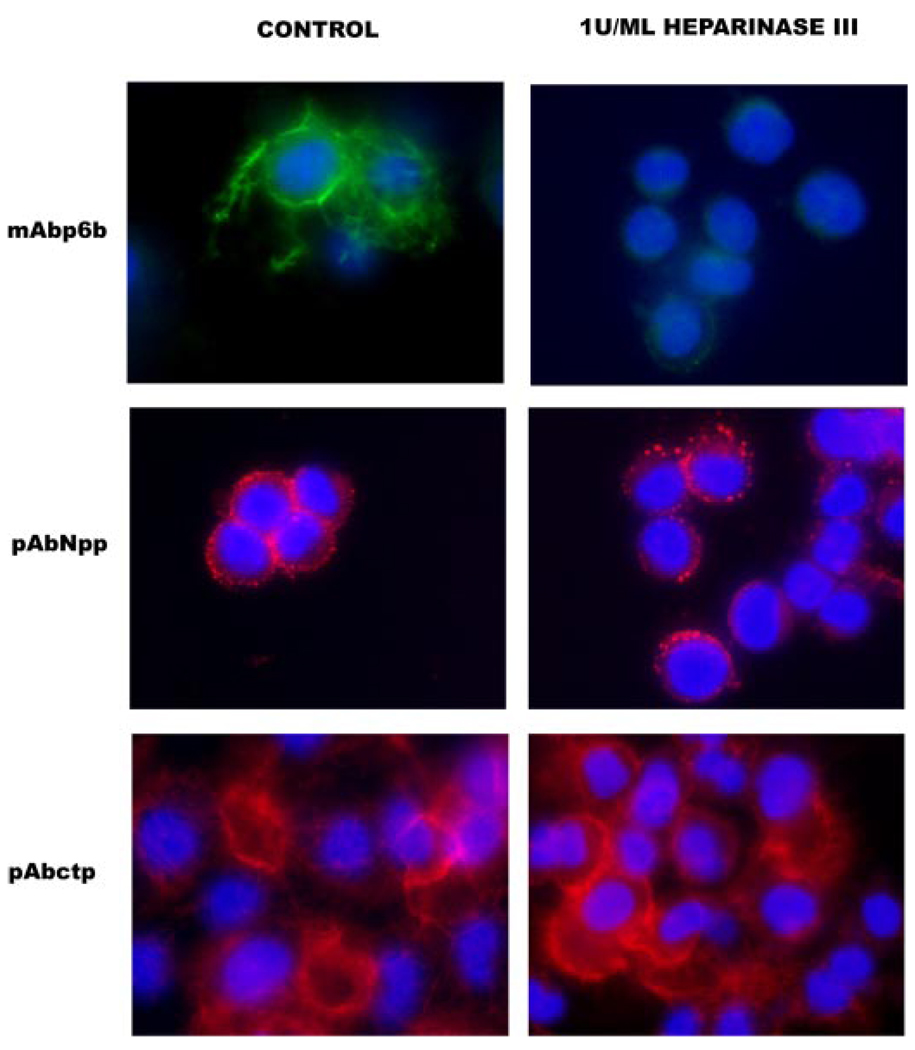
FIGURE 8. Diminished pericellular p6b staining upon heparinase III treatment.
Cells were maintained in culture for 48 h during which pericellular matrix was allowed to assemble in the absence or presence of heparinase III. In the presence of 1 unit/ml heparinase III, mAbp6b showed a decreased level of staining in the pericellular matrix. No alteration was observed in the staining pattern of the major triple helix or Npp of collagen type XI. Nuclei were stained with DAPI (blue). Scale = 10 µm.
Antibodies Used to Investigate Location of Collagen Type XI within Pericellular Matrix
A polyclonal antibody to the rat α1(XI) carboxyl telopeptide of collagen α1(XI) has been described elsewhere (4). The 51-amino acid synthetic peptide comprising the entire p6b sequence was used for the preparation of a monoclonal antibody and described previously (17). A monoclonal antibody to type II collagen was obtained commercially (Neomarkers, Fremont, CA). A polyclonal antibody was raised against the purified 223-amino acid rat recombinant amino propeptide of the collagen α1(XI) chain expressed in an E. coli system as described previously (4).
Immunofluorescence
Cells were fixed on chamber slides in −20 °C methanol for 5 min. The slides were rinsed in 50 mm Tris-buffered saline (TBS) and then permeabilized in TBS-0.5% Triton X-100 (TX) for 10 min. The slides were washed three times for 5 min in TBS-0.1% TX, followed by blocking in TBS-0.1% TX, 2% bovine serum albumin for 10 min. After incubation with the primary antibody, cells were washed five times for 5 min in TBS-0.1% TX. Secondary antibody, rhodamine (TRITC)-conjugated AffiniPure donkey anti-rabbit IgG or fluorescein isothiocyanate-conjugated anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA), diluted 1:100 in TBST, was applied to slides and incubated for 1 h in a humidified chamber at 37 °C. The slides were then washed three times for 5 min in TBS-0.1% TX and incubated in 3.0 µg/ml of DAPI for 10 min (Molecular Probes, Eugene, OR). The slides were washed three times 5 min in TBS-0.1% TX, rinsed in TBS, drained, mounted with Vectashield (Vector Laboratories, Burlingame, CA), and sealed. For microscopic observation and photomicrography of the fluorescently labeled cells, an Olympus BX60 fluorescence microscope equipped with a PM-10AD system was used. The fluorescent molecules were excited with a 100-watt mercury lamp. TRITC- or fluorescein isothiocyanate-labeled molecules were detected with a filter set having 510–560 nm wavelength bandpass, 565 nm dichroic beamsplitter, and 575–645-nm emission filters.
RESULTS
Collagens are modular proteins, comprising distinct domains in addition to the collagenous major triple helix. The α1 chain within the collagen type XI triple helical molecule is shown schematically in Fig. 1 as a linear array of domains. The Npp of the α2 chain is rapidly removed after synthesis while the α3 chain (primarily type IIB) is very short and does not undergo further processing (7, 19).
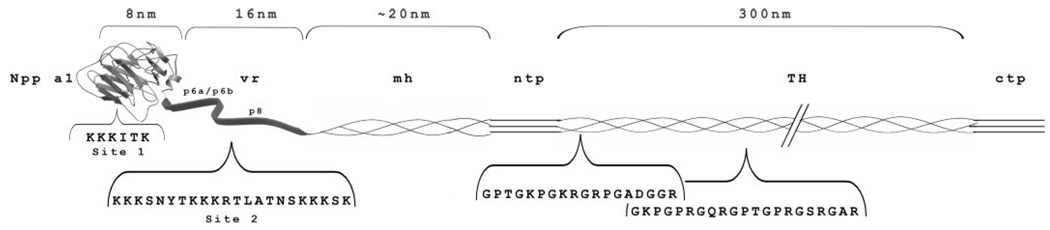
FIGURE 1. Schematic representation of putative heparan sulfate binding sites within the collagen type XI molecule.
