Abstract
Giardia lamblia, which is an important parasitic cause of diarrhea, uses activated forms of glucose to make glycogen and activated forms of mannose to make glycophosphosphoinositol anchors. A necessary step for glucose activation is isomerization of glucose-6-phosphate to glucose-1-phosphate by a phosphoglucomutase (PGM). Similarly, a phosphomannomutase (PMM) converts mannose-6-phosphate to mannose-1-phosphate. While whole genome sequences of Giardia predict two PGM candidates, no PMM candidate is present. The hypothesis tested here is that at least one of the two Giardia PGM candidates has both PGM and PMM activity, as has been described for bacterial PGM orthologs. Nondenaturing gels showed that Giardia has two proteins with PGM activity, one of which also has PMM activity. Phylogenetic analyses showed that one of the two Giardia PGM candidates (Gl-PGM1) shares recent common ancestry with other eukaryotic PGMs, while the other Giardia PGM candidate (Gl-PGM2) is deeply divergent. Both Gl-PGM1 and Gl-PGM2 rescue a Saccharomyces cerevisiae pgm1Δ/pgm2Δ double deletion strain, while only Gl-PGM2 rescues a temperature-sensitive PMM mutant of S. cerevisiae (sec53-ts). Recombinant Gl-PGM1 has PGM activity only, whereas Gl-PGM2 has both PGM and PMM activities. We conclude that Gl-PGM1 behaves as a conventional eukaryotic PGM, while Gl-PGM2 is a novel eukaryotic PGM that also has PMM activity.
Keywords: Giardia, phosphoglucomutase, phosphomannomutase
Introduction
Giardia lamblia (also known as G. duodenalis and G. interstinalis) is a deeply divergent, binucleate, flagellated protist, which causes two million cases of diarrhea per year in the United States (Steiner et al. 1997; Adam 2001; Simpson et al. 2006). Whole genome sequences of two human isolates of Giardia called WB and GS suggest that they separated from shared common ancestor tens of millions of years ago (Morrison et al. 2007; Franzén et al. 2009). Each Giardia genome is small (∼11.7 Mb) and encodes fewer proteins (∼4500) than the vast majority of eukaryotes. Giardia has a large number of genes, which have been received from bacteria by lateral gene transfer (LGT) (Nixon et al. 2002; Andersson et al. 2003; Morrison et al. 2007). Giardia genes obtained by LGT encode numerous glycolytic enzymes, most fermentation enzymes, as well as nitroreductases that reduce and activate metronidazole (Pal et al. 2009).
Compared to other eukaryotes, Giardia has many fewer enzymes involved in protein glycosylation:
The dolichol pyrophosphate-linked precursor to Asn-linked glycans (N-glycans) of Giardia contains just two sugars (GlcNAc2) rather than the 14-sugar precursor present in metazoa, fungi and plants (Helenius and Aebi 2004; Samuelson et al. 2005). Giardia N-glycans are not modified by Golgi enzymes (Ratner et al. 2008).
Giardia is missing the N-glycan-dependent quality control (QC) systems for glycoprotein folding and degradation (Banerjee et al. 2007). While there is positive selection for sites of N-linked glycosylation (sequons) in secreted proteins of the majority of eukaryotes that have N-glycan-dependent QC, there is no selection for sequons in Giardia (Cui et al. 2009).
Giardia has a single nucleotide-sugar transporter that transports GlcNAc (Banerjee et al. 2008), and Giardia is one of the few protists that adds O-linked GlcNAc to nucleo cytosolic proteins (Banerjee et al. 2009).
While many eukaryotes use dolichol-phosphate-mannose to make N-glycan precursors, O-linked glycans and glycophosphosphoinositol (GPI) anchors, Giardia only uses dolichol-phosphate-mannose to make GPI anchors and does not synthesize dolichol-phosphate glucose (Orlean 1990; Das et al. 1991; Samuelson et al. 2005).
The cis-prenyltransferase of Giardia makes dehydrodolichol pyrophosphate containing 12 isoprenoid units like those of bacteria, and the Giardia enzyme shares common ancestry with bacterial and trypanosome enzymes rather than metazoan, fungal and plant enzymes (Grabińska et al. 2010).
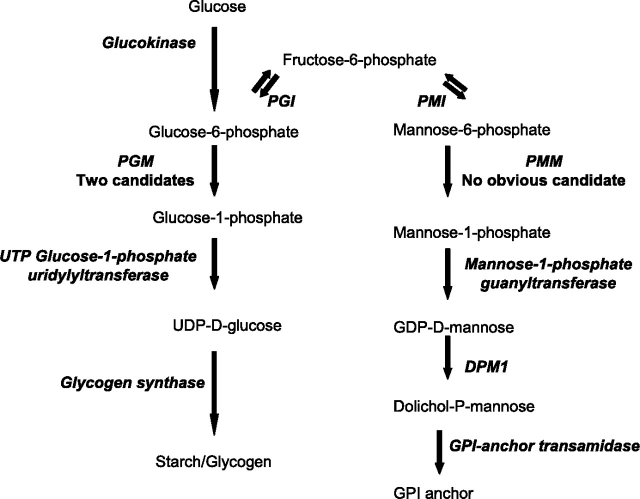
In this report, we are interested in the Giardia phosphoglucomutase (PGM) that makes the activated form of glucose (Glc-1-P) for glycogen synthesis and the phosphomannomutase (PMM) that makes activated mannose (Man-1-P) for GPI anchor synthesis (Figure 1). While candidate Giardia enzymes have been identified for other steps in the synthesis of glycogen and GPI anchors, whole genome sequences of the organism predict two candidate PGM enzymes: Gl-PGM1 (GiardiaDB 50803_17254) and Gl-PGM2 (GiardiaDB 50803_11448) but no candidates for PMM (Henze et al. 2001; Morrison et al. 2007). This result is surprising because PMM orthologs (defined by common ancestry) of higher eukaryotes may have both PMM and PGM activities (Pirard et al. 1999; Cromphout et al. 2006), but the eukaryotic PGM orthologs have not been shown to have PMM activity (Table I) (Quick et al. 1974). In contrast, all bacteria lack a PMM ortholog, while some bacteria have PGM orthologs that have both PGM and PMM activities (Regni et al. 2004, 2006). The reaction of the bacterial PGM/PMM involves a phosphoryl transfer from a phosphoserine on the enzyme to the bound substrate to form a bisphosphorylated intermediate, which is then reoriented 180° for a second phosphoryl transfer from the intermediate back to the enzyme.
