Abstract
Lewis x (Lex) and sialyl Lewis x (SLex)-containing glycans play important roles in numerous physiological and pathological processes. The key enzyme for the final step formation of these Lewis antigens is α1-3-fucosyltransferase. Here we report molecular cloning and functional expression of a novel Helicobacter hepaticus α1-3-fucosyltransferase (HhFT1) which shows activity towards both non-sialylated and sialylated Type II oligosaccharide acceptor substrates. It is a promising catalyst for enzymatic and chemoenzymatic synthesis of Lex, sialyl Lex and their derivatives. Unlike all other α1-3/4-fucosyltransferases characterized so far which belong to Carbohydrate Active Enzyme (CAZy, http://www.cazy.org/) glycosyltransferase family GT10, the HhFT1 shares protein sequence homology with α1-2-fucosyltransferases and belongs to CAZy glycosyltransferase family GT11. The HhFT1 is thus the first α1-3-fucosyltransferase identified in the GT11 family.
Keywords: cloning, fucosyltransferase, Helicobacter hepaticus, Lewis x, sialyl Lewis x
Introduction
Fucosyltransferases (EC 2.4.1.-) are enzymes that catalyze the transfer of monosaccharide l-fucose from its activated sugar nucleotide guanosine-5′-diphosphate β-l-fucose (GDP-Fuc) to acceptors for the formation of fucosides (fucose-containing oligosaccharides and glycoconjugates). Mechanistically, fucosyltransferases are inverting glycosyltransferases (GTs) as α-fucosylated products are formed from β-fucosylated sugar nucleotide donor GDP-Fuc. Based on the types of acceptors and the regio-specificity of the fucosides formed by fucosyltransferase-catalyzed reaction, fucosyltransferases are categorized into α1-2, α1-3 and/or α1-4, α1-6 and O-fucosyltransferases (Ma et al. 2006). Except for O-fucosyltransferases (O-FucT) which catalyze the transfer of a fucose residue from GDP-Fuc directly to the serine or threonine residue on proteins (Ma et al. 2006), all others catalyze the transfer of a fucose residue to galactose (Gal), N-acetylglucosamine (GlcNAc) or another fucose (Fuc) residue (Marques et al. 1998) on oligosaccharides, polysaccharides or glycoconjugates. Furthermore, based on their protein sequence similarity, except for an unusual α1-2FucT in Dictyostelium discoideum (van Der Wel et al. 2001), all other α1-2FucTs are categorized into Carbohydrate Active Enzyme (CAZy, http://www.cazy.org/) (Cantarel et al. 2009) glycosyltransferase family GT11, while all α1-3 and/or α1-4-FucTs characterized so far belong to GT10 (Ma et al. 2006).
l-Fucose is usually a terminal monosaccharide in oligosaccharides and the carbohydrate moieties of glycoconjugates. Protein O-fucosylation on serine or threonine has also been found in many plasma glycoproteins and is believed to be important for regulating protein functions including Notch signaling (Okajima et al. 2008). Fucose-containing structures in eukaryotes are believed to be involved in tissue development, angiogenesis, fertilization, cell adhesion, inflammation and tumor metastasis (Ma et al. 2006; Miyoshi et al. 2008). On the other hand, fucose-containing lipopolysaccharides (LPS) are expressed by some pathogenic bacteria including Helicobacter pylori and Escherichia coli (Guo et al. 2005). They have been suggested to be involved in molecular mimicry, adhesion, colonization and modulating the host immune response (Ma et al. 2006).
Lewis x (Lex) trisaccharide Galβ1-4(Fucα1-3)GlcNAc and sialyl Lewis x (SLex) tetrasaccharide Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAc are among the most important fucose-containing oligosaccharides that play essential roles in numerous physiological and pathological processes. For example, Lex antigens expressed by human pathogenic bacterium H. pylori mimic the host cell antigens and mask the pathogenic bacterium from the host immune detection (Chan et al. 1995; Moran et al. 1996; Monteiro et al. 1998). Lex antigens on parasite Schistosoma mansoni downregulate the host’s protective immune response against the parasite (Wang et al. 2009). SLex is the essential recognition component of E-, P-, and L-selectin ligands (Kannagi 2002; Lowe 2003; Dube and Bertozzi 2005). The interaction of SLex and selectins is believed to mediate lymphocyte homing, initiate leukocyte–endothelial cell adhesion in acute and chronic inflammation (Kannagi 2002; Lowe 2003) and be involved in cancer migration (Ma et al. 2006). SLex is considered a cancer marker for diagnosis and prognosis of cancer metastasis (Kannagi 2004; Magnani 2004). It has been a leading structure for developing anti-inflammatory reagent and cancer vaccine (Simanek et al. 1998; Danishefsky and Allen 2000; Ouerfelli et al. 2005; Seeberger and Werz 2007).
Due to the importance of fucosides in biological systems and their potential application in treating inflammation, bacterial and viral infection and cancer, they have been attractive synthetic targets. Chemical synthesis of fucosidic bond is challenged by its acid lability; thus, fucosyltransferase-catalyzed enzymatic and chemoenzymatic approaches are considered preferred alternatives. Although many fucosyltransferases have been identified from humans, Caenorhabditis elegans, plants and bacteria (Ma et al. 2006), only H. pylori fucosyltransferases and human recombinant α1-3-fucosyltransferase have been used for the preparative-scale synthesis of Lex (Lubineau et al. 1997, 1998; Wang et al. 2009) and SLex (Ichikawa et al. 1992; Bowman et al. 2001; Belot et al. 2002; Pratt and Bertozzi 2004). As the expression levels of reported H. pylori and human α1-3-fucosyltransferases are not high, there is a need for obtaining active and soluble α1-3-fucosyltransferases with high expression level, preferably in E. coli expression hosts, to allow large-scale enzymatic synthesis of fucosides.
H. pylori and Helicobacter hepaticus are two well-studied Gram-negative Helicobacter species. Unlike H. pylori, which is a gastric Helicobacter species, H. hepaticus is an enterohepatic Helicobacter species which colonizes the intestinal tracts and livers of mice to cause chronic hepatic inflammation, liver cancer and inflammatory bowel disease (Fox et al. 1994; Cahill et al. 1997). The association of H. hepaticus infection with liver tumors in mice provides a valuable animal model for studying mechanisms of liver cancers caused by bacterial infection (Rogers and Fox 2004). Like its close relative H. pylori, which can also cause chronic inflammation and carcinoma in the host (Suerbaum and Michetti 2002), H. hepaticus exhibits phase variation in genes encoding putative fucosyltransferases and other glycosyltransferases, which may contribute to LPS modification and antigenic mimicry (Wang et al. 2000) for efficient immune evasion of hosts (Suerbaum et al. 2003). Nevertheless, unlike fucosyltransferases from H. pylori (Chan et al. 1995; Monteiro et al. 1998; Ma et al. 2003, 2005; Rabbani et al 2005; Sanabria-Valentin et al. 2007; Sun et al. 2007, Wang et al. 2009), which have been well studied, no fucosyltransferase from H. hepaticus has been functionally characterized so far. Analysis of H. hepaticus ATCC51449 genome (Suerbaum et al. 2003) identified two genes (HH0072 and HH1776) encoding putative fucosyltransferases in addition to two putative truncated α1-2-fucosyltransferase mutants (encoded by HH0069 and HH0070). We report herein the cloning, functional expression and characterization of the first fucosyltransferase (HhFT1) from H. hepaticus encoded by gene HH0072 as a novel α1-3-fucosyltransferase which can use both sialylated (Neu5Acα2-3Galβ1-4GlcNAc) and non-sialylated (Galβ1-4GlcNAc) type II glycans as acceptors for the synthesis of sialyl Lewis x [Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAc] and Lewis x [Galβ1-4(Fucα1-3)GlcNAc] antigens. It is the first α1-3-fucosyltransferase identified in CAZy glycosyltransferase family GT11, which contains, so far, only α1-2-fucosyltransferases. The cloning and characterization of the second putative H. hepaticus fucosyltransferase (HhFT2) encoded by gene HH1776 are ongoing processes.