Domains of collagen type XI are shown as a schematic, beginning on the left with 1) the 223 residue amino propeptide (Npp); 2) a variable region (vr); 3) the minor triple helix (mh); 4) the amino telopeptide (ntp); 5) the major triple helix (TH); 6) the carboxyl telopeptide (ctp); and 7) the carboxyl propeptide (not shown in figure). The Npp and the variable region together constitute the amino-terminal domain (NTD) referred to in the introduction and under “Experimental Procedures,” “Results,” and “Discussion.” Amino acid sequences of putative heparan sulfate binding sites 1 and 2 are indicated as well as the sequence of previously reported binding sites within the major triple helix. The α2(XI) Npp is rapidly removed after synthesis and is not shown in this schematic diagram. The α3(XI) Npp is also not represented in this diagram.
To study the structure and interactions of the NTD of α1(XI) collagen, sequences encoding the three most prevalent splice variants from exons 2 through 9 of the rat α1(XI) polypeptide were inserted into a prokaryotic expression vector. Design of the polypeptides as recombinant proteins included a COOH-terminal His6 tag and the absence of the native signal peptide. To study the interactions of α1(XI) Npp in the absence of the adjacent variable region, sequences encoding exons 2 through 5 were used. The recombinant proteins were isolated from inclusion bodies, purified by chelation chromatography, and subsequently unfolded and refolded. Purified proteins were characterized by SDS-polyacrylamide gel electrophoresis shown in Fig. 2. Amino acid analysis and analysis of tryptic peptides by electrospray ionization tandem mass spectrometry were used to confirm identity of the purified recombinant proteins. In addition, CD spectra were compared with previously obtained data to evaluate the secondary structural composition of the refolded proteins (data not shown).
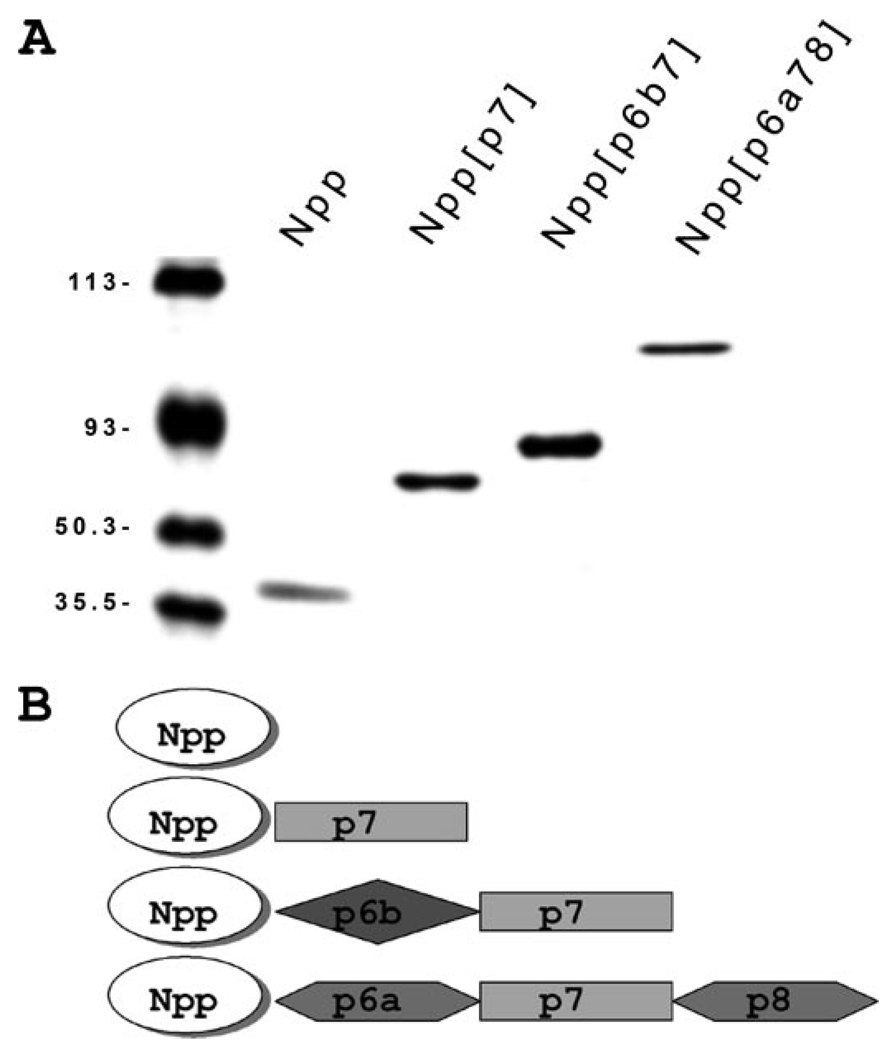
FIGURE 2. Protein expression and purification of recombinant fragments of collagen α1(XI) chain Npp and the three most prevalent amino-terminal domain splice variants.
A, purified recombinant collagen α1(XI) fragments were analyzed by SDS-polyacrylamide gel electrophoresis using a 10.5% acrylamide gel. Protein fragments generated for this study include: the amino propeptide (Npp), a fragment containing p7 of the variable region adjacent to the Npp domain (Npp[p7]), a fragment representing the amino-terminal domain with p6b and p7 adjacent to the Npp domain (Npp[p6b7]), and a fragment containing p6a, p7, and p8 adjacent to the Npp domain (Npp[p6a78]). Note that p6b7 is predicted to contain a heparan sulfate binding site. Note also that p6a78 is the largest, most negatively charged of the splice variants of α1(XI). Molecular weight markers are included in the left-hand lane. B, modular representation of the recombinant fragments used in this study, corresponding to proteins shown in A.
