Abstract
Colon cancer is a common malignant tumor that is associated with increased morbidity and mortality. Nasopharyngeal carcinoma-associated gene 6 (NGX6) is a novel candidate suppressor gene of tumor metastasis, which is down-regulated in colon cancer. This study was designed to investigate the roles of NGX6 on the growth and invasiveness of human colon cancer cell line, HT-29, and to elucidate the molecular mechanism of their action. Results showed that NGX6 could inhibit the invasiveness and extracellular matrix adhesion of HT-29 cells and restore the gap junctional intercellular communication of cells. Moreover, NGX6 could suppress the translocation of β-catenin from nucleus and cytoplasm to plasma membrane, inhibit the activity of TCF4 transcript factor, and down-regulate the expression of Wnt-direct-targeted genes c-myc, cyclin D1 and COX-2. We suggested that NGX6 inhibits cell invasion and adhesion through the suppression of Wnt signal pathway in colon cancer.
Keywords: NGX6, colon carcinoma, Wnt/β-catenin signaling pathway, metastasis
Introduction
Despite advances in multimodal therapies, the 5-year survival rate for colorectal cancer is approximately 55% and has remained essentially unchanging over the last 45 years [1,2]. Because metastatic disease is the major cause of treatment failure, a new therapeutic strategy is essential.
NGX6, a gene inhibitory to metastasis, inhibits proliferation [3] and arrests cell cycle progression at G0/G1 phase of colon cancer cells in vitro [4]. Moreover, it causes the regression of tumorigenesis and metastasis of colon and nasopharyngeal cancer in vivo [3–7]. In normal colorectal tissues, NGX6 is expressed at high levels, but at very low or undetectable level in colorectal carcinoma tissues and cells, especially in colorectal carcinoma tissues with lymph node or distant metastasis [5,6]. Transfection of NGX6 into colorectal and nasopharyngeal carcinoma cells induces the reversion of some malignant phenotypes and down-regulates molecules involved in Wnt/β-catenin signal pathway by DNA microarray analysis [7]. The NGX6 protein has been found to localize mostly on membrane, based on studies using a GFP-NGX6 fusion protein in COS7 and COS1 cells. NGX6 has been found to interact with ezrin by immunoprecipitation assays in COS7 cells [8]. Overexpression of NGX6 led to the inhibition of nuclear translocation of JNK1/2 proteins, and caused the accumulation of p-JNK1 protein in cytoplasm [6]. The expression of NF-κB is inhibited in NGX6-transfected colon carcinoma cells [9]. In this study, we try to investigate the roles of NGX6 on the growth and invasiveness of human colon cancer cell line and explore its possible mechanism.
Materials and Methods
Cell culture and transfection
The human colon cancer cell line HT-29 was a gift from Prof. Minhua Liu (Cancer Institute of Central South University, Hengyang, China). Cells were cultured at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Life Technologies, Grand Island, USA) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 1% nonessential amino acids. HT-29 cells were seeded into 6-well plates in DMEM with 10% FBS before transfection. Stable transfections were performed with Lipofectin (Invitrogen, Carlsbad, USA) according to manufacturer's instructions. Briefly, HT-29 cells were transfected with pcDNA3.1(+)-NGX6 and pcDNA3.1(+) vectors. After 24 h of transfection, cells were splited 1:3, challenged with 500 μg/ml gentamycin sulfate (G418; Invitrogen), and cultured for 2–3 weeks with a medium change of every 3 days, and used to form a stably transfected clone pool.
RT-PCR analysis
Total RNA was isolated using Trizol reagent (Gibco-BRL, Gaithersburg, USA) according to manufacturer's protocol. After being treated with DNase-I (Promega, Madision, USA), 1 mg of total RNA was reversely transcribed into complementary DNA (cDNA) with oligo(dT) using cDNA synthesis kit (Promega). Then 1 ml cDNA product was used as template to amplify specific fragments in a 25 ml reaction mixture. The PCRs were performed using Taq polymerase and reaction buffer (Promega) supplied with 0.2 mM dNTPs and 0.2 mM primers. RT-PCR reaction was carried out as follows: denaturation at 95°C, 5 min; 35 cycles of 94°C, 50 s; 56°C, 50 s; 72°C, 60 s; and final extension at 72°C, 10 min. At the same time, a housekeeping gene, GAPDH1 or GAPDH2, was amplified as an internal control to normalize relative levels of cNDA, in which primers generated a PCR product of 475 or 760 bp. Ten microliters of aliquot of each reaction products were analyzed by 1.0% agarose gel electrophoresis.
Primers corresponding to NGX6 and COX-2 sequences were designed with online primer tool (Primer 3; University of Massachusetts Medical School, USA) and synthesized by Takara (Dalian, China). Gene-specific forward and reverse primers for NGX6 and COX-2 were designed to produce PCR products of 498 and 290 bp. Primer sequences were as follows: NGX6, 5′-CAACAGCCTCAAGATCATCAG (forward), 5′-GAGGAGGGGAGATTCAGTGTGGT-3′ (reverse); COX-2, 5′-CCCAGCACTTCACGCATCA-3′ (forward), 5′-CAGACCAGGCACCAGACCAA-3′ (reverse).