Fig. 1.
Substrates and enzymes, which include PGM and PMM, used by Giardia to make glycogen and GPI anchors. Candidate enzymes (italics) include the following: glucokinase (encoded by GiardiaDB 50803_8826) (Henze et al. 2001; Aurrecoechea et al. 2008), phosphoglucomutases (PGMs) (50803_11448 and 50803_11448), UTP-Glc-1-P uridylyltransferase (50803_29307), glycogen synthase (50803_1903), glucose-6-phosphate isomerase (PGI) (50803_9115), mannose-1-phosphate guanyltransferase (50803_16598), dolichol-phosphate-mannose synthase (DPM1) (50803_3180) (Samuelson et al. 2005) and GPI-anchor transamidase (50803_15267). There is no candidate phosphomannomutase (PMM)
Table I.
List of eukaryotic and bacterial PGM and PMM orthologs and their preferred activity
| PGM | PMM | References | |
|---|---|---|---|
| Eukaryotic PGM orthologs | − | Quick et al. (1974) | |
| Eukaryotic PMM1 | Pirard et al. (1999); Cromphout et al. (2006) | ||
| Eukaryotic PMM2 | − | Pirard et al. (1999); Cromphout et al. (2006) | |
| Bacterial PGM orthologs | Regni et al. (2004, 2006) | ||
| Giardia PGM1 | − | In this study | |
| Giardia PGM2 | In this study |
The major hypothesis tested here is that at least one of the Giardia PGM orthologs has both PGM and PMM activities, as has been described for bacterial PGM orthologs. The minor hypothesis tested was that Giardia obtained this bifunctional enzyme by LGT from bacteria. To test this idea, we used nondenaturing gels to separate PGM activities of Giardia and to determine whether any of the Giardia PGMs also have PMM activity. We used phylogenetic methods to determine whether the Giardia PGMs might have derived from bacteria by LGT. In addition, we tested the ability of the Giardia PGMs from the WB isolate to complement a Saccharomyces cerevisiae pgm1Δ/pgm2Δ double deletion strain and a Saccharomyces sec53-ts mutant that lacks PMM activity at the nonpermissive temperature and measured the kinetics of PGM and PMM activities of recombinant Giardia PGMs.
Results and discussion
Nondenaturing gels suggest that Giardia has two enzymes with PGM activity, one of which also has PMM activity
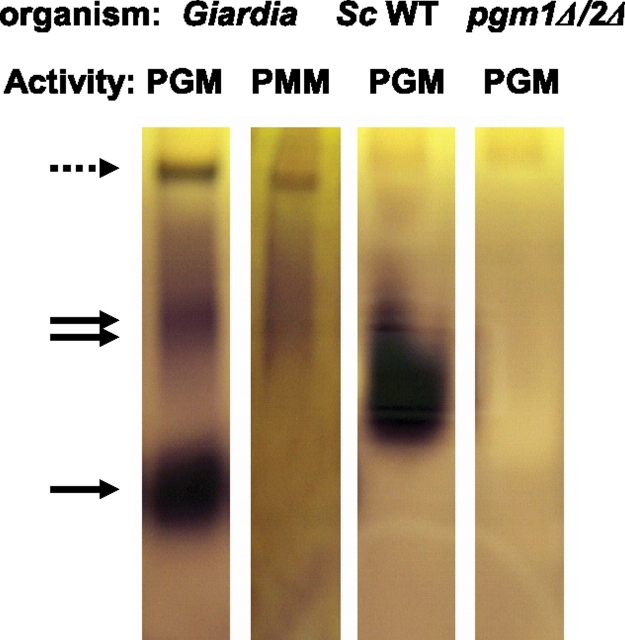
To determine whether Giardia has one or more enzymes with PGM and/or PMM activities, trophozoites of WB were lysed by sonication, and soluble proteins were separated on a nondenaturing polyacrylamide gel. This gel was overlayed with agar containing colorometric reagents to detect PGM and PMM activities. We found that Giardia has two enzymes with PGM activity (Figure 2). The faster migrating Giardia enzyme has much greater PGM activity than the slower migrating enzyme, consistent with either a more active enzyme or a more abundant protein, or both. However, the slower migrating Giardia enzyme also has PMM activity, albeit weaker than the PGM activity. As controls for the PGM assays, wild-type Saccharomyces shows a single wide band of PGM activity, which is absent in a Saccharomyces pgm1Δ/pgm2Δ double deletion strain.
Fig. 2.