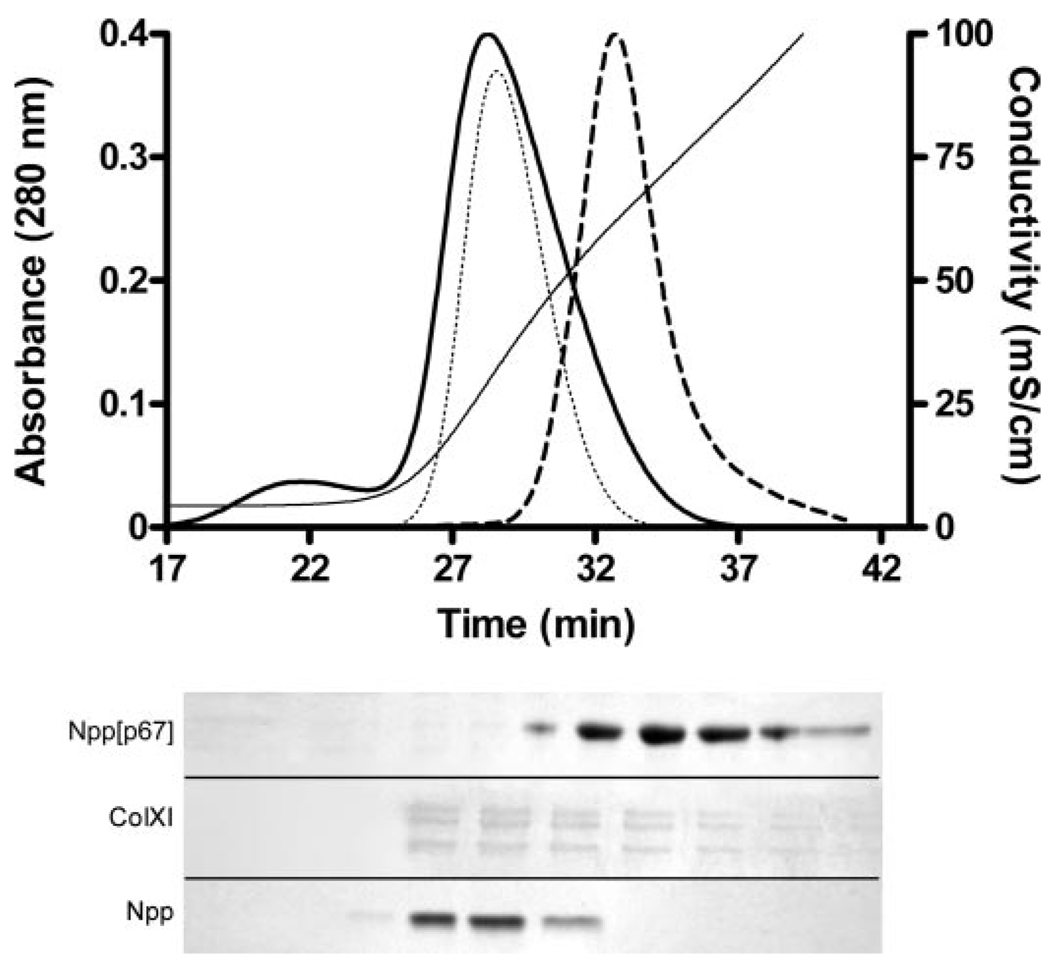
The affinity of the α1(XI) Npp for heparin was determined by heparin-agarose affinity chromatography. Elution with a [NaCl] gradient of increasing concentration was used to determined conditions of dissociation. The Npp domain of collagen α1(XI) was eluted from the heparin-agarose column at approximately the same [NaCl] as was the major triple helix of collagen type XI (Fig. 3). Under the same conditions, the major triple helix of collagen type II is eluted from the column at 0.12 m NaCl and was therefore found in the flow-through under the conditions of the experiment, consistent with previous results (19). The splice variant containing p6b within the variable region was eluted from the heparin-agarose column at a mean [NaCl] of 0.85 m, which is higher than that required for elution of either the major triple helix of collagen type XI or the α1(XI) Npp (Fig. 3). The mean [NaCl] at which elution was observed for the major triple helix, the Npp domain, and the three most prevalent splice variants are summarized in Table 2.
FIGURE 3. Heparin affinity chromatography of fragments of collagen α1(XI).
Upper panel, elution profiles are shown for Npp (dotted line), Npp[p6b7] (dashed line), and the triple helical region of collagen type XI (solid bold line). Absorbance at 280 nm was normalized to the maximum height of the Npp[p6b7] peak. Conductivity during NaCl gradient elution of the protein is indicated on the right-hand axis (solid line). Lower panel, Coomassie Blue-stained SDS-polyacrylamide gels of successive 1-ml fractions collected during elution of the collagen α1(XI) fragments. Alternating fractions are shown for Npp. The position of the splice variant containing p6b within the variable region (Npp[p6b7]), the amino propeptide (Npp), and the major triple helix of type XI collagen after pepsin treatment (ColXI) are indicated. Collagen binds Coomassie Blue more weakly than noncollagenous proteins, explaining in part, the fainter bands in the ColXI gel of the lower panel. The position of the bands in the lower panel are shown in relation to the chromatograms in the upper panel.
TABLE 2. Binding of N-terminal fragments of collagen XI α1 chain to heparin.
Heparin binding was determined by affinity chromatography in 0.05m Tris-HCl, pH 7.4. Binding strength is denoted by NaCl concentration required for displacement of protein from heparin agarose column.
| Isoform/Fragment | [NaCl] | Conductivity |
|---|---|---|
| m | mS/cm | |
| Collagen XI triple helix | 0.449 | 33.0 |
| α1(XI) NTD Npp | 0.404 | 29.7 |
| α1(XI) NTD (Npp[p6b7]) | 0.847 | 62.3 |
| α1(XI) NTD (Npp[p7]) | 0.309 | 22.7 |
| α1(XI) NTD (Npp[p6a78]) | 0.253 | 18.6 |
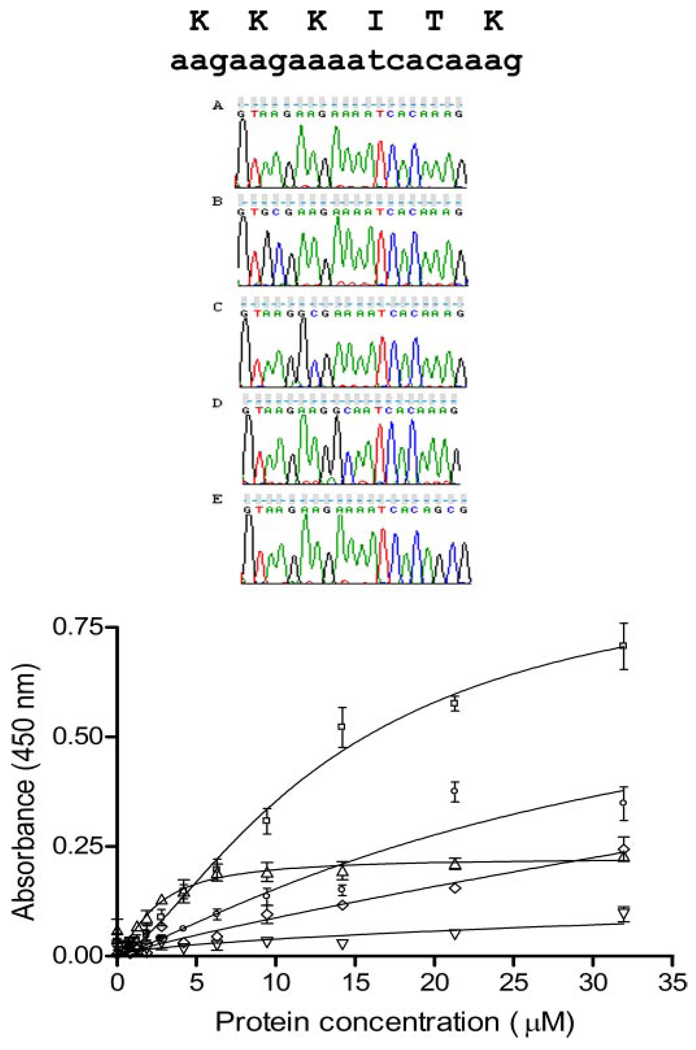
To investigate the potential contribution of individual amino acids within the α1(XI)Npp domain, previously identified candidate amino acid residues (11) were modified. Lysine 147 was modified to an alanine, as was lysine 148, lysine 149, and lysine 152, by site-directed mutagenesis. The resulting recombinant proteins were analyzed for changes in affinity for heparan sulfate by fitting the binding data to the Hill equation as described under “Experimental Procedures.” The conversion of each of three lysine residues to alanine individually resulted in lower affinity for heparan sulfate as determined by a solid phase binding assay (Fig. 4). The dissociation constant between α1(XI)Npp and heparan sulfate increased 2-fold upon modification of lysine 152 to an alanine. Modification of lysine 149 to alanine resulted in a dissociation constant approximately 2 orders of magnitude higher than the original unmodified protein. Substitution of lysine 148 with alanine eliminated specific interaction of α1(XI)Npp with heparan sulfate altogether. Unexpectedly, substitution of lysine 147 with alanine resulted in a 5-fold increase in affinity (Kd) for heparan sulfate with a decrease in Bmax (Table 3).