Results
Sequence comparison of HhFT1, HhFT2 and other bacterial fucosyltransferases
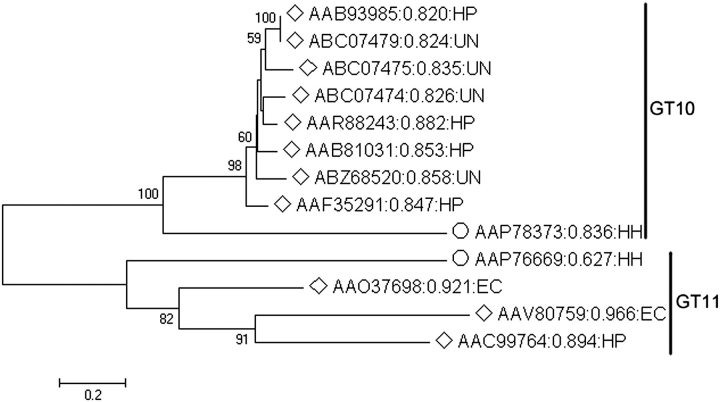
Proteins encoded by HH0072 (GenBank accession number AAP76669) and HH1776 (GenBank accession number AAP78373) in carcinogenic bacterium H. hepaticus ATCC 51449 genome are annotated as conserved hypothetical proteins predicted to be a fucosyltransferase and an α1-3-fucosyltransferase, respectively. Here, we name them HhFT1 and HhFT2, respectively. Based on protein sequence homology, HhFT1 encoded by HH0072 is classified into CAZy glycosyltransferase family GT11 which contains all α1-2-fucosyltransferases characterized so far except for an unusual α1-2FucT in D. discoideum (van Der Wel et al. 2001). HhFT2 encoded by HH1776 is classified into CAZy glycosyltransferase family GT10 which contains all α1-3/4-fucosyltransferases characterized so far. Nevertheless, they are quite divergent from other characterized bacterial fucosyltransferases (Figure 1). The protein which has the closest sequence homology to HhFT1 is an E. coli O128:B12 α1-2-fucosyltransferase WbsJ (GenBank accession number AAO37698) which is a part of O-antigen biosynthesis cluster (Shao et al. 2003). They share 27% identity and 50% similarity. Sequence alignment (Figure 2) indicates that HhFT1 has three conserved motifs shared by GT11 family fucosyltransferases including motif II which is likely a part of the GDP-Fuc binding domain (Shao et al. 2003). The protein which has the closest sequence homology to HhFT2 is a H. pylori α1-4-fucosyltransferse (GenBank accession number AAR88243) (Rabbani et al. 2005), and they share 35% identity and 51% similarity. Sequence-based protein solubility prediction (Smialowski et al. 2007) indicates that HhFT1 has a better solubility score than all other possible fucosyltransferases except for a putative protein from Geobacter lovleyi SZ (GenBank accession number ACD96461). Unlike α1-3FucTs from H. pylori (Ge et al. 1997; Lin et al. 2006), HhFT1 does not have C-terminal tandem repeats, and it is a shorter protein (320 amino acid residues) compared to the full-length Hp1-3FT (478 amino acid residues) (Lin et al. 2006).
Fig. 1.
Phylogenetic analysis of identified (labeled with unfilled diamonds) and putative (labeled with unfilled circles) FucTs from bacteria. Neighbor-Joining (NJ) method was used to create this unrooted tree. Bootstrap values (expressed as percentages of 1000 replications) greater than 50% are indicated above the branches. The protein identification numbers are followed by predicted class probability of insolubility (Smialowski et al. 2007) and species from which the genes were cloned. HP, H. pylori; EC, E. coli; HH, Helicobacter hepaticus; UN, unknown.
Fig. 2.
Alignment of HhFT1 and E. coli O128:B12 α1-2-fucosyltransferase WbsJ (GenBank accession number AAO37698).
Cloning, expression and purification of recombinant protein
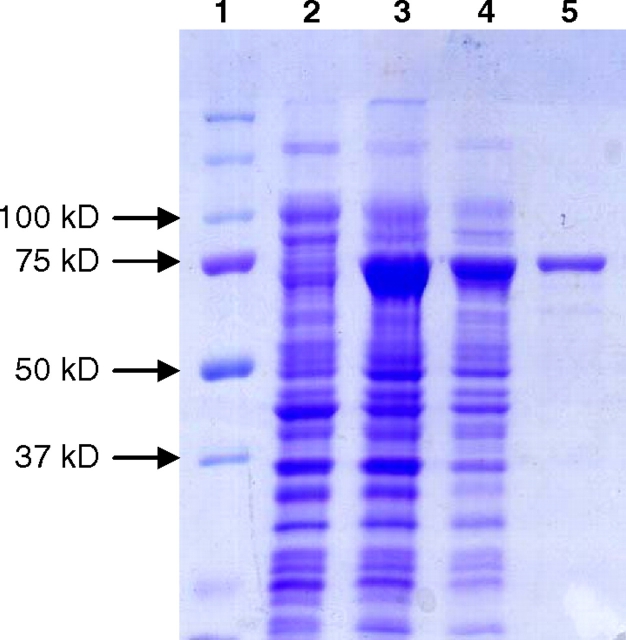
Full-length H. hepaticus HH0072 synthetic gene with codons optimized for E. coli expression was initially cloned into pET15b vector as an N-His6-tagged recombinant protein (His6-HhFT1), but a low amount of soluble protein was obtained in cell lysate (less than 1 mg/L cell culture, data not shown). In order to increase the amount of soluble recombinant protein, pMAL-c4X vector (Kapust and Waugh 1999) was chosen to express HhFT1 as a maltose binding protein (MBP)-fusion protein with the MBP tag at the N-terminus. A His6-tag was also introduced at the C-terminus to allow easy purification of the recombinant protein by Ni2+-affinity columns. The gene and protein sequences of the resulting fusion protein MBP–HhFT1–His6 are shown in Figure 3. The codon-optimized HhFT1 gene contains 27% adenine, 18% cytosine, 22% guanine and 33% thymine as compared to the original sequence containing 29% adenine, 19% cytosine, 21% guanine and 31% thymine (GenBank accession no. AE017125). Expression of the recombinant protein MBP–HhFT1–His6 in E. coli BL21 (DE3) followed by Ni2+-column purification resulted in a yield of 45 mg L−1 cell culture. This expression level is the highest among all reported recombinant fucosyltransferases. As shown in Figure 4, the purified protein exhibited a molecular mass of about 75 kDa in sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) which is close to the calculated molecular weight of 80 kDa.
Fig. 3.
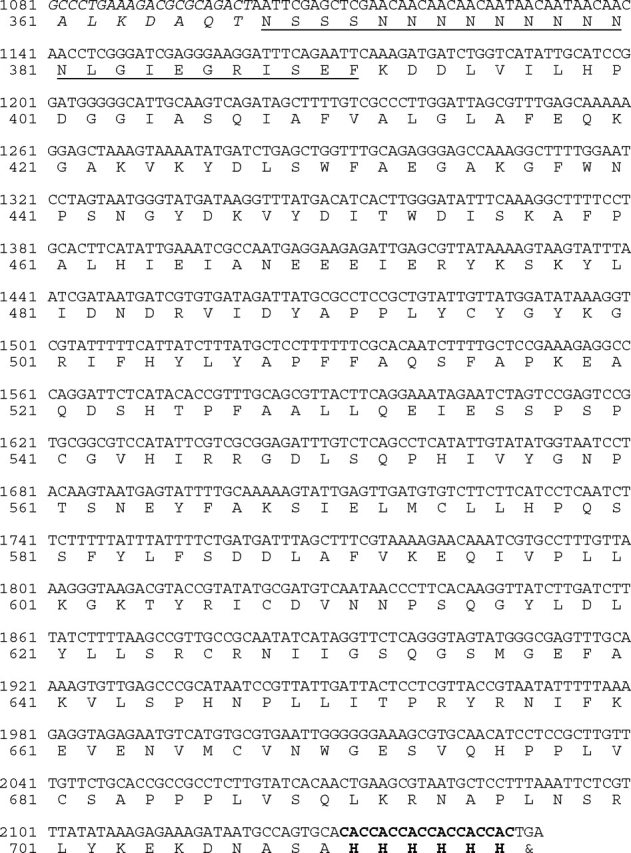
Gene and protein sequences of the codon-optimized MBP–HhFT1–His6. Only the C-terminal sequence of MBP is shown (in italics). The multiple cloning sites of pMAL-c4X vector are underlined. The six histidine residues introduced at the C-terminus during cloning are in bold.
Fig. 4.
SDS-PAGE analysis of MBP–HhFT1–His6 expression and purification. Lanes: 1, protein standards; 2, whole cell extraction before induction; 3, whole cell extraction after induction; 4, cell lysate after induction; 5, Ni2+-column purified protein.
Acceptor substrates of MBP–HhFT1–His6
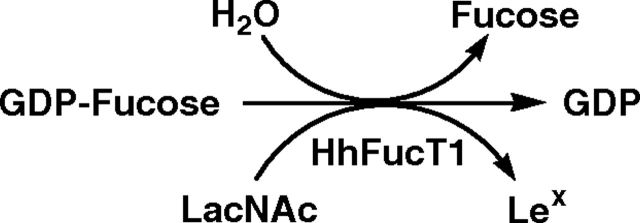
Using GDP-fucose as a donor, both type I (Galβ1-3GlcNAcβProN3) and type II (Galβ1-4GlcNAcβProN3) disaccharides were tested as potential acceptor substrates for the recombinant MBP–HhFT1–His6. Thin-layer chromatography (TLC) analyses indicated that the recombinant enzyme was reactive towards type II Galβ1-4GlcNAcβProN3 substrate, and no product was observed when type I disaccharide Galβ1-3GlcNAcβProN3 was used as the potential acceptor substrate. Further TLC tests indicated that α2-3-sialylated type II oligosaccharide with N-acetylneuraminic acid (Neu5Ac) as the sialic acid form (Neu5Acα2-3Galβ1-4GlcNAcβProN3) was also an acceptor for the enzyme, suggesting that this novel recombinant enzyme is most likely an α1-3-fucosyltransferase in spite of its protein sequence homology to α1-2-fucosyltransferases in CAZy glycosyltransferase family GT11.