A nondenaturing, discontinuous polyacrylamide gel shows that lysates of Giardia contain two proteins with PGM activities, one of which also has PMM activity. Giardia lysates were tested for PGM activity (lane 1) and PMM activity (lane 2). The single arrow identifies a fast migrating Giardia protein, which only has PGM activity. The double arrow identifies a slower migrating Giardia protein, which has both PGM and PMM activities. The dashed arrow is the interface between stacking and running gels. Lysates of wild-type Saccharomyces (Sc WT in lane 3) have PGM activity, which is absent from lysates of the Saccharomyces pgm1Δ/pgm2Δ double deletion strain (lane 4)
In addition to the activity data shown in Figure 2, we used multiple dimensional protein identification technology (MudPIT) to identify proteins from lysed Giardia. These unpublished data have been deposited at GiardiaDB (Aurrecoechea et al. 2008). MudPIT analyses suggested that one of the candidate Giardia PGMs (Gl-PGM1) with 24 peptides identified is more abundant than the other (Gl-PGM2) with just five peptides identified.
We conclude that Giardia has two enzymes with PGM activity, and one of these PGMs that may be less abundant also has PMM activity. In the following sections, we use bioinformatics methods to compare candidate Giardia PGMs to those of other eukaryotes, and we test the ability of the Giardia PGMs to complement a S. cerevisiae pgm1Δ/pgm2Δ double deletion mutant and a Saccharomyces sec53-ts mutant that lacks PMM activity at the nonpermissive temperature.
One of the predicted Giardia phosphoglucomutases (Gl-PGM2) is deeply divergent
The predicted proteins of the WB isolate of Giardia include two PGM orthologs, which we labeled as Gl-PGM1 (GiardiaDB 50803_17254) and Gl-PGM2 (GiardiaDB 50803_11448) (Morrison et al. 2007). Gl-PGM1 is 655-amino acids long and contains throughout its length a PGM domain (Marchler-Bauer et al. 2005). In contrast, the second predicted PGM of Giardia (Gl-PGM2) is shorter than other PGMs (527-amino acids long) and contains only the N-terminal half of the PGM domain. Gl-PGM1 of WB strain and Gl-PGM2 of GS strain have a 37% identity and 54% similarity over a 494 amino acid alignment with 4% gaps (Supplemental Figure 1). Because Gl-PGM2 is much more similar to Gl-PGM1 than to any other PGM (data not shown), it is likely that Gl-PGM2 resulted from gene duplication in Giardia rather than from gene duplication in a common ancestor to Giardia and other eukaryotes.
Gl-PGM1 shows a 31% identity and a 44% similarity over a short (132 amino acid) overlap with the Pseudomonas PGM/PMM that has been extensively characterized (Regni et al. 2004, 2006). While the phosphoryl transfer site including the Ser that is phosphorylated is conserved in Gl-PGM1, phosphate contacts and sugar ring contacts are not conserved (Supplemental Figure 1). Gl-PGM2 cannot be directly aligned with Pseudomonas PGM/PMM or with any of the other PGMs that have been crystallized. However, using Gl-PGM1 as a guide for the alignment, it is clear that the phosphoryl transfer site, including the Ser that is phosphorylated, is not well conserved in Gl-PGM2 (Supplemental Figure 1). Crystallization of the Gl-PGM1 and Gl-PGM2 with various substrates, which is necessary for understanding structure–function relations of the enzymes, is beyond the scope of the present study.
Candidate Giardia PGMs share common ancestry with eukaryotic enzymes
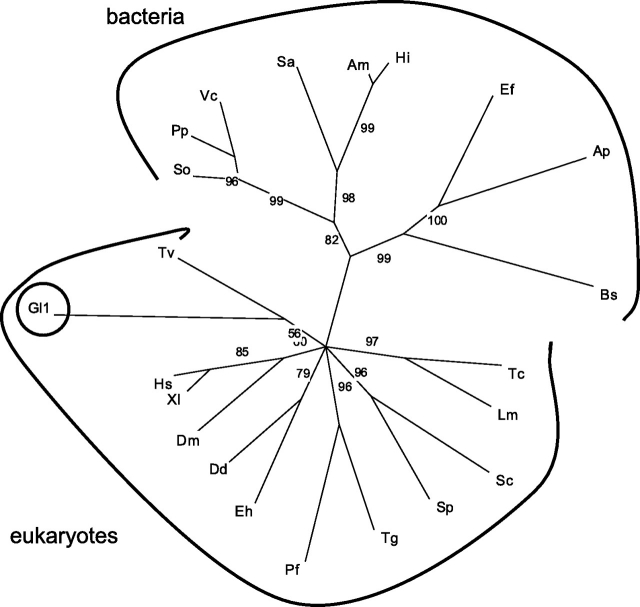
As Gl-PGM2 is so unlike other PGMs and so was unalignable over its C-terminal half with the others, this predicted Giardia PGM was not included in a phylogenetic analysis of PGMs from representative eukaryotes and bacteria (Figure 3). The phylogenetic analyses showed two things. First, all eukaryotes examined (with the exception of Trypanosoma brucei) have at least one PGM, consistent with the importance of this enzyme in the synthesis of activated Glc used to make glycogen, starch and other products (Quick et al. 1974). Second, there is a clear distinction between eukaryotic PGMs, which includes Gl-PGM1, and bacterial PGMs. This result shows that the Giardia PGM was not derived from bacteria by LGT, as we hypothesized (Nixon et al. 2002; Andersson et al. 2003; Morrison et al. 2007).