FIGURE 4. Relative affinities for heparan sulfate after site-directed mutagenesis.
Site-specific mutagenesis of heparan sulfate binding site 1. Upper panel, sequence verification of site-directed change in sequence. A, wild type sequence; B, K147A mutant; C, K148A mutant; D, K149A mutant; E, K152A mutant. Lower panel, heparan sulfate solid phase binding assay. Binding curves are shown for wild type amino-terminal domain fragment (NTD[p7], squares) and four mutants: K147A (triangles), K148A (inverted triangles), K149A (diamonds), K152A (circles). Specific binding is almost eliminated for K148A and K149A and decreased significantly for K152A. In contrast, a decrease in maximal binding with a decrease in nonspecific binding (nonsaturable binding) is observed for K147A with improvement of Kd(see Table 3).
TABLE 3.
Bmax, n, and Kd values for site 1 binding
| NTD[p7] | K147A | K148Aa | K149Aa | K152A | |
|---|---|---|---|---|---|
| Best-fit values | |||||
| Bmax | 0.997 | 0.209 | NSB | NSB | 0.754 |
| n | 1.47 | 2.23 | ND | ND | 1.18 |
| Kd | 13.6 | 2.32 | NSB | NSB | 31.7 |
| Standard error | |||||
| Bmax | 0.118 | 0.013 | ND | ND | 0.455 |
| n | 0.183 | 0.610 | ND | ND | 0.305 |
| Kd | 2.66 | 0.330 | ND | ND | 30.8 |
| 95% confidence intervals | NTD[p7] | K147A | K152A | ||
| Bmax | 0.757 to 1.24 | 0.182 to 0.236 | 0.0 to 1.68 | ||
| Kd | 8.13 to 19.0 | 1.65 to 2.99 | 0.0 to 94.5 | ||
NSB, nonsaturable binding. ND, not determined.
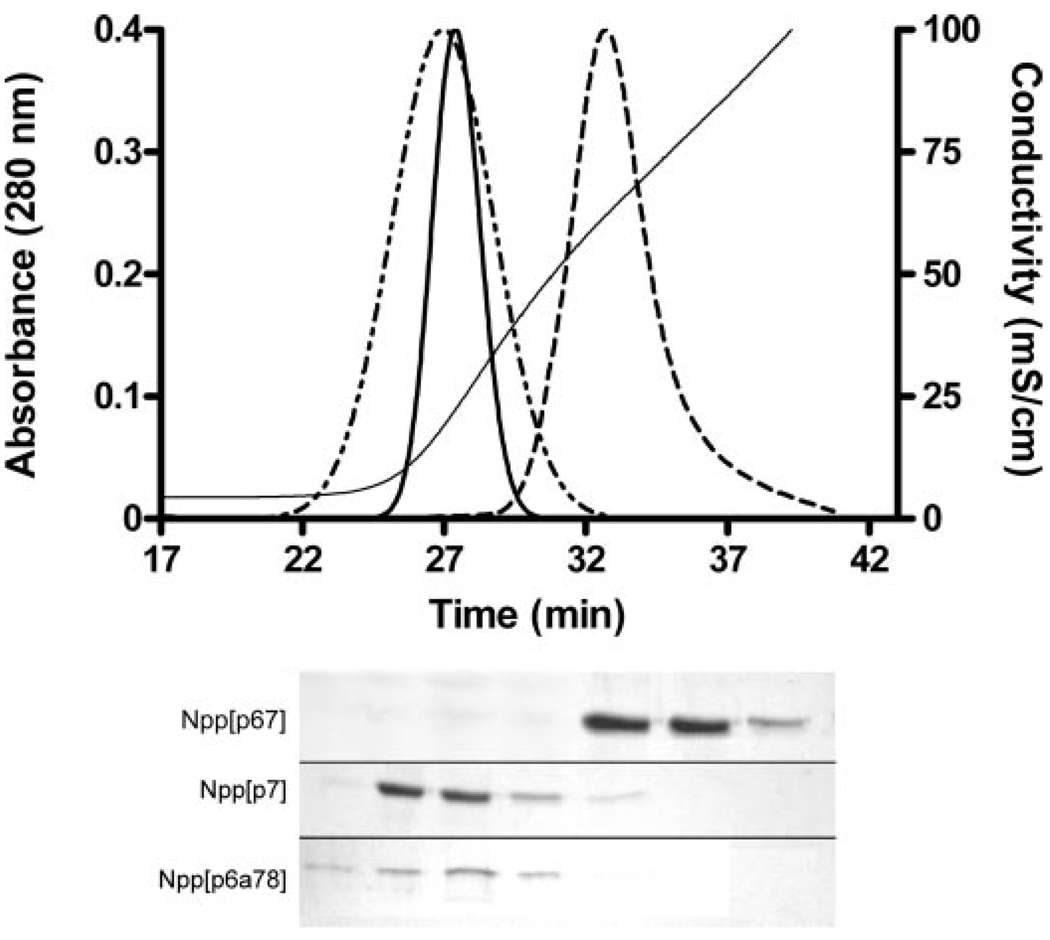
The three most prevalent isoforms of the variable region were generated as recombinant proteins in tandem with the adjacent Npp domain and used to determine the effect of alternative splicing on subsequent potential interactions. The isoform containing p6b eluted at a higher [NaCl] than did the splice variants containing p6a and p8 within the variable region, α1(XI)Npp[p6a78], or the isoform created by skipping exons encoding p6a, p6b, and p8, α1(XI)Npp[p7] (Fig. 5). In comparisosn, the major triple helix of collagen type XI, containing two heparan sulfate binding sites, eluted at a sodium chloride concentration of 0.45 m under the same conditions (Fig. 3). The fragments representing the NTD alternative splice variants Npp[p7] and Npp[p6a78] were eluted from the heparin column at 0.31 m NaCl and 0.25 m NaCl, respectively, summarized in Table 2.