Confirming the α1-3-fucosyltransferase activity of MBP–HhFT1–His6 by one-pot two-enzyme synthesis of Galβ1-4(Fuc1-3)GlcNAcβProN3
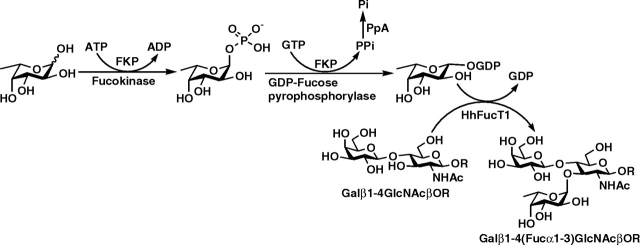
In order to confirm the α1-3-fucosyltransferase activity of MBP–HhFT1–His6, a preparative-scale synthesis of Lewis x trisaccharide Galβ1-4(Fucα1-3)GlcNAcβProN3 from type II disaccharide Galβ1-4GlcNAcβProN3 was carried out using a one-pot two-enzyme reaction (Figure 5) containing the recombinant fucosyltransferase MBP–HhFT1–His6 and a recombinant bifunctional l-fucokinase/GDP-fucose pyrophosphorylase (FKP) from Bacteroides fragilis strain NCTC9343 (Yi et al. 2009). The presence of the bifunctional FKP allows the synthesis of GDP-fucose, the sugar nucleotide donor for MBP–HhFT1–His6, from simple starting materials such as l-fucose, adenosine 5′-triphosphate (ATP) and guanosine-5′-triphosphate (GTP) (Yi et al. 2009). Lewis x (Lex) trisaccharide Galβ1-4(Fucα1-3)GlcNAcβProN3 was obtained in a yield of 63% with the supplement of additional ATP and GTP periodically (see Materials and methods for details). Nuclear magnetic resonance (NMR) and high-resolution mass spectrometry (HRMS) studies confirmed the structure of the trisaccharide product. As shown in Table I, the 13C NMR chemical shifts of the purified product agree well with previously reported data for Lewis x trisaccharide (Wang et al. 2009). More specifically, comparing the chemical shifts of the disaccharide starting material and the trisaccharide product indicates that significant changes occur at C-3 (a downfield shift of 2.35 ppm from 72.70 ppm in the disaccharide starting material to 75.05 ppm in the trisaccharide product) and C-4 (an upfield shift of 3.22 ppm from 78.69 ppm in the disaccharide starting material to 75.47 ppm in the trisaccharide product) of the GlcNAc residue, confirming that fucosylation takes place at C-3 of the GlcNAc residue. High-resolution mass spectrometry spectrum (ESI) obtained shows the desired m/z for molecular ion [M]+. of 612.2425 (calculated 612.2490 for C23H40N4O15).
Fig. 5.
Schematic illustration for the one-pot three-enzyme preparative-scale synthesis of Lewis x trisaccharide Galβ1-4(Fucα1-3)GlcNAcβProN3 from fucose, ATP, GTP and type II disaccharide Galβ1-4GlcNAcβProN3 (LacNAcβProN3). Enzymes used: FKP, a recombinant bifunctional l-fucokinase/GDP-fucose pyrophosphorylase from B. fragilis strain NCTC9343; HhFT1, H. hepaticus α1-3-fucosyltransferase 1; PpA, a recombinant inorganic pyrophosphatase from P. multocida.
Table I.
13C NMR chemical shifts assignment of LacNAcβProN3 (Galβ1-4GlcNAcβProN3) and LexβProN3 [Galβ1-4(Fuc1-3)GlcNAcβProN3]
| Residue | Carbon atom | Chemical shift (ppm) |
|
|---|---|---|---|
| βDGlcNAc | C | LacNAcβProN3 | LexβProN3 |
| 1 | 101.27 | 101.09 | |
| 2 | 55.26 | 55.94 | |
| 3 | 72.70 | 75.05 | |
| 4 | 78.69 | 75.47 | |
| 5 | 74.91 | 73.49 | |
| 6 | 60.26 | 59.88 | |
| C=O | 174.61 | 174.40 | |
| CH3 | 22.37 | 22.36 | |
| βDGal(1-4) | 1 | 103.07 | 101.96 |
| 2 | 71.14 | 71.16 | |
| 3 | 72.54 | 72.58 | |
| 4 | 68.72 | 68.47 | |
| 5 | 75.52 | 75.10 | |
| 6 | 61.19 | 61.63 | |
| αLFuc(1-3) | 1 | 98.78 | |
| 2 | 67.82 | ||
| 3 | 69.33 | ||
| 4 | 72.03 | ||
| 5 | 66.85 | ||
| CH3 | 15.43 | ||
| ProN3 | OCH2CH2CH2N3 | 67.29 | 67.32 |
| OCH2CH2CH2N3 | 28.29 | 28.24 | |
| OCH2CH2CH2N3 | 47.96 | 47.88 | |
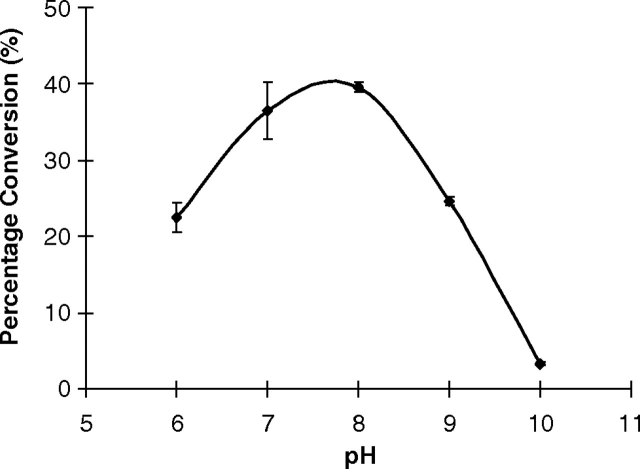
pH Profile of MBP–HhFT1–His6
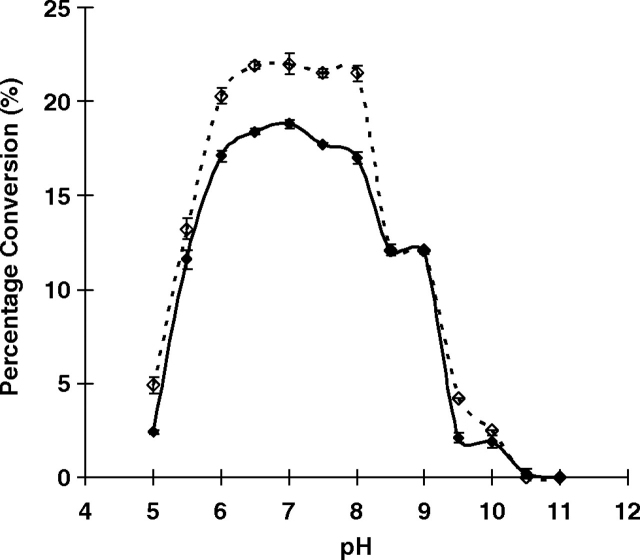
High-performance liquid chromatography (HPLC)-based pH profile study for the fucosyltransferase was carried out using fluorophore 2-anthranilic acid (2AA)-labeled non-sialylated and sialylated type II oligosaccharides (Galβ1-4GlcNAcβPro2AA and Neu5Acα2-3Galβ1-4GlcNAcβPro2AA respectively) as acceptors. A similar pH profile pattern was seen for the MBP–HhFT1–His6 with these two acceptors, although the overall activity was higher when the sialylated acceptor Neu5Acα2-3Galβ1-4GlcNAcβPro2AA was used (Figure 6). The fucosyltransferase activity was optimum in a relatively broad pH range of 6.0–8.0. Medium activity was observed at pH 5.5 and in a pH range of 8.5–9.0. No or minimum activity was observed when the pH fell below 5.0 or higher than 9.5. This pH profile is similar to that reported recently for an E. coli O128:B12 α1-2-fucosyltransferase WbsJ, which is also active in a wide range of pH values (5.5–8.5) (Li et al. 2008).
Fig. 6.
The pH profile of MBP–HhFT1–His6 when LacNAcβPro2AA (filled diamonds) or Neu5Acα2-3LacNAcβPro2AA (dashed line with unfilled diamonds) was used as an acceptor substrate. Buffers used: MES (pH 5.0–6.5), HEPES (pH 7.0–8.0), Tris-HCl (pH 8.5–9.0), N-cyclohexyl-2-hydroxyl-3-aminopropanesulfonic acid (CAPSO) (pH 10), and N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) (pH 10.0–11.0).
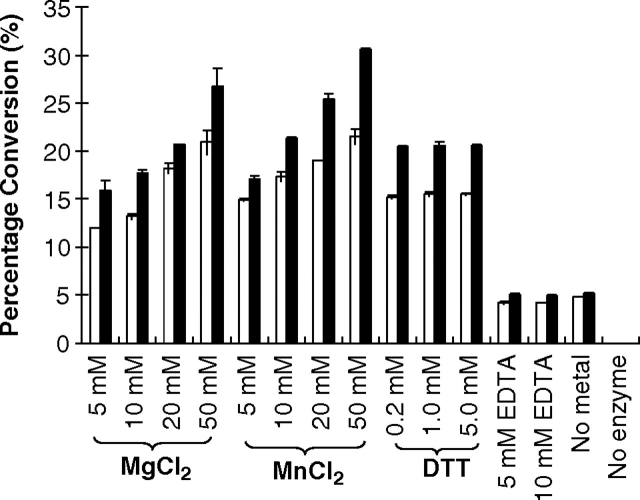
Effects of metal ions, EDTA and dithiothreitol on the fucosyltransferase activity of MBP–HhFT1–His6
The effects of dithiothreitol (DTT), metal ions Mg2+ and Mn2+, and a chelating agent EDTA on the fucosyltransferase activity of MBP–HhFT1–His6 were examined at pH 7.0. As shown in Figure 7, a divalent metal ion is required for the fucosyltransferase activity of the enzyme as the enzyme activity decreased dramatically in the absence of metal ions. This is consistent with the requirement of the divalent metal ion by most glycosyltransferases using nucleotide diphosphate sugars as donor substrates. The enzyme activity increases with the increase of the metal ion concentration up to 50 mM. The divalent metal ions may facilitate the glycosidic bond cleavage in the donor by neutralizing the negative charges on the GDP product. Indeed, protein sequence analysis of HhFT1 indicates a metal-binding D92ND94 motif. It is also noticed that the enzyme activity did not disappear completely without the addition of metal ions or in the presence of a metal chelator EDTA. The reason is unclear, but it is possible that positively charged amino acid residues in the protein may contribute to stabilize the GDP formed (Sun et al. 2007).