Acridine orange/ethidium bromide staining
Acridine orange/ethidium bromide (AO-EB) cocktail (80:l) mixed with 1 ml DMEM were added to culture plates. Fields of stained cells was selected and analyzed using fluorescence microscopy (Nikon Eclipse E800 microscope; Nikon, Kawasaki, Japan). Viable cells stained only by AO appeared bright green with intact subcellular structure; early apoptotic cells stained by AO-EB had bright green areas in nucleus with red-orange chromatin. Late apoptotic cells stained by AO-EB presented red-orange chromatin with condensation [10]. Apoptosis rate was calculated as follow: apoptosis rate = (number of apoptotic cells/number of total cells) × 100%. Data were from triplicate fields.
FACS analysis
Cells were prepared using Annexin-V-FLUOS Staining Kit (Roche, Penzberg, Germany) according to manufacturer's instructions and sample volumes and cell concentrations were adapted for FACS analysis. Briefly, 0.5 ml of incubation buffer per 106 cells was added, and the suspension was analyzed by a flow cytometer at 488 nm excitation wavelength. Electronic compensation of instrument was setted to exclude overlapping of the two emission spectra.
Adhesion assay
Cell adhesion was measured according to the method described by Busk et al. [11]. In brief, the wells on a 96-well plate were coated with 0.1 ml fibronectin (Sigma-Aldrich, St. Louis, USA). In addition, 1 mg/ml of poly-d-lysine and 1% bovine serum albumin (BSA) in PBS were coated into two wells as maximal- and minimal-adhesion controls, respectively. The 96-well plate was incubated at 37°C for 1 h and blocked with 1% BSA at 37°C for 0.5 h. 1 × 105 cells were added to each coated well and incubated for 2 h at 37°C, then wells were washed twice and stained with crystal violet. Absorbance at 595 nm was measured. The proportion of cells adhering to coated wells was calculated as follow: [A595(exp)−A595(BSA)]/[A595(poly-d-lysine)−A595(BSA)]. Data were from triplicate wells.
Cell invasion and migration assay
Cell invasion was measured by Matrigel invasion assay using 6.5-mm, 8-µm-pore Transwell chamber (Transwell; Corning, New York, USA). Matrigel (BD, Franklin Lakes, USA) was diluted in cold distilled water to concentration of 200 mg/ml, then 0.1 ml was added to the upper well of Transwell chamber, and dried in a sterile hood. The Matrigel was reconstituted with medium for 1 h at 37°C before cells were added. Cells were starved overnight in serum-free medium and resuspended at a concentration of 2.5 × 105 cells/ml in serum-free medium containing 0.1% BSA. Cell suspension (0.2 ml) was added to the top of each well, and a 10 mg/ml fibronectin solution was added to the bottom well of the chamber as a chemoattractant. Thirty-six hours later, the cells remaining in the top chamber were carefully removed from the upper surface of filters using a cotton swab. The cells that migrated to the lower surface of filter were fixed with methanol and stained with hematoxylin and eosin. Cell migration was quantitated by counting five random fields per filter at 40× magnification. Data are presented as the mean value of cell per high-power field based on triplicate measurements from two independent experiments.
Western blot analysis
Nuclear proteins were harvested from cells using the NE-PER Nuclear and Cytoplasmic Extraction kit (Pierce, Beijing, China) according to manufacturer's instructions. Protein concentrations were determined using the BCA protein assay method (Pierce). Cell extracts containing 50 μg of protein were separated on 8–12% SDS-PAGE gels and transferred to nitrocellulose membranes (Hyclone, Beijing, China). The membranes were blocked with 5% nonfat milk powder for 1 h and subsequently incubated overnight at 4°C with a mouse anti-β-catenin monoclonal antibody or a rabbit anti-α-tublin polyclonal antibody (Upstate, New York, USA). After three washes, HRP-conjugated anti-mouse or anti-rabbit secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, USA) were used to detect and visualize the results using the ECL Western blotting detection system (Amersham Biosciences, Buckinghamshire, UK). The α-tubulin antibody and C23 antibody (Santa Cruz Biotechnology) were used as a loading control.
Detection of gap junctional intercellular communication
Gap junctional intercellular communication (GJIC) was assessed by the scrape-loading/dye transfer technique using method described previously [12]. Briefly, cells were plated to confluence overnight in a 6-well plate, and immersed in 0.05% Lucifer yellow in PBS. Scrape-loading was performed using a sharp knife. Cells were incubated in dye solution for an additional 3 min at room temperature, rinsed with PBS, and observed. Transfer of fluorescent dye was observed using a phase-contrast inverted microscope (Olympus, Tokyo, Japan) equipped with epifluorescence.