FIGURE 5. Heparin affinity chromatography comparing variable regions of collagen α1(XI).
Upper panel, elution profiles are shown for Npp[p6a78] (dotted and dashed line), Npp[p6b7] (dashed line), and Npp[p7] (solid bold line). Absorbance at 280 nm was normalized to the maximum height of the Npp[p6b7] peak. Conductivity during NaCl gradient elution of the protein is indicated on the right-hand axis (solid line). Lower panel, SDS-polyacrylamide gels of alternating 1-ml fractions collected during elution of the collagen α1(XI) splice variants. The position of the splice variants Npp[p6b7], Npp[p7], and Npp[p6a78] are indicated. The position of the bands in the lower panel are shown in relation to the chromatograms in the upper panel.
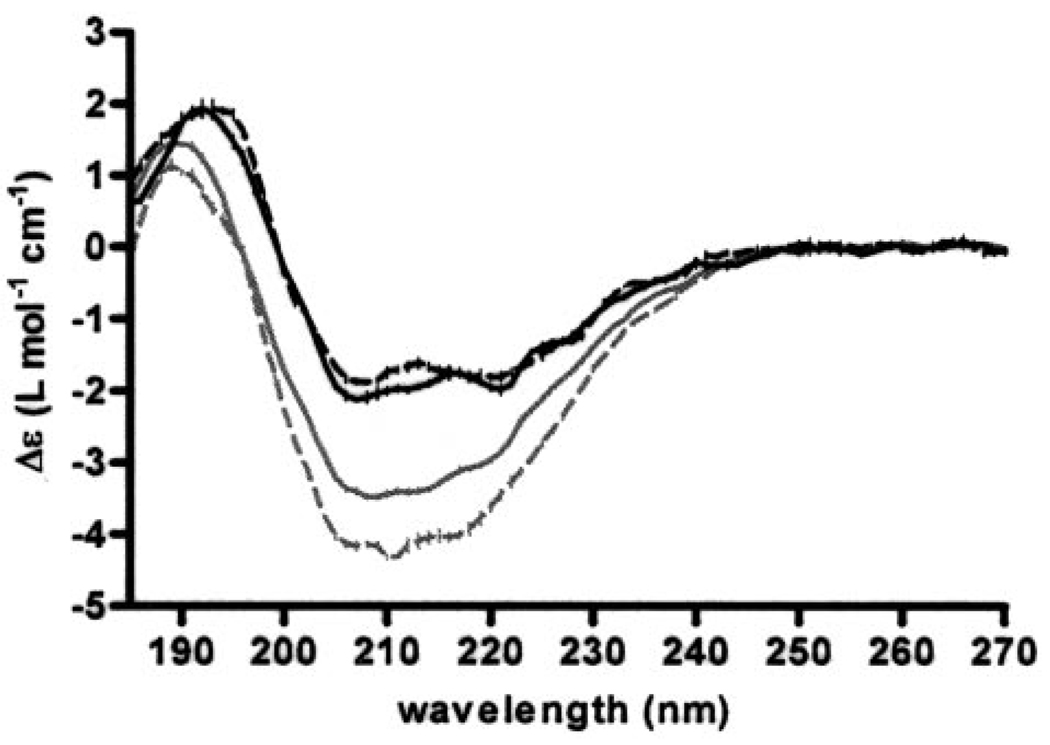
CD spectra of recombinant amino-terminal splice variants with and without the p6b contained within the variable region were measured in the absence and presence of heparan sulfate. Fitting of the far-UV CD spectra indicated overall conservation of secondary structure with subtle increases in periodic secondary structure for both proteins in the presence of heparan sulfate (Fig. 6). The structural change in Npp indicated an increase in the α-helical content, from 17 to 20% in the presence of heparan sulfate. Changes in α-helix and β-sheet content (20 to 22% and 26 to 28%, respectively) were observed for Npp[p6b7] in the presence of heparan sulfate, with a decrease in β-turn (24 to 20%) (Table 4).
FIGURE 6. Circular dichroism.
Far-UV CD spectra of Npp[p7] and Npp[p6b7] are shown. CD spectra showing overlay of Npp[p7], 4.78 mg/ml, with (gray dashed line) and without (gray solid line) 0.25 mg/ml heparan sulfate and overlay of Npp[p6b7], 3.40 mg/ml, with (black dashed line) and without (black solid line) 0.25 mg/ml heparan sulfate. Note subtle change in spectra in the presence of heparan sulfate indicting a minor change in periodic secondary structure of the protein upon binding.
TABLE 4. Circular dichroism.
Far-UV CD spectral fit. The percents of α-helix, β-sheet, and unordered structured were calculated using ContinII with reference protein data set 1. The normalized root mean square deviation (NRMSD) parameter was used to evaluate the goodness of fit of the calculated data to the experimental data.
| α-Helix | β-Sheet | β-Turn | Unordered | NRMSD | |
|---|---|---|---|---|---|
| Npp[p7] | 17% | 23% | 25% | 35% | 0.165 |
| Npp[p7] + HS | 20% | 23% | 25% | 32% | 0.262 |
| Npp[p6b7] | 20% | 26% | 24% | 30% | 0.089 |
| Npp[p6b7] + HS | 22% | 28% | 20% | 30% | 0.184 |
To investigate the role of interactions between collagen type XI and heparan sulfate in a cell culture system, newly synthesized pericellular matrix was allowed to accumulate for 48 h after trypsin/collagenase treatment. A difference in the indirect immunofluorescence staining pattern was detected for the splice variant containing p6b when grown in the presence of excess heparan sulfate, while no change was observed in the localization of the majority of collagen type XI molecules as indicated by the c-telopeptide epitope, as well as the collagen type II (Fig. 7). In addition, treatment with heparinase III to reduce the quantity of heparan sulfate moieties within the pericellular matrix resulted in decreased staining for p6b without significant alteration to the staining pattern of the major triple helix or the amino propeptide of collagen type XI (Fig. 8).
Results from the analysis of the amino acid sequence of the putative heparan sulfate binding sites of collagen type XI are presented in Table 5. The putative heparan sulfate binding site of collagen α1(XI) Npp, denoted as site 1, is conserved among several species including chick, human, bovine, dog, zebrafish, rabbit, and rat (Table 5A). The putative binding site 1 also exists within the related collagen α1(V) and collagen α2(XI) chains but not in the case of the collagen α3 and α4(V) chain (Table 5B). The putative binding site within the variable region, denoted as site 2 of collagen α1(XI) chain, is highly conserved among species (Table 5C). No analogous site was identified in collagen α1(V) or α2(XI) chains. A heparin binding site within the corresponding region of collagen α3 and α4(V) has been reported (24, 25), however, the amino acid sequence is not homologous to the collagen α1(XI) variable region chain.