Fig. 3.
Phylogenetic analyses of phosphoglucomutases suggest that the Giardia PGM gene is deeply divergent. In the phylogenetic tree, which was constructed by maximum likelihood methods, the branch lengths are proportionate to differences between sequences, while the nodes indicate bootstrap support. Nodes with <50% bootstrap support were collapsed. The tree includes just one of the predicted PGMs of Giardia (Gl-PGM1) because the other Gl-PGM2 aligns so poorly with the rest of the PGMs. Representative protists include Dictyostelium discoideum (Dd), Entamoeba histolytica (Eh), Leishmania major (Lm), Plasmodium falciparum (Pf), Toxoplasma gondii (Tg), Trichomonas (Tv) and Trypanosoma cruzi (Tc). Other representative eukaryotes include Drosophila melanogaster (Dm), Homo sapiens (Hs), Schizosaccharomyces pombe (Sp), Saccharomyces cerevisiae (Sc) and Xenopus laevis (Xl). Representative eubacteria include Actinobacillus minor (Am), Anaerococcus prevotii (Ap), Borrelia spielmanii (Bs), Enterococcus faecalis (Ef), Haemophilus influenzae (Hi), Photobacterium profundum (Pp), Shewanella oneidensis (So), Streptomyces avermitilis (Sa) and Vibrio cholerae (Vc). No archaeal ortholog was identified
Evidence for secondary loss of the Giardia PMM ortholog
Giardia is the only eukaryote examined that is missing a candidate PMM (see Supplemental Figure 2 for the PMM tree). As whole genome sequences were examined from two different Giardia isolates (WB and GS), it is highly unlikely that the absence of a Giardia PMM ortholog is due to an artifact in library construction or sequencing (Morrison et al. 2007; Franzén et al. 2009). As PMM orthologs are present in other deeply divergent eukaryotes (e.g. Trichomonas and Trypanosoma), it is likely that secondary loss rather than primary absence best explains why Giardia is missing a PMM gene (Simpson et al. 2006). The absences of Alg enzymes that synthesize N-glycan precursors and of N-glycan-dependent QC proteins in the endoplasmic reticulum have also been best explained by secondary loss of the genes that encode these proteins (Samuelson et al. 2005; Banerjee et al. 2007).
Functional complementation of Saccharomyces pgm1Δ/pgm2Δ double deletion strain with both Gl-PGM1 and Gl-PGM2
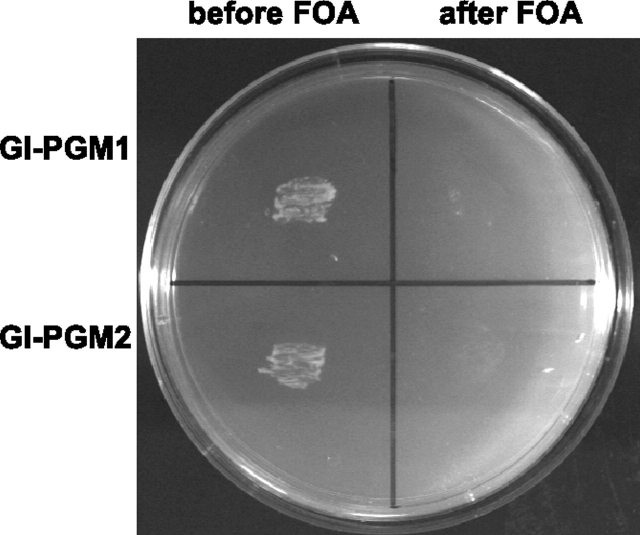
To determine whether the Gl-PGM1 and Gl-PGM2 genes encode functional PGMs, we tested their ability to complement the phenotype of a S. cerevisiae pgm1Δ/pgm2Δ double deletion strain. The complete coding sequences of Gl-PGM1 and Gl-PGM2 were expressed in the Saccharomyces pgm1Δ/pgm2Δ double deletion strain using the pVV214 vector, which contains the URA3 gene that encodes orotidine 5-phosphate decarboxylase. The double deletion strain is incapable of metabolizing galactose (Gal) and presents a lethal phenotype when Gal is used as the sole carbon source (Daran et al. 1997). The deletion strain bearing either Gl-PGM1 or Gl-PGM2 grows normally on Gal (Figure 4), indicating that both Giardia PGM genes encode proteins with PGM activity (Table I).
Fig. 4.
Functional complementation of Saccharomyces pgm1Δ/pgm2Δ double deletion mutant by Giardia phosphoglucomutases (Gl-PGM1 and Gl-PGM2). A Saccharomyces pgm1Δ/pgm2Δ double deletion mutant was transformed with pVV214 vector containing the full-length of Gl-PGM1 gene (top half of plate) or the Gl-PGM2 gene (bottom half of plate). Transformed yeasts were selected in synthetic medium-URA containing 2% glucose and then plated onto synthetic medium-URA supplemented with 2% (w/v) galactose (Gal) as the only carbon source (shown here). In the absence of a functional PGM, the Saccharomyces pgm1Δ/pgm2Δ double deletion mutant does not grow on Gal plates (data not shown). Before FOA treatment, both Giardia PGMs complement the Saccharomyces pgm1Δ/pgm2Δ double deletion mutant, so there is growth on Gal (left side of plate). After FOA treatment, which removes the plasmids containing the Giardia PGM genes, the Saccharomyces pgm1Δ/pgm2Δ cannot grow on Gal (right side of plate) but grow on Glc (data not shown)
As a control, we took advantage of URA3 marker in the transformants and grew them on plates with 1% (w/v) 5-fluoroorotic acid (FOA). FOA is converted to a toxic compound 5-fluorouracil by URA3, so the presence of URA3 on a plasmid is lethal in the presence of FOA. In this way, URA3 plasmids were removed from the transformants. As expected, the FOA-treated transformants, which no longer contain the Giardia PGM genes, do not grow on Gal plates (Figure 4).