Fig. 7.
Effects of metal ions (Mg2+ and Mn2+), EDTA, and DTT (in the presence of 20 mM Mg2+) on the fucosyltransferase activity of MBP–HhFT1–His6 when LacNAcβPro2AA (white columns) or Neu5Acα2-3LacNAcβPro2AA (black columns) was used as an acceptor substrate.
There are seven cysteine residues in the HhFT1 protein sequence, so the effect of DTT on the fucosyltransferase activity of the enzyme was studied. The addition of DTT did not change the enzyme activity significantly, indicating that disulfide formation is not required for the fucosyltransferase activity of MBP–HhFT1–His6.
Kinetics
Kinetic studies carried out for the fucosyltransferase activity of MBP–HhFT1–His6 using sialylated and non-sialylated type II acceptors indicate that both glycans are similarly good acceptors for the enzyme although the efficiency for sialylated glycan Neu5Acα2-3LacNAcβPro2AA (kcat/Km = 0.63) is a little better than the non-sialylated glycan LacNAcβPro2AA (kcat/Km = 0.43) due to a smaller Km and a better kcat for the sialylated acceptor (Table II). This is different from a C-terminal truncated α1-3-fucosyltransferase (HpFTΔ45) cloned from H. pylori (NCTC 11639) (Lin et al. 2006) which prefers N-acetyllactosamine (LacNAc) over Neu5Acα2-3LacNAc as an acceptor. Nevertheless, the Km value of LacNAcβPro2AA (45 mM) for MBP–HhFT1–His6 is about 50-fold higher than that (0.71–0.89 mM) of LacNAc for truncated α1-3-fucosyltransferases cloned from H. pylori (NCTC 11639) (Lin et al. 2006), indicating a much weaker binding of the type II glycan to the HhFT1 than the H. pylori FucT.
Table II.
Kinetic parameters of recombinant MBP–HhFT1–His6
| Substrates | Km (mM) | kcat (min-1) | kcat/Km (mM-1 min-1) | |
|---|---|---|---|---|
| Fucosyltransferase | LacNAcβPro2AA | 45 ± 2 | 19 ± 1 | 0.43 |
| GDP-fucose | 2.4 ± 0.4 | 2.3 ± 0.1 | 1.0 | |
| Neu5Acα2-3LacNAcβPro2AA | 40 ± 1 | 25 ± 1 | 0.63 | |
| GDP-fucose | 3.3 ± 0.6 | 3.6 ± 0.2 | 1.1 | |
| GDP-Fucose hydrolysis | GDP-fucose | 13 ± 2 | 48 ± 2 | 3.7 |
GDP-fucose hydrolysis activity of the MBP–HhFT1–His6
In the absence of a glycan acceptor, water can be considered as an acceptor for the fucosyltransferase activity of the enzyme. This leads to the GDP-fucose hydrolysis. Significant GDP-fucose hydrolysis activity was observed for MBP–HhFT1–His6 with an optimal activity at pH 8.0 (Figure 8). At pH 7.0, the hydrolysis of GDP-fucose (kcat/Km = 3.7) catalyzed by the enzyme is about 3-fold more efficient than the fucosyltransferase activity of the enzyme when either non-sialylated (kcat/Km = 1.0) or sialylated type II glycans (kcat/Km = 1.1) was used as an acceptor (Table II).
Fig. 8.
The pH profile of GDP-fucose hydrolysis catalyzed by MBP–HhFT1–His6. Buffers used: MES, pH 6.0; HEPES, pH 7.0, 8.0; Tris-HCl, pH 9.0; and CAPS, pH 10.0.
Fucosidase activity studies
No fucosidase activity was observed for the MBP–HhFT1–His6 when LexβProN3 was used as a substrate for TLC assays or when 4-methylumbelliferyl β-Lewis x (LexβMU) and Neu5Acα2-3LexβMU were used in HPLC-based assays.
Product inhibition studies using GDP or LexβProN3 as an inhibitor
No product inhibition effect was observed for GDP or LexβProN3 by HPLC-based fluorescent assays.
Discussion
In this study, we report the molecular cloning, functional expression and characterization of a novel H. hepaticus α1-3-fucosyltransferase. Based on its protein sequence homology, HhFT1 is classified into glycosyltransferase family 11 (GT11) in the Carbohydrate Active enZyme database (CAZy, http://www.cazy.org/) (Campbell et al. 1997; Coutinho et al. 2003). All members of GT11 family that have been functionally determined so far are α1-2 fucosyltransferases. These include WbsJ from E. coli O128:B12 (Li et al. 2008), a potential α1-2FT WbwK from E. coli O86:B7 (Guo et al. 2005), HpFT2 from H. pylori (Wang et al. 1999), CE2FT-1 and CE2FT-2 (Zheng et al. 2008) from C. elegans, FUT1 and FUT2 from human (Larsen et al. 1990; Kelly et al. 1995; Rouquier et al. 1995; Kudo et al. 1996; Koda et al. 1997), mouse (Lin et al. 2000; Domino et al. 2001), rat (Piau et al. 1994; Bureau et al. 2001), bovine (Barreaud et al. 2000; Saunier et al. 2001), and primates (Apoil et al. 2000), as well as Sec1 from bovine (Barreaud et al. 2000; Saunier et al. 2001), mouse (Lin et al. 2000; Domino et al. 2001) and primates (Borges et al. 2008). The enzyme has the highest sequence homology to HhFT1 is WbsJ, an E. coli O128:B12 α1-2-fucosyltransferase which is a part of O-antigen biosynthesis cluster (Shao et al. 2003). It shares 27% identity and 50% similarity to HhFT1.
In comparison, all α1-3 and/or α1-4-fucosyltransferases characterized so far are listed in GT10 family. HhFT1 thus represents the first α1-3-fucosyltransferase that belongs to the CAZy GT11 family. This also indicates that, while protein sequence homology can be used as a guide to predict the type of the enzyme, experimental confirmation is necessary to characterize the function of the protein.
All fucosyltransferases are inverting glycosyltransferases and are classified into eight glycosyltransferase (GT) families in CAZy website: GT10, GT11, GT23, GT3, GT56, GT65, GT68 and GT74 (Campbell et al. 1997; Coutinho et al. 2003; Cantarel et al. 2009). The only known fucosyltransferase crystal structures are for H. pylori α1-3FT in the GT10 family (Sun et al. 2007) and Bradyrhizobium sp. WM9 α1-6FT NodZ (EC 2.4.1.-) (Brzezinski et al. 2007) and human GlcNAc α1-6FT (FUT8) (EC 2.4.1.68) (Ihara et al. 2007) in the GT23 family. All of these fucosyltransferases have a glycosyltransferase B (GT-B) fold containing two separated Rossmann domains (Breton et al. 2006). Protein x-ray crystal structures have not been reported for any member of the GT11 family. The good solubility and expression level (45 mg L−1 cell culture) of MBP–HhFT1–His6 present an excellent opportunity for structural characterization of GT11 fucosyltransferases.
Similar to H. pylori, the hypothetic genes involved in LPS synthesis of H. hepaticus ATCC 51449 are scattered throughout the whole genome. Homologues of HP0826 (β1-4-galactosyltransferase gene), HP0360 (galE), HP0326 (neuA) as well as GDP-fucose biosynthetic genes rfbM (HP0043), rfbD (HP0044) and wbcJ (HP0045), which are involved in the synthesis of Lewis antigen in H. pylori 26695 (Moran 2008), are also harbored by H. hepaticus ATCC 51449 (HH0323, HH0380, HH0900, HH0675, HH0172 and HH0173, respectively). Analysis of H. hepaticus ATCC 51449 genome has identified two CMP-Neu5Ac pathway genes, neuB (HH0908) and neuC (HH0082) (Daines et al. 2000), although no sialyltransferase has been discovered by searching the whole H. hepaticus ATCC 51449 genome using known sialyltransferase sequences from uniproKB database (http://www.uniprot.org/). A putative sialidase (HH1672) is also found in the genome of H. hepaticus ATCC 51449.