TABLE 5. Amino acid sequence conservation analysis.
Amino acid sequence analysis of putative heparan sulfate binding sites in related proteins is shown.
| A. Species | Npp site 1 XBBBXXBX |
|---|---|
| Chick | CKKKITKF |
| Human | CKKKTTKP |
| Bovine | CKKKTTKP |
| Dog | CKKKTTKP |
| Zebrafish | CKKKLTKP |
| Rabbit | CKKKVTKF |
| Rat | CKKKITKP |
| B. V/XI chains |
Npp site 1 XBBBXXBX |
| α1(XI) | CKKKITKP |
| α1(V) | CKKKITKF |
| α2(XI) | CKKRVTRP |
| C. Species |
Variable region site 2 α1(XI) BBBXXXXBBBBXXXXXXBBBXBBXXXXBXXBXXXBBBBBXXXXXBXBX |
| Rat | KKKSNYTKKKRTLATNSKKKSKMSTTPKSEKFASKKKKRNQATAKAKL |
| Mouse | KKKSNYTKKKRTLTTNSKKKTKKFTSPKSEKFASKKKKRNPATAKAKL |
| Human | KKKSNFKKKMRTVATKSKEKSKKFTPPKSEKFSSKKKKSYQASAKAKL |
| Bovine | KKKFSFKMKMKTVATNSKKKSKKFTPPKSEKFASKKKKTYQAAAMAKL |
| Chicken | KKKAMVKKKKRTVATSSKDKSQKATTKKSEKYASKKKKSYQAATKDKL |
DISCUSSION
This work has identified two heparan sulfate binding sites within the collagen α1(XI) chain. The new sites are in addition to the previously reported glycosaminoglycan binding sites of the pepsinized triple helical domain (23).
Although the amino-terminal domain of the α1(XI) collagen chain is in close association with the α2 and α3 (XI) chains within a triple helical molecule immediately after synthesis and secretion, the Npp of the α2 chain is rapidly removed after synthesis while the α3 chain (primarily type IIB) is very short and does not undergo further processing (7, 19). In contrast, the amino-terminal domain of collagen α1(XI) is retained on the surface of the collagen fibril. To better understand the function of the α1(XI) amino-terminal domain isoforms which extend from the surface of the collagen fibrils of cartilage, the three most prevalent splice variants of collagen α1(XI) amino-terminal domain were generated as recombinant proteins.
The triple helical domain of collagen XI bound to heparin agarose under the conditions of this experiment and eluted at a mean [NaCl] of 0.45 m. This finding is consistent with published results (23). The Npp fragment also bound to heparin agarose and eluted at a mean [NaCl] of 0.40 m, demonstrating similar affinity for heparin. The Npp domain is proteolytically removed from the major triple helix at varying rates and to varying extents during matrix assembly in an isoform-dependent manner. Furthermore, the Npp domain can be detected as a separate protein in the extracellular matrix (40). Interaction between the Npp domain and heparan sulfate may play a role in the function of the Npp domain in the extracellular matrix while it is still attached to the collagen fibril and perhaps in an as yet undefined way after proteolytic removal from the collagen fibril surface.
The interaction with heparan sulfate mediated by site 1 (within the Npp domain) was further investigated using site directed mutagenesis to determine the role of individual amino acids within the predicted binding site. Previous analysis of the surface distribution of amino acid side chains and molecular modeling with a Lamarkian genetic algorithm (41) energy evaluation of an Npp-heparin interaction indicated a putative site of interaction centered around amino acid residues 147–152 (11). Based on our results, amino acid residues Lys-148, Lys-149, and Lys-152 appear to have a moderate to strong influence on the ability of α1(XI)Npp to interact with heparan sulfate, as the dissociation constant decreased after substitution of each of these residues. In contrast, a K147A substitution resulted in a 5-fold increase in affinity (Kd) for heparan sulfate. One possible explanation for this observation is that the K147A change may allow the adjacent downstream amino acid residues increased accessibility for heparan sulfate interaction. Cysteine 146 is immediately adjacent to lysine 147 and forms a disulfide bond with cysteine residue 200. The region immediately surrounding the Cys-147-Cys-200 disulfide bond may be constrained with low flexibility. The mutation K147A may increase accessibility of the neighboring amino acid residues to facilitate interaction with heparan sulfate. Positive cooperativity was observed for this binding event, as indicated in Table 3 by the value for n greater than 1. A decrease in Bmax was observed for the K147A mutation, potentially indicating a decrease in the number of binding sites for heparan sulfate.
The fragments representing the NTD with alternative splice variants Npp[p7] and Npp[p6a78] eluted from the heparin column at a slightly lower NaCl concentration than did the major triple helix of collagen type XI. Both exhibited a similar affinity for heparin as the Npp domain alone, indicating that adjacent protein domains do not significantly alter or modify interactions between collagen α1(XI) Npp and heparan sulfate. A comparison of collagen type XI triple helical molecules to collagen type II triple helical molecules with respect to their affinity for heparin demonstrates that the interaction between the collagen type XI domains and heparin are all stronger than the interaction of collagen type II and heparin, indicating the potential for physiological impact of the differences in affinities. Collagen type II can be eluted at 0.12 m NaCl in part because it lacks the heparin binding domains found in the triple helical region of collagen type XI and therefore does not bind to the heparin agarose column in 0.15 m NaCl, the experimental conditions used for these studies. In contrast, the triple helical collagen type XI molecule binds and is eluted at 0.45 m NaCl under the experimental conditions used in this study. Additional structural studies may be required to understand the complete effect of adjacent domains on the binding site within the Npp domain, which, however, are beyond the scope of this study. The recombinant fragment containing both putative sites 1 and 2, Npp[p6b7], interacted with heparin-agarose more strongly than any of the other splice variants, strongly supporting the hypothesis of an additional high affinity binding site within the p6b region.
Subtle changes in CD spectra were observed in the presence of heparan sulfate, suggesting a small conformational change upon interaction. Additionally, our results suggest that the variable region may contain ordered structure (26).
The putative binding sites identified in this study share common features with other members of the collagen V/XI family. Sequence analysis indicated potential interaction with heparan sulfate for other chains of the collagen type V/XI family that correspond to sites 1 and 2 of the NTDα1(XI) described in this study. Based upon sequence similarity, all type V/XI collagen α chains except the α3 and α4(V) chains are predicted to have a heparan sulfate interaction site located within the Npp domain (site 1). On the other hand, a high affinity binding site corresponding to site 2 has been identified within the α3 and α4(V) chain (24, 25), indicating that interaction with heparan sulfate may be a common feature of all of the collagen V/XI family members and may contribute to their shared function in collagen fibril diameter regulation. An additional function may exist for the high affinity binding site within p6b.