Functional complementation of Saccharomyces temperature-sensitive PMM mutant with Gl-PGM2 but not with Gl-PGM1
To determine whether the Gl-PGM1 and Gl-PGM2 genes encode functional PMMs, we tested their ability to complement a S. cerevisiae temperature-sensitive PMM mutant (HMSF33 sec53-6 strain). In addition, as a positive control, we tested the complementation of a functional Saccharomyces sec53 gene. While the Saccharomyces sec53-ts mutant grows normally at 25°C (the permissive temperature) (Figure 5A), the mutant transformed with an empty vector (negative control) does not grow at 37°C (the restrictive or nonpermissive temperature) because the PMM is not functional (Figure 5B). Similarly, the Saccharomyces sec53-ts mutant transformed with Gl-PGM1 does not grow at 37°C. In contrast, the Saccharomyces sec53-ts mutant transformed with either the Gl-PGM2 gene or a functional Saccharomyces sec53 gene (positive control) grows at 37°C. These results, which indicate that Gl-PGM2 has PMM activity while Gl-PGM1 does not (Table I), are supported by kinetic data for the recombinant Gl-PGM1 and Gl-PGM2 that are presented in the next section.
Fig. 5.
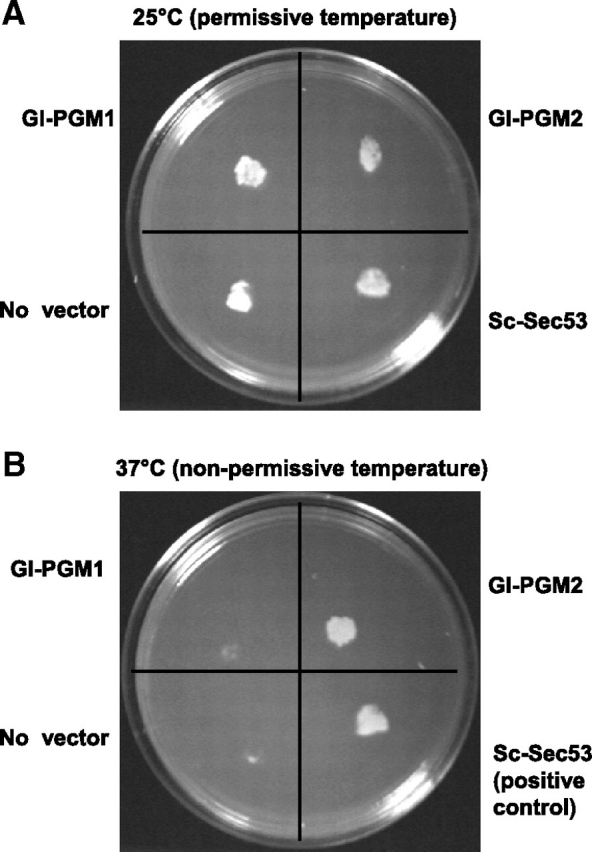
Functional complementation of Saccharomyces PMM temperature-sensitive PMM mutant (HMSF33 sec53-6 strain) by Gl-PGM2 but not by Gl-PGM1. The Saccharomyces sec53 gene encodes phosphomannomutase (PMM). (A) At 25°C (the permissive temperature), the mutant strain with no vector or transformed with Gl-PGM1, Gl-PGM2 or a functional PMM gene (Sc-Sec53) all grow normally. (B) At 37°C (nonpermissive or restrictive temperature), the mutant and the mutant transformed with Gl-PGM1 cannot grow. In contrast, the mutant transformed with Gl-PGM2 or with the functional PMM (Sc-Sec53) both grow at 37°C. These results show that the Gl-PGM2 has PMM activity but the Gl-PGM1 does not
Enzyme activities of recombinant Gl-PGM1 and Gl-PGM2
Recombinant Gl-PGM1 that has an N-terminal polyHis tag was expressed in E. coli and purified using Ni-sepharose affinity column chromatography. Recombinant Gl-PGM2 that has a C-terminal polyHis tag was expressed in wild-type Saccharomyces and purified by Ni-sepharose affinity column chromatography. Gl-PGM1 has only PGM activity (Km 8.2 ± 2.0 for Glc-1-P and specific activity is 87 ± 7 μmol/min/mg) (Figure 6A). Gl-PGM2 has both PGM and PMM activities (Km 3.0 ± 0.6 μM, specific activity 76 ± 4 μmol/min/mg for Glc-1-P and Km 5.6 ± 1.7 μM, specific activity 25 ± 2 μmol/min/mg for Man-1-P) (Figure 6B). These results are in agreement with the data from total lysates of Giardia separated on nondenaturing gels (Figure 2), which showed that one Giardia enzyme has PGM activity and no PMM activity, while the other Giardia enzyme has both PGM and PMM activities. These results are also in agreement with the Saccharomyces complementation assays that showed Gl-PGM1 has PGM but not PMM activity, while Gl-PGM2 has both PGM and PMM activities.
Fig. 6.