Immunofluorescence staining
Cells were fixed in cold methanol for 2 min. They were permeabilized using 0.2% Triton X-100 and blocked using normal goat serum as recommended (Molecular Probes, Eugene, USA). The mouse primary antibody (Upstate) was added and incubated at room temperature for 2 h followed by washing with PBS. The secondary antibodies, FITC-conjugated sheep anti-mouse IgG and Cy3-conjugated sheep anti-rabbit IgG (Sigma-Aldrich), were added and incubated for 1 h. The coverslips were washed four times with PBS and mounted using gel/mount mounting media (Biomeda, Foster City, USA). Control experiments were carried out without primary or secondary antibodies. Nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole). Stained cells were viewed with a Zeiss LSM510 laser scanning fluorescence microscope (Carl Zeiss SMS, Jena, Germany) with a 40× objective and a 10× eyepiece.
Luciferase reporter assay
On the day before transfection, 5 × 104 HT-29 cells were seeded into 12-well plates. Reporter construct of 0.3 μg and each expression construct of 0.3 μg were transfected using Superfect transfection reagent (Qiagen, Valencia, USA) according to manufacturer's instructions. TCF4-luciferase construct of 0.005 μg was transfected as an internal control. The total DNA amount was kept at 1.5 μg/well. Corresponding empty vectors were used to make up the difference between treatments. Luciferase activity was measured 48 h later using a Monolight 3010 Luminometer (PharMingen, San Diego, USA). The dual luciferase assay kit was purchased from Promega. All experiments were done in triplicates and performed at least three times. Data were expressed as the mean ± SE.
Statistical analysis
Data were presented as the means ± SE. Differences in the variables between groups were tested by the one-way ANOVA test using SPSS 11.0. Differences were considered significant when P < 0.05.
Results
Construction of NGX6 over-expressed stable cell line
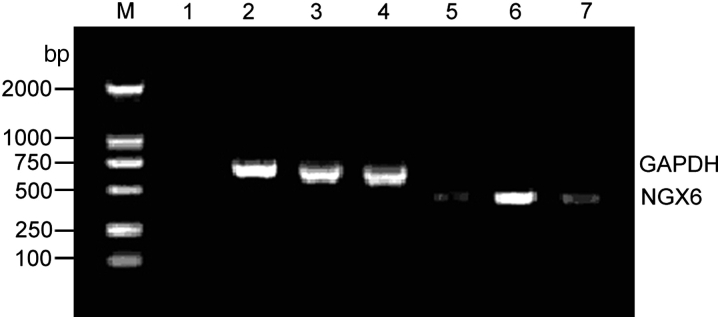
The expression of NGX6 gene was low in HT-29 cells. HT-29 cells were stably transfected with pcDNA3.1(+)-NGX6 and pcDNA3.1(+) vectors. We examined the level of NGX6 mRNA in HT-29 cells by RT-PCR. It showed that the expression of NGX6 gene is higher in pcDNA3.1(+)-NGX6/HT-29 cells than that in HT-29 cells and pcDNA3.1(+)/HT-29 cells (Fig. 1).
Figure 1.
RT-PCR analysis for the expression of NGX6 mRNA in HT-29 cells The RT-PCR products with NGX6 primers produced 489 bp fragments and GAPDH primers produced 700 bp fragments. Lane M, DL-2000 marker; lane 1, negative control; lanes 2 and 5, HT-29 cells; lanes 3 and 6, pcDNA3.1(+)/NGX6/HT-29; lanes 4 and 7, pcDNA3.1(+)/HT-29. It showed that expression of NGX6 mRNA reached the highest level in pcDNA3.1(+)/NGX6/HT-29 cells.
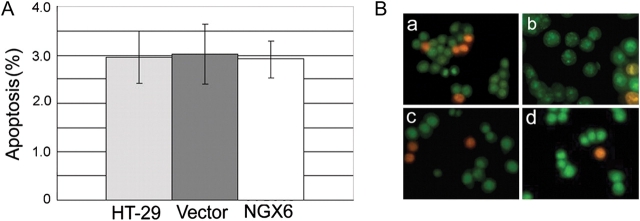
NGX6 has no effect on apoptosis in human HT-29 cells
Results of flow cytometry analysis [Fig. 2(A)] and AO-EB dual staining [Fig. 2(B)] showed that no apoptosis was detected in HT-29 cells and the expression of NGX6 did not induce apoptosis in these cells (P > 0.05).
Figure 2.
Effect of NGX6 on the apoptosis of human HT-29 cells by annexin-V-FLUOS (A) and AO-EB (B) staining There are no differences of apoptosis rate in three groups and it is showed that NGX6 has no effects on the induction of apoptosis in HT-29 cells. (a), HT-29; (b), pcDNA3.1(+)/HT-29; (c), pcDNA3.1(+)/NGX6/HT-29; (d), DMSO. All experiments were repeated three times.