GDP-fucose can be efficiently hydrolyzed by MBP–HhFT1–His6, which competes with the fucosyltransferase activity of the enzyme, leading to low yields due to the consumption of the donor substrate by water (Figure 9). This can be overcome by adding additional GDP-fucose periodically during the reaction process. In situ generation of GDP-fucose from GTP and ATP using a bifunctional FKP from B. fragilis strain NCTC9343 (Yi et al. 2009) also helps to improve the yield. As most glycosyltransferases can catalyze the hydrolysis of the corresponding sugar nucleotide donor substrates, periodical addition of sugar nucleotides and in situ generation of sugar nucleotides can be used as general strategies to improve the yields of glycosyltransferase-catalyzed reactions. These will be extremely useful approaches when weak acceptors are used and water competes significantly for the same donor.
Fig. 9.
Schematic illustration of water as a competing acceptor for the fucosyltransferase activity of MBP–HhFT1–His6 causing hydrolysis of GDP-fucose.
Materials and methods
Materials
E. coli electrocompetent DH5α and chemically competent BL21 (DE3) cells were from Invitrogen (Carlsbad, CA, USA). Vector plasmid pET15b was purchased from Novagen (EMD Biosciences Inc. Madison, WI, USA). QIAprep spin miniprep kit and QIAquick gel extraction kit were from Qiagen (Valencia, CA, USA). Herculase-enhanced DNA polymerase was from Stratagene (La Jolla, CA, USA). T4 DNA ligase and 1 kb DNA ladder were from Promega (Madison, WI, USA). NdeI, BamHI, EcoRI, HindIII restriction enzymes and vector plasmid pMAL-c4X were from New England Biolabs (Beverly, MA, USA). Ni2+-NTA agarose (nickel-nitrilotriacetic acid agarose) was from 5 PRIME (Gaithersburg, MD, USA). Bicinchoninic acid (BCA) protein assay kit was from Pierce Biotechnology Inc. (Rockford, IL).
Chemical synthesis of GDP-fucose
GDP-fucose was synthesized by following a reported procedure (Timmons and Jakeman 2007). Detailed procedures are presented in the supporting information.
Synthesis of LacNAcβPro2AA and Neu5Acα2-3LacNAcβPro2AA as acceptors for HhFT1
Fluorescent LacNAcβPro2AA was synthesized from LacNAcβProNH2 (obtained by reduction of azidoproyl LacNAc, LacNAcβProN3) (Chokhawala et al. 2008) and 2-(methoxycarbonyl) succinanilic acid NHS ester (2AA-OSu). Its α2-3-sialylated form Neu5Acα2-3LacNAcβPro2AA was synthesized from LacNAcβPro2AA using a one-pot two-enzyme sialylation reaction (Yu et al. 2009) catalyzed by a Neisseria meningitidis CMP-sialic acid synthetase (NmCSS) (Yu et al. 2004) and a multifunctional Pasteurella multocida α2-3-sialyltransferase (PmST1) (Yu et al. 2005). Detailed procedures are presented in the supporting information.
Cloning
Full-length H. hepaticus HH0072 synthetic gene with codons optimized for E. coli expression was customer synthesized by Biomatik (Wilmington, DE, USA) and provided in a pGH vector. It was cloned as an N-His6-tagged or an N-MBP-tagged with a C-His6-tagged fusion protein. The primers used for cloning the N-His6-tagged protein in pET15b vector were: forward primer 5′-GATCCATATGAAAGATGATCTGGTCATATTGCAT-3′ (NdeI restriction site is underlined) and reverse primer 5′-AAGGGATCCTCATGCACTGGCATTATCTTTCTCTT-3′ (BamHI restriction site is underlined). To clone the N-MBP-tagged with C-His6-tagged protein in pMAL-c4X vector, the forward primer used was 5′-GATCGAATTCAAAGATGATCTGGTCATATTGCAT-3′ (EcoRI restriction site is underlined), and the reverse primer used was 5′-AAGAAGCTTTCAGTGGTGGTGGTGGTGGTGTGCACTGGCATTATCTTTCTCTT-3′ (HindIII restriction site is underlined, and the sequence that encodes the hexahistidine tag is italicized). Polymerase chain reactions (PCRs) for amplifying the target gene were performed in a 50-µL reaction mixture containing plasmid DNA (10 ng), forward and reverse primers (0.2 µM each), 1× Herculase buffer, dNTP mixture (0.2 mM), and 5 U (1 µL) of Herculase-enhanced DNA polymerase. The reaction mixture was subjected to 30 cycles of amplification at an annealing temperature of 55°C. The resulting PCR product was purified and double digested with NdeI and BamHI or EcoRI and HindIII restriction enzymes. The purified and digested PCR product was ligated with the predigested pET15b or pMAL-c4X vector and transformed into electrocompetent E. coli DH5α cells. Selected clones were grown for minipreps and characterized by restriction mapping. DNA sequencing was performed by the Davis Sequencing Facility in the University of California-Davis.
Overexpression
Positive plasmids were selected and transformed into E. coli BL21 (DE3) chemical competent cells. The plasmid-bearing E. coli strains were cultured in LB-rich medium (10 g L−1 tryptone, 5 g L−1 yeast extract and 10 g L−1 NaCl) supplemented with ampicillin (100 µg mL−1). Overexpression of the target protein was achieved by inducing the E. coli culture with 0.1 mM of isopropyl-1-thio-β-d-galactopyranoside (IPTG) when OD600 reached 0.8 followed by incubating at 20°C for 20 h with vigorous shaking at 250 rpm in a C25KC incubator shaker (New Brunswick Scientific, Edison, NJ).
Purification
His6-tagged target proteins were purified from cell lysate. To obtain the cell lysate, cell pellet harvested by centrifugation at 4000 rpm for 2 h was resuspended in lysis buffer (pH 8.0, 100 mM tris(hydroxymethyl)aminomethane (Tris)-HCl containing 0.1% Triton X-100) (20 mL L−1 cell culture). Lysozyme (50 µg mL−1) and DNaseI (3 µg mL−1) were then added, and the mixture was incubated at 37°C for 60 min with vigorous shaking. Cell lysate was obtained by centrifugation at 12,000 rpm for 30 min as the supernatant. Purification of His-tagged proteins from the lysate was achieved using a Ni2+-resin column. The column was pre-equilibrated with 10 column volumes of binding buffer (5 mM imidazole, 0.5 M NaCl, 50 mM Tris-HCl, pH 7.5) before the lysate was loaded. After washing with 8 column volumes of binding buffer and 10 column volumes of washing buffer (40 mM imidazole, 0.5 M NaCl, 50 mM Tris-HCl, pH 7.5), the protein was eluted with an elute buffer (200 mM imidazole, 0.5 M NaCl, 50 mM Tris-HCl, pH 7.5). The fractions containing the purified enzymes were collected and stored at 4°C.
Quantification of purified protein
The concentration of purified enzyme was obtained in a 96-well plate using a BCA Protein Assay Kit (Pierce Biotechnology, Rockford, IL) with bovine serum albumin as a protein standard. The absorbance of samples was measured at 562 nm by a BioTek SynergyTM HT Multi-Mode Microplate Reader.
Sodium dodecylsulfate-polyacrylamide gel electrophoresis
SDS-PAGE was performed in a 12% Tris-glycine gel using a Bio-Rad Mini-protein III cell gel electrophoresis unit (Bio-Rad) at DC = 150 V. Bio-Rad Precision Plus Protein Standards (10–250 kDa) were used as molecular weight standards. Gels were stained with Coomassie Blue.
TLC analysis of the substrate specificity
LacNAcβProN3 (Galβ1-4GlcNAcβProN3, type II, 10 mM) or Galβ1-3GlcNAcβProN3 (type I, 10 mM) was used as a potential acceptor in the presence of GDP-fucose (10 mM) in a 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (pH 7.0, 100 mM) to analyze the acceptor substrate specificity of the fucosyltransferase activity of the recombinant enzyme (9.0 µg) at 37°C. The crude reaction mixture was analyzed by TLC.
One-pot two-enzyme synthesis of Galβ1-4(Fucα1-3)GlcNAcβProN3 using MBP–HhFT1–His6
HhFT1 acceptor Galβ1-4GlcNAcβProN3 (30 mg), l-fucose (21 mg, 2 equivalents), ATP (71 mg, 2 equivalents), GTP (67 mg, 2 equivalents) and MgCl2 (45 mg, 22 mM) were dissolved in H2O. A stock solution of 2-(N-morpholino)ethanesulfonic acid (MES) buffer (0.5 M, pH 6.2, 1 mL) was added. After the addition of a recombinant FKP from B. fragilis strain NCTC9343 (Yi et al. 2009) (2.3 mg, FKP is a bifunctional l-fucokinase/GDP-fucose pyrophosphorylase which catalyzes the synthesis of GDP-fucose from l-fucose, ATP and GTP via a fucose-1-phosphate (Fuc-1-P) intermediate) and MBP–HhFT1–His6 (0.9 mg), water was added to bring the volume of the reaction mixture to 10 mL. The reaction was carried out by incubating the solution in an incubator shaker for 72 h at 37 °C. The supplements of additional 0.5 equivalents of ATP (18 mg) and GTP (17 mg) were added periodically in every 12 h (2.5 equivalent totals). The reaction was quenched by adding cold ethanol (10 mL) and was then centrifuged to remove precipitates. A BioGel P-2 filtration and a silica gel column (EtOAc:MeOH:H2O, 7:2:1) were used to purify the product to afford Galβ1-4(Fucα1-3)GlcNAcβProN3 (25 mg, 63 %).
pH profile by HPLC
Assays were performed in a total volume of 10 µL in a buffer (200 mM) with pH varying from 5.0 to 11.0 containing GDP-fucose (12 mM) with LacNAcβPro2AA (10 mM) or GDP-fucose (10 mM) with Neu5Acα2-3LacNAcβPro2AA (5 mM) and the recombinant enzyme (9.0 µg). Reactions were allowed to proceed for 60 min (when LacNAcβPro2AA was used as an acceptor) or 30 min (when Neu5Acα2-3LacNAcβPro2AA was used as an acceptor) at 37°C before being quenched by adding ice-cold 12% acetonitrile (1990 µL) to make 200-fold dilutions. The samples were then kept on ice until aliquots of 5 µL were injected and analyzed by a Shimadzu LC-2010A system equipped with a membrane online degasser, a temperature control unit and a fluorescence detector. A reverse-phase Premier C18 column (250 × 4.6 mm i.d., 5 µm particle size, Shimadzu) protected with a C18 guard column cartridge was used. The mobile phase was 18% acetonitrile when LacNAcβPro2AA was used as an acceptor or 6.9% acetonitrile with 6.9% methanol when Neu5Acα2-3LacNAcβPro2AA was used as an acceptor. Fluorescence of the products LacNAcProβ2AA and Neu5Acα2-3LacNAcβPro2AA and the acceptor substrates were detected by excitation at 315 nm and emission at 400 nm. All assays were carried out in duplicate.