Immunofluorescence studies demonstrated changes in the staining pattern using an antibody directed to the p6b portion of the variable region. In the presence of excess free heparan sulfate, proteins within the pericellular matrix that contained the epitope for the p6b antibody were diminished. In addition, when heparan sulfate groups were removed by heparinase III treatment, staining for p6b was reduced. These results suggest that the immunolocalization of the p6b epitope and collagen molecules containing the p6b epitope within the pericellular matrix is dependent upon heparan sulfate moieties. These data may be explained by both the displacement of proteolytically cleaved p6b-containing N-propeptides and the intact pN-type XI collagen molecules. Western blot data (data not shown) analyzing the proteins released into the medium from cell culture support the interpretation that both the proteolytic cleavage products (55 kDa) and the intact collagen molecules (>150 kDa) are displaced from the pericellular matrix but specifically those that contain the p6b epitope.
Collagen types V/XI can be detected on the surface of the heterotypic collagen fibrils (16, 21) and have been implicated in the regulation of collagen fibril nucleation and growth. We suggest that the mechanism for this function relies on both steric hindrance as well as interaction with extracellular matrix components. The amino-terminal domain remains on the collagen fibril for an extended period of time in the case of collagen α1(XI) (19). An interaction between the NTD of α1(XI) and heparan sulfate groups may contribute to the mechanism of fibrillogenesis and to the establishment of interactions that are critical for tissue integrity. A role in both fibrillogenesis and localization of extracellular matrix components may be dependent upon interactions with heparan sulfate.
Collagen types V/XI are localized preferentially to the pericellular matrix. Heparan sulfate-collagen interactions may indicate the potential for cell surface heparan sulfate proteoglycan interaction with candidate molecules including syndecans and glypicans, which are expressed during chondrogenesis (42–46). Alternatively, interactions with other heparan sulfate proteoglycans such as perlecan may be indicated. Skeletal defects have been reported in perlecan-null and mutant mice (47–50), and interactions with collagen fibrils may provide details of the mechanism for regulating collagen fibril formation in tissue. Perlecan-deficient mice develop fetal dwarfism and a disorganized growth plate, which is also true of collagen type XI deficient mice. Multiple binding sites within the same collagen molecule, as indicated by our current study, may facilitate simultaneous interaction with different heparan sulfate proteoglycans, potentially contributing to the coordination of cell-matrix interactions.
Although these studies were carried out with heparin and heparan sulfate, the physiological ligand may include other glycosaminoglycans. Future studies will determine the relative affinities for sites 1 and 2 with other glycosaminoglycans. The demonstration of isoform-specific affinities for heparan sulfate clarifies our understanding of unique splice variant functions within tissues. Type V/XI collagen molecules are also found as quantitatively minor constituents of collagen fibrils in noncartilage extracellular matrix of several other tissues including the vitreous of the eye, neuro-epithelium of the brain, odontoblasts, trabecular bones, atrioventricular valve of the heart, the tongue, the intestine, and the otic vesicle (15). Interestingly, the collagen α1(XI) isoforms that contain site 2 have been found specifically expressed in a limited region of the cartilage immediately underlying the perichondrium, yet excluded from the developing articular surface. Specific localization of the p6b containing splice variants together with the high affinity for heparan sulfate further supports the hypothesis of distinct functions for each of the unique splice variants of the amino-terminal domain of α1(XI) collagen.
Acknowledgments
We acknowledge the technical support of Christina Blasick and Noriko Hazeki-Taylor as well as Katey Irwin for assistance in figure preparation.
Footnotes
This work was supported in part by a grant from the Arthritis Foundation, National Institutes of Health/NIAMS Grants RO1AR47985 and KO2AR48672, National Institutes of Health/National Center for Research Resources Grant P20RR16454, and by funding from the M. J. Murdock Foundation and the Lori and Duane Stueckle Dean’s Distinguished Professorship. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: Npp, amino propeptide; VR, variable region; NTD, amino-terminal domain; PBS, phosphate-buffered saline; HS, horse serum; TBS, Tris-buffered saline; TX, Triton X-100; TRITC, tetramethylrhodamine isothiocyanate; DAPI, 4′,6-diamidino-2-phenylindole.
REFERENCES
- 1.Boot-Handford RP, Tuckwell DS, Plumb DA, Rock CF, Poulsom R. J. Biol. Chem. 2003;278:31067–31077. doi: 10.1074/jbc.M212889200. [DOI] [PubMed] [Google Scholar]
- 2.Pace JM, Corrado M, Missero C, Byers PH. Matrix Biol. 2003;22:3–14. doi: 10.1016/s0945-053x(03)00007-6. [DOI] [PubMed] [Google Scholar]
- 3.Veit G, Kobbe B, Keene DR, Paulsson M, Koch M, Wagener R. J. Biol. Chem. 2006;281:3494–3504. doi: 10.1074/jbc.M509333200. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Laccerda DA, Warman ML, Beier DR, Yoshioka H, Ninomiya Y, Oxford JT, Morris NP, Andrikopoulos K, Ramirez F, Wardell BB, Lifferth GD, Teuscher C, Woodward SR, Taylor BA, Seegmiller RE, Olsen BR. Cell. 1995;80:423–430. doi: 10.1016/0092-8674(95)90492-1. [DOI] [PubMed] [Google Scholar]
- 5.Blaschke UK, Eikenberry EF, Hulmes DJS, Galla H-J, Bruckner P. J. Biol. Chem. 2000;275:10370–10378. doi: 10.1074/jbc.275.14.10370. [DOI] [PubMed] [Google Scholar]
- 6.Smith GN, Jr, Hasty KA, Brandt KD. Matrix. 1989;9:186–192. doi: 10.1016/s0934-8832(89)80049-6. [DOI] [PubMed] [Google Scholar]
- 7.Ryan MC, Sieraski M, Sandell LJ. Genomics. 1990;8:41–48. doi: 10.1016/0888-7543(90)90224-i. [DOI] [PubMed] [Google Scholar]