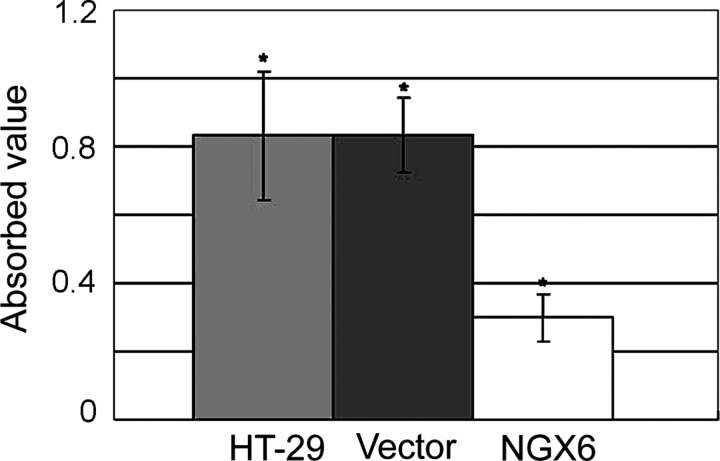
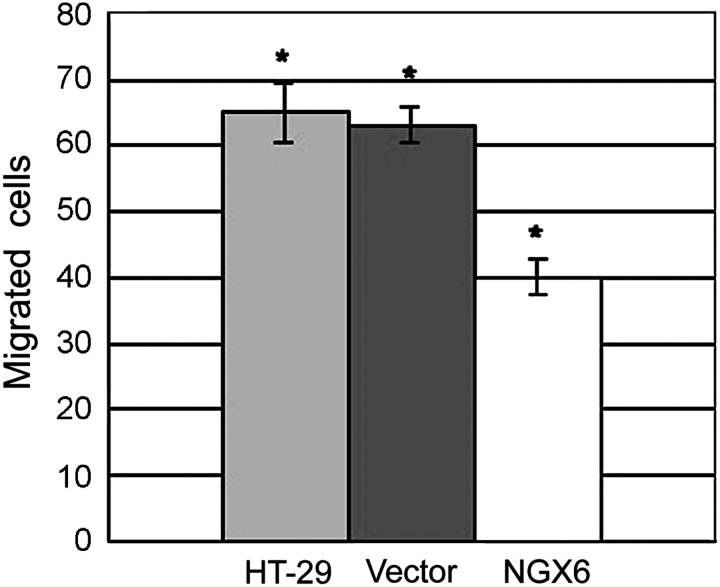
NGX6 inhibits invasion and adhesion ability of human HT-29 cells
As shown in Fig. 3, the adhesion ability of HT-29 cells transfected with pcDNA3.1(+)/NGX6 was significantly decreased, compared with that of HT-29 cells and pcDNA3.1(+)/HT-29 cells (P < 0.05). Invasion assays were performed as described in the ‘Materials and Methods’ section. As shown in Fig. 4, cell invasion was markedly reduced in HT-29 cells overexpressing NGX6, compared with in pcDNA3.1(+)/HT-29 and HT-29 cells (P < 0.01).
Figure 3.
Effect of NGX6 on the adhesion ability of human HT-29 Data indicated that NGX6 suppress the adhesion ability of HT-29 cells. *P < 0.01 compared with HT-29 and pcDNA3.1(+)/HT-29 cells. All experiments were repeated three times.
Figure 4.
Effect of NGX6 on the invasion ability of human HT-29 cells The migration and invasion in chemotaxic cells decreased after induction, compared with pcDNA3.1(+)/HT-29 and HT-29 cells. NGX6 did suppress the invasion ability of HT-29 cells (P < 0.01). All experiments were repeated three times.
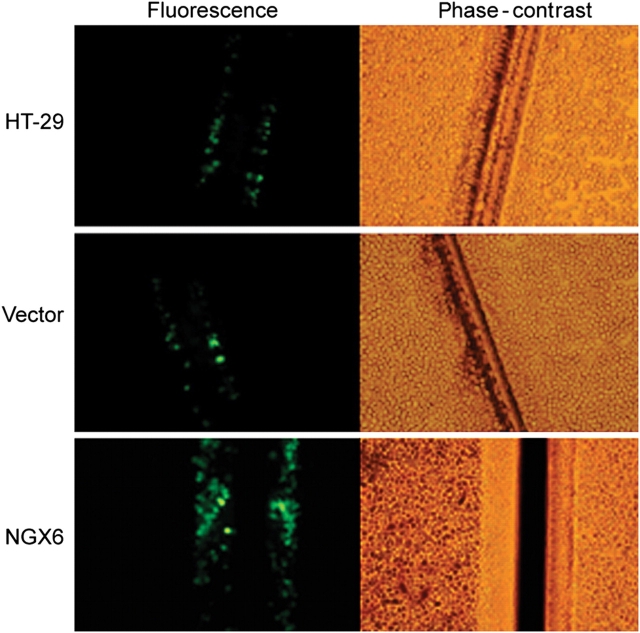
NGX6 confers GJIC ability
The existence of functional GJIC was demonstrated by the transfer of Lucifer yellow. Lucifer yellow does not diffuse through intact cell membranes, but its molecular mass is still small enough to allow it to pass through gap junctions in knife-injured cells. Cells capable of GJIC showed transfer of Lucifer yellow from injured border to interior cells, while cells incapable of GJIC did not show dye transfer. In this study, we found that HT-29 and pcDNA3.1(+)/HT-29 cells did not show GJIC, while the overexpression of NGX6 could confer GJIC on HT-29 cells (Fig. 5).
Figure 5.