Effects of metal Ions, EDTA and DTT
EDTA (5 and 10 mM), different concentrations (5, 10, 20, and 50 mM) of MgCl2 or MnCl2, and various concentrations of DTT (0.2, 1, and 5 mM) with 20 mM MgCl2 were used in HEPE buffer (pH 7.0, 100 mM) to analyze their effects on the fucosyltransferase activity of the recombinant enzyme (9.0 µg). A reaction without EDTA, DTT and metal ions was used as a control. The concentrations of the substrates and other reaction conditions were the same as described above for the pH profile assays.
Kinetics by HPLC assay
The assays were carried out in a total volume of 10 µL in HEPES buffer (100 mM, pH 7.0) containing MgCl2 (20 mM), GDP-fucose, acceptor substrate (LacNAcβPro2AA or Neu5Acα2-3LacNAcβPro2AA) and the recombinant protein (9.0 µg). Reactions were allowed to proceed for 20 min at 37°C. Apparent kinetic parameters were obtained by varying the GDP-fucose concentration from 2.0 to 50.0 mM (2.0, 3.0, 5.0, 10.0, 20.0, and 50.0 mM) and a fixed concentration of LacNAcβPro2AA or Neu5Acα2-3LacNAcβPro2AA (5 mM); or a fixed concentration of GDP-fucose (10 mM) and varied concentrations of LacNAcβPro2AA or Neu5Acα2-3LacNAcβPro2AA (1, 1.5, 2.5, 5.0, 10.0, and 25.0 mM). Apparent kinetic parameters were obtained by fitting the data (the average values of duplicate assay results) into the Michaelis–Menten equation using Grafit 5.0.
pH profile of GDP-fucose hydrolysis by capillary electrophoresis assays
Assays were performed in a total volume of 10 µL in a buffer (200 mM) with pH varying from 6.0 to 10.0 containing GDP-fucose (10 mM), MgCl2 (20 mM) and the recombinant enzyme (9.0 µg). Reactions were allowed to proceed for 30 min at 37°C before being quenched by adding ice-cold water (90 µL) to make 10-fold dilutions. The samples were then kept on ice until aliquots of 6 µL were withdrawn and analyzed by a Beckman P/ACE MDQ capillary electrophoresis system (60 cm × 75 µm i.d.) with a PDA detector. The ratio of the absorbance for GDP-fucose and GDP at 254 nm was determined at different concentrations (2.5, 5 and 10 mM). All assays were carried out in duplicate.
Kinetics of GDP-fucose hydrolysis by capillary electrophoresis assays
The enzymatic assays were carried out in a total volume of 10 µL in HEPES buffer (100 mM, pH 7.0) containing MgCl2 (20 mM), GDP-fucose and the recombinant protein (9.0 µg). Reactions were allowed to proceed for 20 min at 37°C. Apparent kinetic parameters were obtained by varying the GDP-fucose concentration from 2.0 to 50.0 mM (2.0, 3.0, 5.0, 10.0, 20.0 and 50.0 mM). Apparent kinetic parameters were obtained by fitting the data (the average values of duplicate assay results) into the Michaelis–Menten equation using Grafit 5.0.
Fucosidase activity studies
Assays were carried out in a total volume of 10 µL in HEPES buffer (100 mM, pH 7.0) containing MgCl2 (20 mM), LexβProN3 (10 mM) and various amounts of the recombinant protein (9.0, 18.0 or 27.0 µg). After 1 h reaction, the crude reaction mixture was analyzed by TLC. In addition, both LexβMU and Neu5Acα2-3LexβMU (5 mM) were used as substrates to test the fucosidase activity of the recombinant enzyme in a buffer (200 mM) containing MgCl2 (20 mM) with pH varying from 5.0 to 8.0.
Product inhibition studies using GDP or LexβProN3 as an inhibitor
Different concentrations (1, 5 and 10 mM) of LexβProN3 or GDP (10 mM) were added to the reaction mixture to analyze its effect on the fucosyltransferase activity of the recombinant enzyme. Reaction without LexβProN3 or GDP in HEPES buffer (pH 7.0, 100 mM) containing MgCl2 (20 mM), LacNAcβPro2AA (5 mM), GDP-Fucose (10 mM) and the recombinant enzyme (9.0 µg) was used as a control.
Funding
This work was supported by the Alfred P. Sloan Research Fellowship and Camille Dreyfus Teacher-Scholarship (to X. Chen) and NIH R01HD061935 (to P.G. Wang).
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Conflict of interest statement
None declared.
Abbreviations
- ATP
adenosine 5′-triphosphate
- BCA
bicinchoninic acid
- CAPS
N-cyclohexyl-3-aminopropanesulfonic acid
- CAPSO
N-cyclohexyl-2-hydroxyl-3-aminopropanesulfonic acid
- CAZy
Carbohydrate Active Enzyme
- DTT
dithiothreitol
- FKP
l-fucokinase/GDP-fucose pyrophosphorylase
- FT
fucosyltransferase
- Fuc
fucose
- Gal
galactose
- GDP-Fuc
guanosine-5′-diphosphate l-fucose
- GlcNAc
N-acetylglucosamine
- GT
glycosyltransferase
- GTP
guanosine-5′-triphosphate
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HhFT1
Helicobacter hepaticus α1-3-fucosyltransferase
- HhFT2
H. hepaticus fucosyltransferase
- HPLC
high-performance liquid chromatography
- LacNAc
N-acetyllactosamine
- Lex
Lewis x
- LPS
lipopolysaccharides
- MBP
maltose binding protein
- MES
2-(N-morpholino)ethanesulfonic acid
- Neu5Ac
N-acetylneuraminic acid
- NMR
nuclear magnetic resonance
- PCR
polymerase chain reactions
- SDS-PAGE
sodium dodecylsulfate-polyacrylamide gel electrophoresis
- SLex
sialyl Lewis x
- TLC
thin-layer chromatography
- Tris
tris(hydroxymethyl)aminomethane
References
- Apoil PA, Roubinet F, Despiau S, Mollicone R, Oriol R, Blancher A. Evolution of alpha 2-fucosyltransferase genes in primates: relation between an intronic Alu-Y element and red cell expression of ABH antigens. Mol Biol Evol. 2000;17:337–351. doi: 10.1093/oxfordjournals.molbev.a026314. [DOI] [PubMed] [Google Scholar]
- Barreaud JP, Saunier K, Souchaire J, Delourme D, Oulmouden A, Oriol R, Leveziel H, Julien R, Petit JM. Three bovine alpha2-fucosyltransferase genes encode enzymes that preferentially transfer fucose on Galbeta1-3GalNAc acceptor substrates. Glycobiology. 2000;10:611–621. doi: 10.1093/glycob/10.6.611. [DOI] [PubMed] [Google Scholar]
- Belot F, Rabuka D, Fukuda M, Hindsgaul O. Chemoenzymatic synthesis of sulfated O-linked oligosaccharides: epitopes for MECA-79. Tetrahedron Lett. 2002;43:7743–7747. [Google Scholar]
- Borges BN, Paiva TS, Harada ML. Evolution of the SEC1 gene in New World monkey lineages (Primates, Platyrrhini) Genet Mol Res. 2008;7:663–678. doi: 10.4238/vol7-3gmr452. [DOI] [PubMed] [Google Scholar]
- Bowman KG, Cook BN, de Graffenried CL, Bertozzi CR. Biosynthesis of l-selectin ligands: sulfation of sialyl Lewis x-related oligosaccharides by a family of GlcNAc-6-sulfotransferases. Biochemistry. 2001;40:5382–5391. doi: 10.1021/bi001750o. [DOI] [PubMed] [Google Scholar]
- Breton C, Snajdrova L, Jeanneau C, Koca J, Imberty A. Structures and mechanisms of glycosyltransferases. Glycobiology. 2006;16:29R–37R. doi: 10.1093/glycob/cwj016. [DOI] [PubMed] [Google Scholar]
- Brzezinski K, Stepkowski T, Panjikar S, Bujacz G, Jaskolski M. High-resolution structure of NodZ fucosyltransferase involved in the biosynthesis of the nodulation factor. Acta Biochim Pol. 2007;54:537–549. [PubMed] [Google Scholar]
- Bureau V, Marionneau S, Cailleau-Thomas A, Le Moullac-Vaidye B, Liehr T, Le Pendu J. Comparison of the three rat GDP-l-fucose:beta-d-galactoside 2-alpha-l-fucosyltransferases FTA, FTB and FTC. Eur J Biochem. 2001;268:1006–1019. doi: 10.1046/j.1432-1327.2001.01962.x. [DOI] [PubMed] [Google Scholar]
- Cahill RJ, Foltz CJ, Fox JG, Dangler CA, Powrie F, Schauer DB. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun. 1997;65:3126–3131. doi: 10.1128/iai.65.8.3126-3131.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JA, Davies GJ, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326(Pt 3):929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NW, Stangier K, Sherburne R, Taylor DE, Zhang Y, Dovichi NJ, Palcic MM. The biosynthesis of Lewis X in Helicobacter pylori. Glycobiology. 1995;5:683–688. doi: 10.1093/glycob/5.7.683. [DOI] [PubMed] [Google Scholar]