- 8.Bork P. FEBS Lett. 1992;307:49–54. doi: 10.1016/0014-5793(92)80900-2. [DOI] [PubMed] [Google Scholar]
- 9.Bork P, Downing AK, Kieffer B, Campbell ID. Q. Rev. Biophys. 1996;29:119–167. doi: 10.1017/s0033583500005783. [DOI] [PubMed] [Google Scholar]
- 10.Mayne R, Brewton RG. Curr. Opin. Cell Biol. 1993;5:883–890. doi: 10.1016/0955-0674(93)90039-s. [DOI] [PubMed] [Google Scholar]
- 11.Fallahi A, Kroll B, Warner LM, Oxford R, Irwin KM, Mercer LM, Shadle SE, Oxford JT. Protein Sci. 2005;14:1526–1537. doi: 10.1110/ps.051363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neame PJ, Young CN, Treep JT. J. Biol. Chem. 1990;265:20401–20408. [PubMed] [Google Scholar]
- 13.Oxford JT, Doege KJ, Morris NP. J. Biol. Chem. 1995;270:9478–9485. doi: 10.1074/jbc.270.16.9478. [DOI] [PubMed] [Google Scholar]
- 14.Zhidkova NI, Justice SK, Mayne R. J. Biol. Chem. 1995;270:9486–9493. doi: 10.1074/jbc.270.16.9486. [DOI] [PubMed] [Google Scholar]
- 15.Yoshioka H, Iyama K, Inoguchi K, Khaleduzzaman M, Ninomiya Y, Ramirez F. Dev. Dyn. 1995;204:41–47. doi: 10.1002/aja.1002040106. [DOI] [PubMed] [Google Scholar]
- 16.Morris NP, Oxford JT, Davies GBM, Smoody BF, Keene DR. J. Histochem. Cytochem. 2000;48:725–741. doi: 10.1177/002215540004800601. [DOI] [PubMed] [Google Scholar]
- 17.Davies GBM, Oxford JT, Hausafus LC, Smoody BF, Morris NP. Dev. Dyn. 1998;213:12–26. doi: 10.1002/(SICI)1097-0177(199809)213:1<12::AID-AJA2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 18.Tsumaki N, Kimura T. J. Biol. Chem. 1995;270:2372–2378. doi: 10.1074/jbc.270.5.2372. [DOI] [PubMed] [Google Scholar]
- 19.Thom JR, Morris NP. J. Biol. Chem. 1991;266:7262–7269. [PubMed] [Google Scholar]
- 20.Medeck RJ, Sosa S, Morris N, Oxford JT. Biochem. J. 2003;376:361–368. doi: 10.1042/BJ20030894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keene DR, Oxford JT, Morris NP. J. Histochem. Cytochem. 1995;43:967–979. doi: 10.1177/43.10.7560887. [DOI] [PubMed] [Google Scholar]
- 22.Smith GN, Jr, Brandt KD. Collagen Rel. Res. 1987;7:315–321. doi: 10.1016/s0174-173x(87)80024-9. [DOI] [PubMed] [Google Scholar]
- 23.Vaughan-Thomas A, Young RD, Phillips AC, Duance VC. J. Biol. Chem. 2001;276:5303–5309. doi: 10.1074/jbc.M008764200. [DOI] [PubMed] [Google Scholar]
- 24.Chernousov MA, Rothblum K, Tyler WA, Stahl RC, Carey DJ. J. Biol. Chem. 2000;275:28208–28215. doi: 10.1074/jbc.M003922200. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi K, Matsue N, Sumiyoshi H, Fujimoto N, Iyama K, Yanagisawa S, Yoshioka H. Matrix Biol. 2005;24:283–294. doi: 10.1016/j.matbio.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 26.Gregory KE, Oxford JT, Chen Y, Gambee JE, Gygi SP, Aebersold R, Neame PJ, Mechling DE, Bachinger HP, Morris NP. J. Biol. Chem. 2000;275:11498–11506. doi: 10.1074/jbc.275.15.11498. [DOI] [PubMed] [Google Scholar]
- 27.Mayne R, van der Rest M, Bruckner P, Schmid T. In: Extracellular Matrix: A Practical Approach. Haralson MA, Hassell JR, editors. Oxford University Press: Oxford; 1995. pp. 73–97. [Google Scholar]
- 28.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. In: The Proteomics Protocols Handbook. Walker JM, editor. Totowa, NJ: Humana Press; 2005. pp. 571–607. [Google Scholar]
- 29.Lees JG, Smith BR, Wien F, Miles AJ, Wallace BA. Anal. Biochem. 2004;332:285–289. doi: 10.1016/j.ab.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Savitsky A, Golay MJE. Anal. Chem. 1964;36:1627–1639. [Google Scholar]
- 31.Lobley A, Wallace BA. Biophys. J. 2001;80:373a. [Google Scholar]
- 32.Lobley A, Whitmore L, Wallace BA. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 33.Whitmore L, Wallace BA. Nucleic Acids Res. 2004;32:668–673. doi: 10.1093/nar/gkh077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Provencher SW, Glockner J. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 35.Van Stokkum IHM, Spoelder HJW, Bloemendal M, Van Grondelle R, Groen FCA. Anal. Biochem. 1990;191:110–118. doi: 10.1016/0003-2697(90)90396-q. [DOI] [PubMed] [Google Scholar]
- 36.Sreerama N, Woody RW. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 37.Sreerama N, Venyaminov SY, Woody RW. Anal. Biochem. 2000;287:243–251. doi: 10.1006/abio.2000.4879. [DOI] [PubMed] [Google Scholar]
- 38.Mao D, Wachter E, Wallace BA. Biochemistry. 1982;21:4960–4968. doi: 10.1021/bi00263a020. [DOI] [PubMed] [Google Scholar]
- 39.Choi HU, Meyer K, Swarm R. Proc. Natl. Acad. Sci. U. S. A. 1971;68:877–879. doi: 10.1073/pnas.68.5.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oxford JT, DeScala J, Morris N, Gregory K, Medeck R, Irwin K, Oxford R, Brown R, Mercer L, Cusack S. J. Biol. Chem. 2004;279:10939–10945. doi: 10.1074/jbc.M310291200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 42.David G. FASEB J. 1993;7:1023–1030. doi: 10.1096/fasebj.7.11.8370471. [DOI] [PubMed] [Google Scholar]
- 43.Solursh M, Reiter RS, Jensen KL, Kato M, Bernfield M. Dev. Biol. 1990;140:83–92. doi: 10.1016/0012-1606(90)90055-n. [DOI] [PubMed] [Google Scholar]
- 44.Rapraeger AC. Semin. Cell Dev. Biol. 2001;12:107–116. doi: 10.1006/scdb.2000.0239. [DOI] [PubMed] [Google Scholar]
- 45.Paine-Saunders S, Viviano BL, Zupicich J, Skarnes WC, Saunders S. Dev. Biol. 2000;225:179–187. doi: 10.1006/dbio.2000.9831. [DOI] [PubMed] [Google Scholar]
- 46.Grover J, Roughley PJ. Biochem. J. 1995;309:963–968. doi: 10.1042/bj3090963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Costell M, Gustafsson E, Aszodi A, Morgelin M, Bloch W, Hunziker E, Addicks K, Timpl R, Fassler R. Cell Biol. 1999;147:1109–1122. doi: 10.1083/jcb.147.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arikawa-Hirasawa E, Watanabe H, Takami H, Hassell JR, Yamada Y. Nat. Genet. 1999;23:354–358. doi: 10.1038/15537. [DOI] [PubMed] [Google Scholar]
- 49.Costell M, Carmona R, Gustafsson E, Gonzalez-Iriarte M, Fassler R, Munoz-Chapuli R. Circ. Res. 2002;91:158–164. doi: 10.1161/01.res.0000026056.81424.da. [DOI] [PubMed] [Google Scholar]
- 50.Arikawa-Hirasawa E, Wilcox WR, Le AH, Silverman N, Govindraj P, Hassell JR, Yamada Y. Nat. Genet. 2001;27:431–434. doi: 10.1038/86941. [DOI] [PubMed] [Google Scholar]