Effect of NGX6 on gap GJIC in human HT-29 cells HT-29, pcDNA3.1(+)/HT-29, and pcDNA3.1(+)/NGX6/HT-29 cells were cultured in 6-well plates. After incubation, GJIC was assessed with scrape-loading/dye transfer technique. Three minutes after scrape-loading, the NGX6 over-expressing cells allowed Lucifer yellow transfer into contiguous cells while very little dye transfer occurred in HT-29 and pcDNA3.1(+)/HT-29 cells. Data showed that NGX6 conferred GJIC on HT-29 cells.
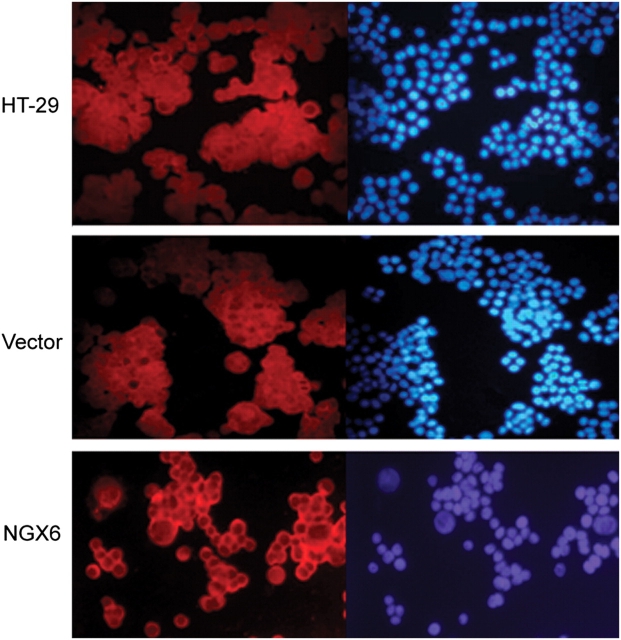
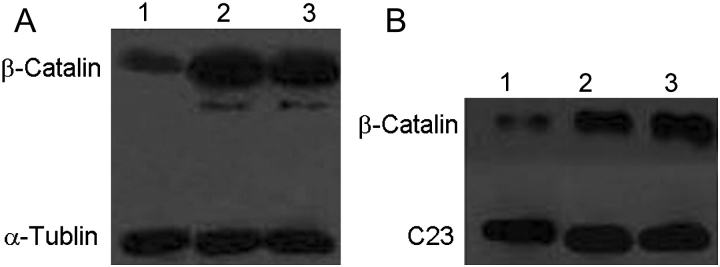
NGX6 decreases expression of β-catenin and inhibits its nuclear translocation
β-catenin proteins presented diffuse staining throughout the cytoplasm, nucleus, and cell membrane in HT-29 and pcDNA3.1(+)/HT-29 cells, while in pcDNA3.1(+)/NGX6/HT-29 cells they localized mainly on cell membrane with low-level staining in cytoplasm and nucleus (Fig. 6). To validate the immunofluorescence results, the expression of β-catenin protein was analyzed by western blotting. α-tubulin or C23 were used as an internal control [Fig. 7A,B)]. In total proteins or nucleus proteins, the expression of β-catenin protein in pcDNA3.1(+)/NGX6/HT-29 cells was lower than that in pcDNA3.1(+)/HT-29 cells and HT-29 cells. Thus, NGX6 inhibits the expression and nuclear translocation of β-catenin.
Figure 6.
β-Catenin is relocated to cell membrane caused by NGX6 HT-29, pcDNA3.1(+)/HT-29, and pcDNA3.1(+)/NGX6/HT-29 cells were cultured in multichamber slides. After incubation, cells were fixed and then detected for β-Catenin expression by Cy3-labeled immunofluorescence staining. DAPI-stained cells were used as an internal control. It showed that β-catenin protein localizes mainly on cell membranes in pcDNA3.1(+)/NGX6/HT-29 cells, and low-level staining observed in cytoplasm and nucleus. β-Catenin proteins present diffuse staining throughout cytoplasm, nucleus, and cell membrane in HT-29 and pcDNA3.1(+)/HT-29 cells.
Figure 7.
Effect of NGX6 on β-catenin expression in human HT-29 cells (A) Effect of NGX6 on β-catenin expression in total cell proteins. Lane 1, pcDNA3.1(+)/NGX6/HT-29; 2, pcDNA3.1(+)/HT-29; 3, HT-29. (B) Effect of NGX6 on β-catenin expression in nuclear protein. Lane 1, HT-29; 2, pcDNA3.1(+)/HT-29; 3, pcDNA3.1(+)/NGX6/HT-29.