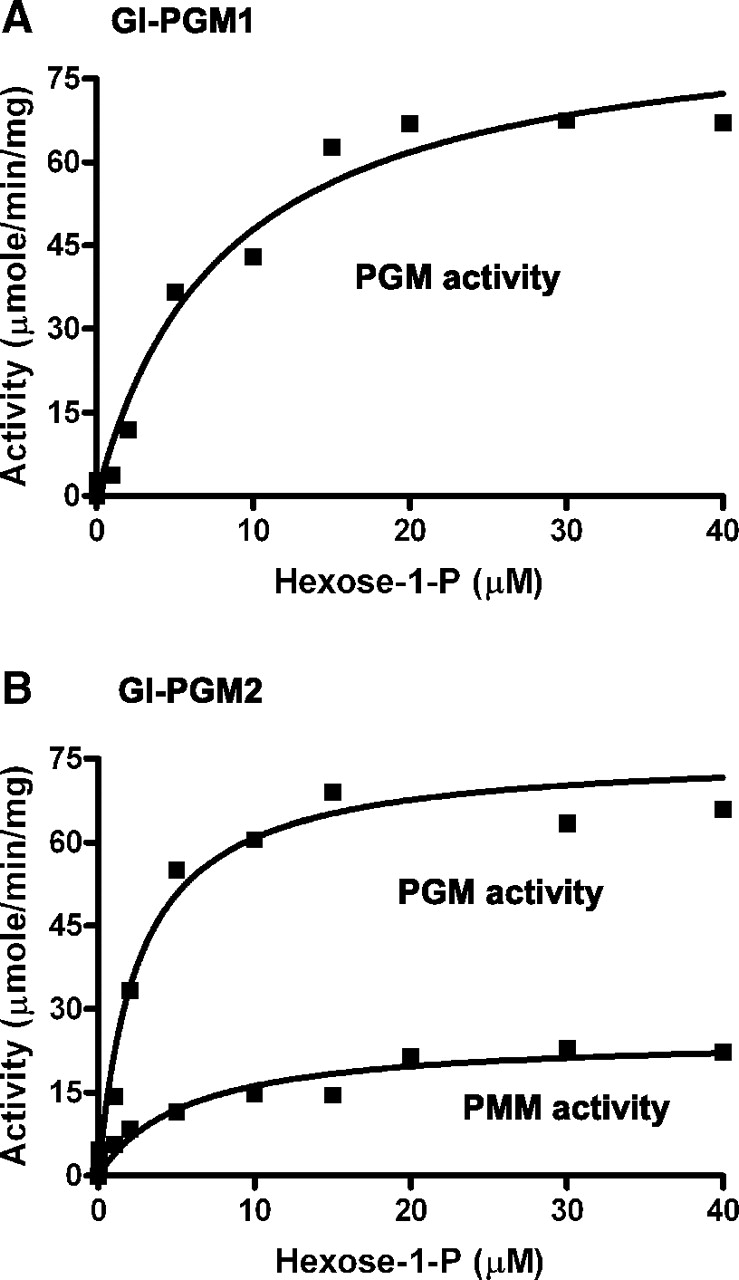
Recombinant Gl-PGM1 has PGM activity only, while recombinant Gl-PGM2 has both PGM and PMM activities (kinetics are given in the text). PGM activity was measured at room temperature by using α-glucose-1-phosphate (Glc-1-P) as the substrate. The initial rate of formation of α-glucose-6-phosphate was measured by converting it to phosphoglucolactone in the presence glucose-6-phosphate dehydrogenase. NADPH generated in this reaction was monitored by change in absorbance at 340 nm, and this gives a measure of PGM activity. Similarly, PMM activity was measured at room temperature in the presence of α-mannose-1-phosphate (Man-1-P), phosphogluco isomerase (PGI) and phosphomanno isomerase (PMI)
Conclusions
The mystery solved here is how Giardia, which lacks a eukaryotic PMM ortholog, is able to make activated mannose that is used to make GPI anchors. As predicted, one of the two Giardia PGM orthologs (Gl-PGM2), which is relatively less abundant in MudPIT analyses, has both PGM and PMM activities (Table I). While this dual PGM/PMM activity is similar to that of bacterial PGM orthologs, it does not appear that the Giardia enzymes were obtained by LGT (Nixon et al. 2002; Andersson et al. 2003; Regni et al. 2006; Morrison et al. 2007). Therefore, the presence of bifunctional PGM/PMM enzymes in Giardia and bacteria likely represents an example of convergent evolution. PMM orthologs of eukaryotes, which are not phylogenetically related to PGMs, have both PGM and PMM activities (Pirard et al. 1999; Cromphout et al. 2006). Broadening of the hexose specificity of these phosphohexosemutases has happened, therefore, on at least three occasions (PGMs acquiring PMM activity in Giardia and in bacteria and a eukaryotic PMM acquiring PGM activity) (Table I). As noted above, a structural understanding of the PGM/PMM activity of Gl-PGM2 will only be possible when the enzyme is crystallized with its various substrates and intermediates (Regni et al. 2004, 2006).