- Chokhawala HA, Huang S, Lau K, Yu H, Cheng J, Thon V, Hurtado-Ziola N, Guerrero JA, Varki A, Chen X. Combinatorial chemoenzymatic synthesis and high-throughput screening of sialosides. ACS Chem Biol. 2008;3:567–576. doi: 10.1021/cb800127n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- Daines DA, Wright LF, Chaffin DO, Rubens CE, Silver RP. NeuD plays a role in the synthesis of sialic acid in Escherichia coli K1. FEMS Microbiol Lett. 2000;189:281–284. doi: 10.1111/j.1574-6968.2000.tb09244.x. [DOI] [PubMed] [Google Scholar]
- Danishefsky SJ, Allen JR. From the laboratory to the clinic: a retrospective on fully synthetic carbohydrate-based anticancer vaccines frequently used abbreviations are listed in the appendix. Angew Chem Int Ed Engl. 2000;39:836–863. doi: 10.1002/(sici)1521-3773(20000303)39:5<836::aid-anie836>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Domino SE, Zhang L, Lowe JB. Molecular cloning, genomic mapping, and expression of two secretor blood group alpha (1, 2)fucosyltransferase genes differentially regulated in mouse uterine epithelium and gastrointestinal tract. J Biol Chem. 2001;276:23748–23756. doi: 10.1074/jbc.M100735200. [DOI] [PubMed] [Google Scholar]
- Dube DH, Bertozzi CR. Glycans in cancer and inflammation-potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- Fox JG, Dewhirst FE, Tully JG, Paster BJ, Yan L, Taylor NS, Collins MJ, Jr, Gorelick PL, Ward JM. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Chan NW, Palcic MM, Taylor DE. Cloning and heterologous expression of an alpha1, 3-fucosyltransferase gene from the gastric pathogen Helicobacter pylori. J Biol Chem. 1997;272:21357–21363. doi: 10.1074/jbc.272.34.21357. [DOI] [PubMed] [Google Scholar]
- Guo H, Yi W, Shao J, Lu Y, Zhang W, Song J, Wang PG. Molecular analysis of the O-antigen gene cluster of Escherichia coli O86:B7 and characterization of the chain length determinant gene (wzz) Appl Environ Microbiol. 2005;71:7995–8001. doi: 10.1128/AEM.71.12.7995-8001.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa Y, Lin YC, Dumas DP, Shen GJ, Garciajunceda E, Williams MA, Bayer R, Ketcham C, Walker LE, Paulson JC, et al. Chemical-enzymatic synthesis and conformational-analysis of sialyl Lewis-X and derivatives. J Am Chem Soc. 1992;114:9283–9298. [Google Scholar]
- Ihara H, Ikeda Y, Toma S, Wang X, Suzuki T, Gu J, Miyoshi E, Tsukihara T, Honke K, Matsumoto A, et al. Crystal structure of mammalian alpha1, 6-fucosyltransferase, FUT8. Glycobiology. 2007;17:455–466. doi: 10.1093/glycob/cwl079. [DOI] [PubMed] [Google Scholar]
- Kannagi R. Regulatory roles of carbohydrate ligands for selectins in the homing of lymphocytes. Curr Opin Struct Biol. 2002;12:599–608. doi: 10.1016/s0959-440x(02)00365-2. [DOI] [PubMed] [Google Scholar]
- Kannagi R. Molecular mechanism for cancer-associated induction of sialyl Lewis X and sialyl Lewis A expression-the Warburg effect revisited. Glycoconj J. 2004;20:353–364. doi: 10.1023/B:GLYC.0000033631.35357.41. [DOI] [PubMed] [Google Scholar]
- Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999;8:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RJ, Rouquier S, Giorgi D, Lennon GG, Lowe JB. Sequence and expression of a candidate for the human Secretor blood group alpha(1, 2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J Biol Chem. 1995;270:4640–4649. doi: 10.1074/jbc.270.9.4640. [DOI] [PubMed] [Google Scholar]
- Koda Y, Soejima M, Wang B, Kimura H. Structure and expression of the gene encoding secretor-type galactoside 2-alpha-l-fucosyltransferase (FUT2) Eur J Biochem. 1997;246:750–755. doi: 10.1111/j.1432-1033.1997.t01-1-00750.x. [DOI] [PubMed] [Google Scholar]
- Kudo T, Iwasaki H, Nishihara S, Shinya N, Ando T, Narimatsu I, Narimatsu H. Molecular genetic analysis of the human Lewis histo-blood group system. II. Secretor gene inactivation by a novel single missense mutation A385T in Japanese nonsecretor individuals. J Biol Chem. 1996;271:9830–9837. doi: 10.1074/jbc.271.16.9830. [DOI] [PubMed] [Google Scholar]
- Larsen RD, Ernst LK, Nair RP, Lowe JB. Molecular cloning, sequence, and expression of a human GDP-l-fucose:beta-d-galactoside 2-alpha-l-fucosyltransferase cDNA that can form the H blood group antigen. Proc Natl Acad Sci USA. 1990;87:6674–6678. doi: 10.1073/pnas.87.17.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Liu XW, Shao J, Shen J, Jia Q, Yi W, Song JK, Woodward R, Chow CS, Wang PG. Characterization of a novel alpha 1, 2-fucosyltransferase of Escherichia coli O128:B12 and functional investigation of its common motif. Biochemistry. 2008;47:378–387. doi: 10.1021/bi701345v. [DOI] [PubMed] [Google Scholar]
- Lin B, Hayashi Y, Saito M, Sakakibara Y, Yanagisawa M, Iwamori M. GDP-fucose: beta-galactoside alpha1, 2-fucosyltransferase, MFUT-II, and not MFUT-I or -III, is induced in a restricted region of the digestive tract of germ-free mice by host-microbe interactions and cycloheximide. Biochim Biophys Acta. 2000;1487:275–285. doi: 10.1016/s1388-1981(00)00103-7. [DOI] [PubMed] [Google Scholar]
- Lin SW, Yuan TM, Li JR, Lin CH. Carboxyl terminus of Helicobacter pylori alpha1, 3-fucosyltransferase determines the structure and stability. Biochemistry. 2006;45:8108–8116. doi: 10.1021/bi0601297. [DOI] [PubMed] [Google Scholar]
- Lowe JB. Glycan-dependent leukocyte adhesion and recruitment in inflammation. Curr Opin Cell Biol. 2003;15:531–538. doi: 10.1016/j.ceb.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Lubineau A, Auge C, Le Goff N, Le Narvor C. Chemoenzymatic synthesis of a 3IV, 6III-disulfated Lewis(x) pentasaccharide, a candidate ligand for human l-selectin. Carbohydr Res. 1997;305:501–509. doi: 10.1016/s0008-6215(97)10043-x. [DOI] [PubMed] [Google Scholar]
- Lubineau A, Le Narvor C, Auge C, Gallet PF, Petit JM, Julien R. Chemo-enzymatic synthesis of a selectin ligand using recombinant yeast cells. J Mol Catalysis B: Enzymatic. 1998;5:229–234. [Google Scholar]
- Ma B, Lau LH, Palcic MM, Hazes B, Taylor ED. A single aromatic amino acid at the carboxyl terminus of Helicobacter pylori alpha1, 3/4 fucosyltransferase determines substrate specificity. J Biol Chem. 2005;280:36848–36856. doi: 10.1074/jbc.M504415200. [DOI] [PubMed] [Google Scholar]
- Ma B, Simala-Grant JL, Taylor ED. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 2006;16:158R–184R. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- Ma B, Wang G, Palcic MM, Hazes B, Taylor DE. C-terminal amino acids of Helicobacter pylori alpha1, 3/4 fucosyltransferases determine type I and type II transfer. J Biol Chem. 2003;278:21893–21900. doi: 10.1074/jbc.M301704200. [DOI] [PubMed] [Google Scholar]
- Magnani JL. The discovery, biology, and drug development of sialyl Lea and sialyl Lex. Arch Biochem Biophys. 2004;426:122–131. doi: 10.1016/j.abb.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Marques ET, Jr, Weiss JB, Strand M. Molecular characterization of a fucosyltransferase encoded by Schistosoma mansoni. Mol Biochem Parasitol. 1998;93:237–250. doi: 10.1016/s0166-6851(98)00033-4. [DOI] [PubMed] [Google Scholar]
- Miyoshi E, Moriwaki K, Nakagawa T. Biological function of fucosylation in cancer biology. J Biochem. 2008;143:725–729. doi: 10.1093/jb/mvn011. [DOI] [PubMed] [Google Scholar]