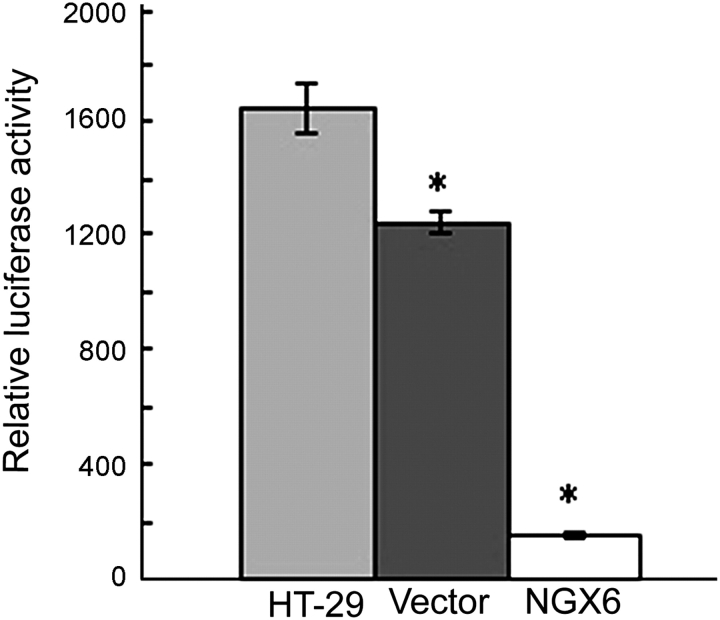
NGX6 reduces the transcriptional activation of β-catenin/TCF and the expression of Wnt direct-targeted genes in human HT-29 cells
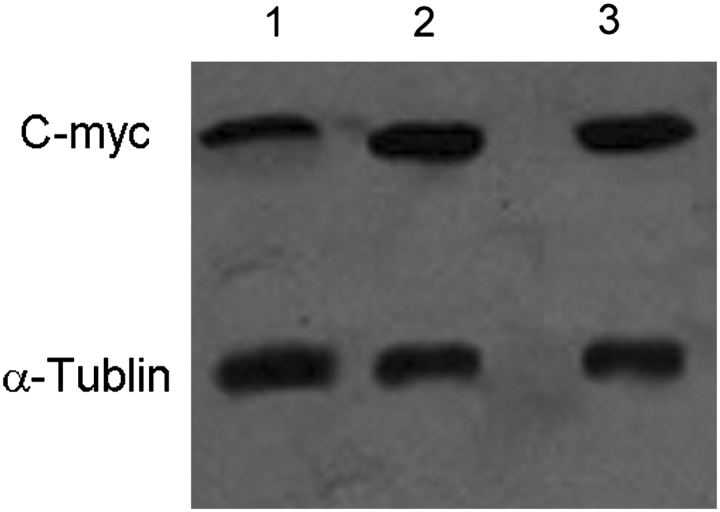
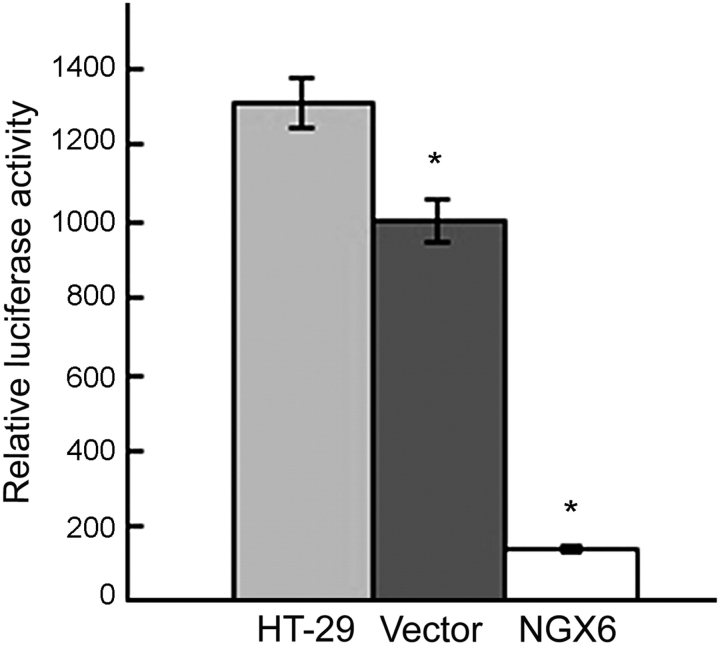
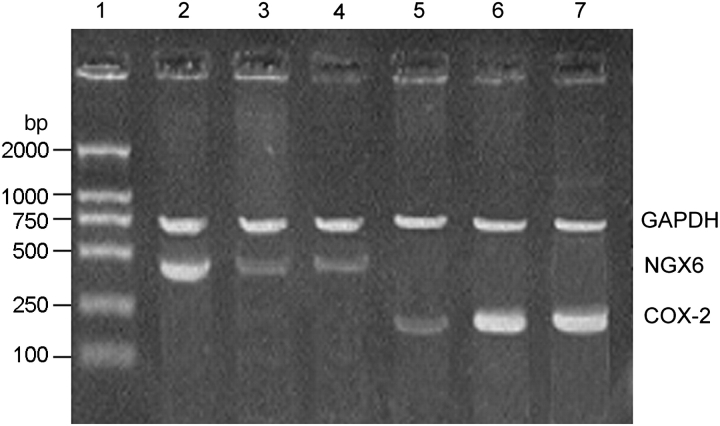
The activity of TCF4 transcription factor was analyzed by luciferase reporter assay. The activity of TCF4 in pcDNA3.1(+)/NGX6/HT-29 cells was lower than that in pcDNA3.1(+)/HT-29 cells and HT-29 cells (Fig. 8, P < 0.05). The expression of c-myc protein was analyzed by western blot and α-tubulin was used as an internal control (Fig. 9). The expression of c-myc protein was down-expressed in pcDNA3.1(+)/NGX6/HT-29 cells, compared with that in pcDNA3.1(+)/HT-29 and HT-29 cells. The activation of cyclin D1 in Wnt signal pathway in pcDNA3.1(+)/NGX6/HT-29 cells was lower than that in pcDNA3.1(+)/HT-29 and HT-29 cells (Fig. 10, P < 0.05). It was determined that COX-2 mRNA level was down-regulated in pcDNA3.1(+)-NGX6/HT-29 cell, compared with that in HT-29 and pcDNA3.1(+)/HT-29 cells by RT-PCR (Fig. 11).