The majority of the “substituted” enzymes in the glycolytic and fermentation pathways of Giardia appear to have derived by LGT (Nixon et al. 2002; Andersson et al. 2003; Regni et al. 2006; Morrison et al. 2007). In addition, the Giardia cis-prenyltransferase that makes dehydrodolichol pyrophosphate was obtained from bacteria by LGT (Grabińska et al. 2010). Here, we infer a plausible historical sequence in which a single Giardia PGM gene was duplicated, one of the PGM paralogs developed PMM activity, and the ancestral eukaryotic PMM ortholog was lost. Why this occurred in Giardia is interesting but likely unanswerable. As T. brucei is missing a PGM ortholog, we expect that its predicted PMM also has PGM activity. Changing substrate specificity is not infrequent in enzymes of protists. For example, Trichomonas has a dolichol-phosphate-glucose synthase ortholog that has dolichol-phosphate-mannose synthase activity, as well as a malate dehydrogenase ortholog that has lactate dehydrogenase activity (Wu et al. 1999; Grabińska et al. 2008).
Materials and methods
Parasite and yeast manipulations
Trophozoites of the WB isolate of G. lamblia were grown axenically in TYI-S media supplemented with 10% serum and 1 mg/mL bile. S. cerevisiae strains used in this study were as follows: BY4741 (MATα; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0), Δpep4 (as BY4741), pep4::kanMX4, Δprb1 (as BY4741, prb1::kanMX4), PGM1Δ (MATa/MATα; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; met15Δ0/met15Δ0; ura3Δ0/ura3Δ0), PGM2Δ (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) and temperature-sensitive PMM mutant (HMSF33 sec53-6) (ATCC). Strains were grown in a synthetic minimal (SD) medium containing 0.67% yeast nitrogen base without amino acids (Difco, Detroit, MI) and 0.5% d-glucose (WAKO, Osaka, Japan), supplemented with an amino acids mixture.
Separation of PGM and PMM activities using nondenaturing gels
Giardia trophozoites (∼106/mL × 50 mL) were chilled on ice for 30 min, concentrated at 2000 rpm for 10 min, lysed by sonication in 10 mM Hepes, pH 7, 10 mM MgCl2 and 25 mM NaCl, supplemented with protease inhibitor cocktail (Sigma, St. Louis, MI) and spun at 13,000 rpm for 30 min to remove insoluble proteins. Saccharomyces (250 mL) were grown in yeast extract peptone dextrose media to O.D. 3 and concentrated by centrifugation at 2000 × g. Yeasts were resuspended in lysis buffer (10 mM Hepes, pH 7, 10 mM MgCl2 and 0.5 mM phenylmethylsulfonyl fluoride) and lysed by sonication in acid-washed glass beads (Sigma). Giardia and yeast cell protein extracts were subjected to electrophoresis toward the anode for about 5 h at 25 mA at 4°C on a 4–12% nondenaturing, discontinuous polyacrylamide gel. The stacking gel buffer consisted of 125 mM Tris (pH 6.8), the resolving gel buffer 375 mM Tris (pH 8.8) and the running buffer Tris–glycine (pH 8.3).
After electrophoresis, the gel was rinsed with deionized water and placed on top of three sheets of Whatman 3MM filter paper saturated with buffer (80 mM imidazole, 5 mM MgSO4) in a Petri dish. An agar gel used as an overlay was prepared, as described (Bevan and Douglas 1969). We melted 0.4 g of Difco Noble agar in 30 mL of 1 mM ethylenediaminetetraacetic acid solution, to which the following ingredients were added and mixed in order: 10 mL of premix (2 mL of 0.8 M imidazole hydrochloride [pH 7.4], 2 mL of 0.1 M MgSO4, 2 mL of 40 mM Glc-1-P, 2 mL of 140 μM Glc-1,6-P, 2 mL of 10 mM NADP+-), 0.4 mL of Glc-6-P dehydrogenase (200 U/mL), 0.4 mL of 60 mM nitroblue tetrazolium and 0.1 mL of 400 mM phenazine methosulfate. This mixture was immediately dispensed evenly over the polyacrylamide gel.
After the overlay agar solidified, the dish was covered with aluminum foil and kept in the dark at 30°C for 2 h to allow development of the purple color indicative of PGM activity. Color development did not occur if Glc-1-P was omitted from the reaction mixture. The stained gel was stored in 7.5% acetic acid until dried or directly photographed with a digital camera. For PMM activity, Glc-1-P was replaced by Man-1-P in the agar premix, and phosphoglucose isomerase (PGI) and phosphomannose isomerase (PMI) were added to the mixture. Again a purple color development indicated PMM activity.
Bioinformatic methods
The predicted proteins of G. lamblia of the WB strain (first genome project) and GS strain (second genome project), which have been deposited in the NR data at GenBank or GiardiaDB, were searched with Psi-Blast using the PGM and PMM protein sequences from S. cerevisiae (Altschul et al. 1997; Gao et al. 2001; Morrison et al. 2007; Aurrecoechea et al. 2008; Franzén et al. 2009). Similar methods were used to search the predicted proteins of representative protists (Dictyostelium discoideum, Entamoeba histolytica, Leishmania major, Plasmodium falciparum, Toxoplasma gondii, Trichomonas, T. brucei and Trypanosoma cruzi) and metazoans (e.g. Homo sapiens, Xenopus laevis and Drosophila melanogaster) in the NR database at the NCBI or at specific databases (e.g. PlasmoDB or GeneDB). Predicted PGMs and PMMs were examined for conserved domains using the CD search at the NCBI (Marchler-Bauer et al. 2005). The mutases were aligned using multiple sequence comparison by log-expectation (Edgar 2004). The alignment was manually refined, and gaps were removed using BioEdit. The finished alignment was used to construct the phylogenetic tree using TREE-PUZZLE, a program to reconstruct phylogenetic trees from molecular sequence data by the maximum likelihood method (Schmidt et al. 2002).