- Monteiro MA, Chan KH, Rasko DA, Taylor DE, Zheng PY, Appelmelk BJ, Wirth HP, Yang M, Blaser MJ, Hynes SO, et al. Simultaneous expression of type 1 and type 2 Lewis blood group antigens by Helicobacter pylori lipopolysaccharides. Molecular mimicry between H. pylori lipopolysaccharides and human gastric epithelial cell surface glycoforms. J Biol Chem. 1998;273:11533–11543. doi: 10.1074/jbc.273.19.11533. [DOI] [PubMed] [Google Scholar]
- Moran AP. Relevance of fucosylation and Lewis antigen expression in the bacterial gastroduodenal pathogen Helicobacter pylori. Carbohydr Res. 2008;343:1952–1965. doi: 10.1016/j.carres.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Moran AP, Prendergast MM, Appelmelk BJ. Molecular mimicry of host structures by bacterial lipopolysaccharides and its contribution to disease. FEMS Immunol Med Microbiol. 1996;16:105–115. doi: 10.1111/j.1574-695X.1996.tb00127.x. [DOI] [PubMed] [Google Scholar]
- Okajima T, Matsuura A, Matsuda T. Biological functions of glycosyltransferase genes involved in O-fucose glycan synthesis. J Biochem. 2008;144:1–6. doi: 10.1093/jb/mvn016. [DOI] [PubMed] [Google Scholar]
- Ouerfelli O, Warren JD, Wilson RM, Danishefsky SJ. Synthetic carbohydrate-based antitumor vaccines: challenges and opportunities. Expert Rev Vaccines. 2005;4:677–685. doi: 10.1586/14760584.4.5.677. [DOI] [PubMed] [Google Scholar]
- Piau JP, Labarriere N, Dabouis G, Denis MG. Evidence for two distinct alpha(1, 2)-fucosyltransferase genes differentially expressed throughout the rat colon. Biochem J. 1994;300(Pt 3):623–626. doi: 10.1042/bj3000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt MR, Bertozzi CR. Syntheses of 6-sulfo sialyl Lewis X glycans corresponding to the l-selectin ligand “sulfoadhesin”. Org Lett. 2004;6:2345–2348. doi: 10.1021/ol0493195. [DOI] [PubMed] [Google Scholar]
- Rabbani S, Miksa V, Wipf B, Ernst B. Molecular cloning and functional expression of a novel Helicobacter pylori alpha-1, 4 fucosyltransferase. Glycobiology. 2005;15:1076–1083. doi: 10.1093/glycob/cwj004. [DOI] [PubMed] [Google Scholar]
- Rogers AB, Fox JG. Inflammation and cancer. I. Rodent models of infectious gastrointestinal and liver cancer. Am J Physiol Gastrointest Liver Physiol. 2004;286:G361–G366. doi: 10.1152/ajpgi.00499.2003. [DOI] [PubMed] [Google Scholar]
- Rouquier S, Lowe JB, Kelly RJ, Fertitta AL, Lennon GG, Giorgi D. Molecular cloning of a human genomic region containing the H blood group alpha(1, 2)fucosyltransferase gene and two H locus-related DNA restriction fragments. Isolation of a candidate for the human Secretor blood group locus. J Biol Chem. 1995;270:4632–4639. doi: 10.1074/jbc.270.9.4632. [DOI] [PubMed] [Google Scholar]
- Sanabria-Valentin E, Colbert MT, Blaser MJ. Role of futC slipped strand mispairing in Helicobacter pylori Lewisy phase variation. Microbes Infect. 2007;9:1553–1560. doi: 10.1016/j.micinf.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunier K, Barreaud JP, Eggen A, Oriol R, Leveziel H, Julien R, Petit JM. Organization of the bovine alpha 2-fucosyltransferase gene cluster suggests that the Sec1 gene might have been shaped through a nonautonomous L1-retrotransposition event within the same locus. Mol Biol Evol. 2001;18:2083–2091. doi: 10.1093/oxfordjournals.molbev.a003749. [DOI] [PubMed] [Google Scholar]
- Seeberger PH, Werz DB. Synthesis and medical applications of oligosaccharides. Nature. 2007;446:1046–1051. doi: 10.1038/nature05819. [DOI] [PubMed] [Google Scholar]
- Shao J, Li M, Jia Q, Lu Y, Wang PG. Sequence of Escherichia coli O128 antigen biosynthesis cluster and functional identification of an alpha-1, 2-fucosyltransferase. FEBS Lett. 2003;553:99–103. doi: 10.1016/s0014-5793(03)00980-3. [DOI] [PubMed] [Google Scholar]
- Simanek EE, McGarvey GJ, Jablonowski JA, Wong CH. Selectin-carbohydrate interactions: From natural ligands to designed mimics. Chem Rev. 1998;98:833–862. doi: 10.1021/cr940226i. [DOI] [PubMed] [Google Scholar]
- Smialowski P, Martin-Galiano AJ, Mikolajka A, Girschick T, Holak TA, Frishman D. Protein solubility: sequence based prediction and experimental verification. Bioinformatics. 2007;23:2536–2542. doi: 10.1093/bioinformatics/btl623. [DOI] [PubMed] [Google Scholar]
- Suerbaum S, Josenhans C, et al. The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. Proc Natl Acad Sci USA. 2003;100:7901–7906. doi: 10.1073/pnas.1332093100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- Sun HY, Lin SW, Ko TP, Pan JF, Liu CL, Lin CN, Wang AH, Lin CH. Structure and mechanism of Helicobacter pylori fucosyltransferase. A basis for lipopolysaccharide variation and inhibitor design. J Biol Chem. 2007;282:9973–9982. doi: 10.1074/jbc.M610285200. [DOI] [PubMed] [Google Scholar]
- Timmons SC, Jakeman DL. Stereoselective chemical synthesis of sugar nucleotides via direct displacement of acylated glycosyl bromides. Org Lett. 2007;9:1227–1230. doi: 10.1021/ol063068d. [DOI] [PubMed] [Google Scholar]
- van Der Wel H, Morris HR, Panico M, Paxton T, North SJ, Dell A, Thomson JM, West CM. A non-Golgi alpha 1, 2-fucosyltransferase that modifies Skp1 in the cytoplasm of Dictyostelium. J Biol Chem. 2001;276:33952–33963. doi: 10.1074/jbc.M102555200. [DOI] [PubMed] [Google Scholar]
- Wang G, Boulton PG, Chan NW, Palcic MM, Taylor DE. Novel Helicobacter pylori alpha1, 2-fucosyltransferase, a key enzyme in the synthesis of Lewis antigens. Microbiology. 1999;145(Pt 11):3245–3253. doi: 10.1099/00221287-145-11-3245. [DOI] [PubMed] [Google Scholar]
- Wang G, Ge Z, Rasko DA, Taylor DE. Lewis antigens in Helicobacter pylori: biosynthesis and phase variation. Mol Microbiol. 2000;36:1187–1196. doi: 10.1046/j.1365-2958.2000.01934.x. [DOI] [PubMed] [Google Scholar]
- Wang W, Hu T, Frantom PA, Zheng T, Gerwe B, Del Amo DS, Garret S, Seidel RD, 3rd, Wu P. Chemoenzymatic synthesis of GDP-l-fucose and the Lewis X glycan derivatives. Proc Natl Acad Sci USA. 2009;106:16096–16101. doi: 10.1073/pnas.0908248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Liu X, Li Y, Li J, Xia C, Zhou G, Zhang W, Zhao W, Chen X, Wang PG. Remodeling bacterial polysaccharides by metabolic pathway engineering. Proc Natl Acad Sci USA. 2009;106:4207–4212. doi: 10.1073/pnas.0812432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Cheng J, Ding L, Khedri Z, Chen Y, Chin S, Lau K, Tiwari VK, Chen X. Chemoenzymatic synthesis of GD3 oligosaccharides and other disialyl glycans containing natural and non-natural sialic acids. J Am Chem Soc. 2009;131:18467–18477. doi: 10.1021/ja907750r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chokhawala H, Karpel R, Yu H, Wu B, Zhang J, Zhang Y, Jia Q, Chen X. A multifunctional Pasteurella multocida sialyltransferase: a powerful tool for the synthesis of sialoside libraries. J Am Chem Soc. 2005;127:17618–17619. doi: 10.1021/ja0561690. [DOI] [PubMed] [Google Scholar]
- Yu H, Yu H, Karpel R, Chen X. Chemoenzymatic synthesis of CMP-sialic acid derivatives by a one-pot two-enzyme system: comparison of substrate flexibility of three microbial CMP-sialic acid synthetases. Bioorg Med Chem. 2004;12:6427–6435. doi: 10.1016/j.bmc.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Van Die I, Cummings RD. A novel alpha1, 2-fucosyltransferase (CE2FT-2) in Caenorhabditis elegans generates H-type 3 glycan structures. Glycobiology. 2008;18:290–302. doi: 10.1093/glycob/cwn007. [DOI] [PubMed] [Google Scholar]