Figure 8.
Effect of NGX6 on TCF4 activity in human HT-29 cells The activity of TCF4 in pcDNA3.1(+)/NGX6/HT-29 cells was lower than that in pcDNA3.1(+)/HT-29 and HT-29 cells by luciferase reporter assay (P < 0.05).
Figure 9.
Effect of NGX6 on c-myc expression in total protein of human HT-29 cells Lane 1, pcDNA3.1(+)/NGX6/HT-29; 2, pcDNA3.1(+)/HT-29; 3, HT-29. Data show that NGX6 inhibits c-myc expression in total proteins.
Figure 10.
Effect of NGX6 on cyclin D1 activity in human HT-29 cells The activity of cyclin D1 in pcDNA3.1(+)/NGX6/HT-29 cells was lower than that in pcDNA3.1(+)/HT-29 cells and in HT-29 cells by luciferase reporter assay (P < 0.05).
Figure 11.
RT-PCR analysis for COX-2 mRNA in HT-29 cells The RT-PCR products with COX-2 primers produced 290 bp fragments, NGX6 primers produced 489 bp fragments and GAPDH primers produced 700 bp fragments. Lane 1, DL-2000 marker; lanes 2 and 5, pcDNA3.1(+)/NGX6/HT-29; lanes 3 and 6, HT-29; lane 4 and 7, pcDNA3.1(+)/HT-29.
Discussion
In this study, we examined the effects of NGX6 on apoptosis, adhesion, invasiveness and GJIC of colorectal cancer cells. We tried to elucidate the molecular basis of these effects. Although the over-expression of NGX6 did not induce apoptosis in these cells, we found that NGX6 gene could inhibit invasion and extracellular matrix adhesion and confer GJIC ability in HT-29 cells.
β-Catenin plays crucial roles in embryogenesis, cells malignant transformation, and tumor metastasis [13]. β-catenin proteins on cell membrane can form β-catenin/E-cadherin complex, which is involved in adhesive junction and plays an important role in maintaining polarity and integrity of epithelial cells [14]. Results in this study showed that NGX6 could inhibit β-catenin expression and nuclear translocation by immunofluorescence staining and western blot analysis. Thus, we draw the conclusion that NGX6 inhibit the invasion and metastasis of colorectal cancer cells. Its possible mechanism might be that NGX6 gene blocks β-catenin nuclear translocation. In the previous study [15], we found that NGX6 protein was associated with ezrin by its cytoplasm region, and it could down-regulate ezrin expression. Elzagheid A [16] reported that ezrin expression significantly predicted both the 5-year disease specific survival (P = 0.035) and 5-year metastases. High ezrin expression level was also related to high E-cadherin (cytoplasmic) expression. The NGX6 protein, which effects ezrin expression, may inhibit β-catenin nuclear translocation by enhancing the binding of β-catenin and E-cadherin, and reducing the amount of cytoplasmic β-catenin.
We also found that NGX6 could inhibit the transcriptional activation of β-catenin/TCF in Wnt signal pathway in HT-29 cells by performing luciferase reporter assays. Moreover, NGX6 could down-regulate the expression of c-myc, cyclin D1 and COX-2 genes by western blot, luciferase reporter gene assays and RT-PCR. These results showed over-expression of NGX6 suppressed Wnt/β-catenin signal pathway in colon cancer cells.
These results suggest that NGX6 may play a role in inhibiting metastasis and invasiveness of colon cancer cells and suppress Wnt/β-catenin signal pathway in cells. These data provide experimental basis for chemopreventive and chemotherapeutic therapy of colorectal cancer.
Funding
This work was supported by grants from the State Key Science Research Program of China (2006CB910503), the National Natural Science Foundation of China (39670344, 30770972), the China Postdoctoral Science Foundation (20060400265), the Natural Sciences Foundations of Hunan Province (06JJ20080, 07JJ3057), and the Research Fund for the Doctoral Program of Higher Education of China (20070533031).
References
- 1.Toribara NW, Kim YS. Premalignancy and malignancy of the colon. In: Sleisenger M, Fordtran J, editors. Gastrointestinal Disease. Philadelphia: W.B. Saunders; 1992. pp. 1–2. [Google Scholar]
- 2.Cohen AM, Minsdy BD, Friedman MA. Rectal cancer. In: DeVita V, Hellman S, Rosenberg S, editors. Cancer: Principles and Practice of Oncology. Philadelphia: Lippincott; 1993. [Google Scholar]
- 3.Wang XY, Shen S, Liu HY, Zhang XM, Peng C, Huan H, Liu F, et al. The effect of NGX6 on cell biology. World Chin J. 2004;12:574–579. [Google Scholar]
- 4.WangX Y, Shen S, Liu F, Wang XY, Shen SR, Liu F, Li XL, Fang SQ. Effects of NGX6 gene on cell cycle in colon cancer cell line HT-29. Prog Biochem Biophys. 2006;33:45–49. [Google Scholar]
- 5.Zhang XM, Wang XY, Sheng SR, Wang JR, Li J. Expression of tumor related genes NGX6, NAG-7, BRD7 in gastric and colorectal cancer. World J Gastroenterol. 2003;9:1729–1733. doi: 10.3748/wjg.v9.i8.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng Y, Li H, Wu M, Wang X, Fan S, Liu F, Xiang B, et al. NGX6 inhibits AP-1 and Ets-1 expression and down-regulates cyclin D1 in human colorectal cancer. Acta Biochim Biophys Sin. 2009;41:504–514. doi: 10.1093/abbs/gmp039. doi:10.1093/abbs/gmp039. [DOI] [PubMed] [Google Scholar]
- 7.Liu F, Wang XY, Lian P, Xiao ZM, Shen SR, Ma J, Xiong W. Effect of NGX6 on gene expression profile of colon carcinoma cell line HT-29. Chin J Cancer. 2005;24:1064–1070. [PubMed] [Google Scholar]
- 8.Ma J, Zhou J, Fan S, Wang L, Li X, Yan Q, Zhou M, et al. Role of a novel EGF-like domain-containing gene NGX6 in cell adhesion modulation in nasopharyngeal carcinoma cells. Carcinogenesis. 2005;26:281–291. doi: 10.1093/carcin/bgh312. doi:10.1093/carcin/bgh312. [DOI] [PubMed] [Google Scholar]
- 9.Lian P, Guo Q, Peng Y, Xiao ZM, Liu F, Wang XY, Shen SR, et al. The role of NGX6 gene on apoptosis of human colon cancer. Prog Biochem Biophys. 2008;35:1154–1160. [Google Scholar]
- 10.Wu M, Huang C, Gan K, Huang H, Chen Q, Ouyang J, Tang Y, et al. RRC4, a putative tumor suppressor gene, requires a functional leucine-rich repeat cassette domain to inhibit proliferation of glioma cells in vitro by modulating the extracellular signal-regulated kinase/protein kinase B/nuclear factor-(B pathway. Mol Biol Cell. 2006;17:3534–3542. doi: 10.1091/mbc.E05-11-1082. doi:10.1091/mbc.E05-11-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busk M, Pytela R, Sheppard D. Characterization of the integrin alpha vbeta 6 as a fibronectin-binding protein. J Biol Chem. 1992;267:5790–5796. [PubMed] [Google Scholar]
- 12.Wu M, Gan K, Huang C, Tang Y, Chen Q, Tang K, Li X, et al. RRC4 controls in vitro invasion of glioblastoma cells through inhibiting RPTP-zeta expression. J Neurooncol. 2006;80:133–142. doi: 10.1007/s11060-006-9173-6. doi:10.1007/s11060-006-9173-6. [DOI] [PubMed] [Google Scholar]
- 13.Fuchs SY, Ougolkov AV, Spiegelman VS, Minamoto T. Oncogenic beta-catenin signaling networks in colorectal cancer. Cell Cycle. 2005;4:1522–1539. doi: 10.4161/cc.4.11.2129. [DOI] [PubMed] [Google Scholar]
- 14.Bremnes RM, Veve R, Gabrielson E, Hirsch FR, Baron A, Bemis L, Gemmill RM, et al. High-throughput tissue microarray analysis used to evaluate biology and prognostic significance of the E-cadherin pathway in non-small-cell lung cancer. J Clin Oncol. 2002;20:2417–2428. doi: 10.1200/JCO.2002.08.159. doi:10.1200/JCO.2002.08.159. [DOI] [PubMed] [Google Scholar]
- 15.Hawcroft G, D'Amico M, Albanese C, Markham AF, Pestell RG, Hull MA. Indomethacin induces differential expression of β-catenin, γ-catenin and T-cell factor target genes in human colorectal cancer cells. Carcinogenesis. 2002;23:107–114. doi: 10.1093/carcin/23.1.107. doi:10.1093/carcin/23.1.107. [DOI] [PubMed] [Google Scholar]
- 16.Dihlmann S, Siermann A, von Knebel Doeberitz M. The nonsteroidal anti-inflammatory drugs aspirin and indomethacin attenuate β-catenin/TCF-4 signaling. Oncogene. 2001;20:645–653. doi: 10.1038/sj.onc.1204123. doi:10.1038/sj.onc.1204123. [DOI] [PubMed] [Google Scholar]