Construction of S. cerevisiae pgm1Δ/pgm2Δ double deletion strain
PGM1Δ (MATa/MATα; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; met15Δ0/met15Δ0; ura3Δ0/ura3Δ0) diploid strain was sporulated and selected for mating type Matα. The kanamycin cassette in PGM2 was replaced by His5-rt6 cassette. Diploid strain obtained by crossing PGM1Δ (PGM1::kanMX4) with PGM2Δ (PGM2::His-rt6) was sporulated. Screening for appropriate markers and inability to grow on Gal identified spores bearing pgm1 and pgm2 alleles.
Complementation of Saccharomyces mutants with Giardia PGMs
WB strain Giardia were chilled on ice for 30 min and concentrated at 2000 rpm for 10 min, and genomic DNA was isolated using Promega kit. Candidate PGM orthologs were amplified from the genomic DNA by polymerase chain reaction using the following primers. For Gl-PGM1 (GiardiDB 50803_17254), the forward primer was ATGGAGGAGAGAGCAAGAGAT, and the reverse primer (that includes a stop codon) was TTAAGGCTCCTTGTCATTGAC. For Gl-PGM2 (GiardiDB 50803_11448), the forward primer was ATGATCAATTTAAAGGACAAAGCAG, and the reverse primer (that includes a stop codon) was CTAAACGCTTTCAACTTGGC. GatewayTM recombinational technology was used to clone the open reading frames into pVV214 vector for expression of Giardia PGMs in Saccharomyces mutants. First, the amplified genes were cloned into entry pCR8/GW/TOPO TA (Invitrogen, Carlsbad, CA) vector. The entry vector bearing Gl-PGM1 or Gl-PGM2 was then recombined with the destination vector pVV214. These plasmids were transformed into S. cerevisiae pgm1Δ/pgm2Δ double deletion strain (described above) or into the S. cerevisiae temperature-sensitive PMM mutant (HMSF33 sec53-6).
Saccharomyces pgm1Δ/pgm2Δ double deletion mutant transformed with vectors containing Gl-PGM1 or Gl-PGM2 were selected in synthetic medium-URA containing 2% glucose and then plated onto synthetic medium-URA supplemented with 2% (w/v) Gal as the only carbon source. In the absence of a functional PGM, the Saccharomyces pgm1Δ/pgm2Δ double deletion mutant does not grow on Gal plates (data not shown). As a control, we took advantage of URA3 marker in the Saccharomyces pgm1Δ/pgm2Δ double deletion mutant transformants and grew them on plates with 1% (w/v) 5-FOA. FOA is converted to a toxic compound 5-fluorouracil by URA3, so the presence of URA3 on a plasmid is lethal in the presence of FOA. In this way, URA3 plasmids were removed from the transformants.
Alternatively, vectors containing Gl-PGM1 or Gl-PGM2 were transformed into the S. cerevisiae temperature-sensitive PMM mutant (HMSF33 sec53-6) in synthetic medium-URA containing 2% glucose at the permissive temperature (25°C). These mutants do not grow at 37°C unless they are transformed with an exogenous PMM-encoding gene. A positive control was a wild-type Saccharomyces sec53 (PMM) gene.
Expression of recombinant Giardia PGM1 in bacteria and PGM2 in yeast
pDESTM17 Gateway vector (Invitrogen) is an adaptive destination vector used to clone and express recombinant protein with N-terminal polyHis tag in E. coli. The entry vector bearing Gl-PGM1 was recombined with pDESTM17 vector and transformed into the BL21-AI strain of E. coli. Bacteria were induced with 0.2% arabinose for 4 h at 37°C according to the protocol provided by Invitrogen. The recombinant Gl-PGM1 was purified using Ni-sepharose resin (Novagen, Madison, WI). Expression and purification of Gl-PGM1 were confirmed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis.
pYES2-Dest52 Gateway vector (Invitrogen) makes recombinant proteins with a C-terminal V5 epitope tag followed by polyHis tag under the Saccharomyces Gal1 promoter. A modified entry vector bearing Gl-PGM2 without the C-terminal stop codon was recombined with pYES2-Dest52 and transformed into S. cerevisiae strain BY4741 (MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). Transformed yeasts were induced with 2% Gal for 16 h at 30°C and then yeasts were lysed with glass beads. Recombinant Gl-PGM2 was purified on the Ni-sepharose resin and confirmed by western blot of purified protein using a horse-radish peroxidase-labeled antibody to the V5 epitope tag.
Enzyme assays
All enzymatic activities were assayed spectrophotometrically at 340 nm by the reduction of NADP+- to NADPH in a reaction mixture incubated at 30°C and containing, unless otherwise stated, 50 mM Hepes, pH 7.1, 5 mM MgCl2, 0.25 mM NADP+- and 10 μg/mL yeast glucose 6-phosphate dehydrogenase. PGM activity was measured in the presence of 500 μM Glc-1-P and 10 μM Glc-1,6-P, and PMM activity was measured in the presence of 10 μM Glc-1,6-P, 100 μM Man-1-P, 10 μg/mL PGI and 3.5 μg/mL PMI.
Supplementary Data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Acknowledgments
This work was supported by National Institutes of Health grants. We thank Dr. Kariona Grabińska for help with yeast manipulations. National Institutes of Health grants AI44070 (to J.S.) and GM31318 (to P.W.R.).
Glossary
Abbreviations
- FOA
5-fluoroorotic acid
- Glc-1-P
glucose 1-phosphate
- Glc-1,6-P
glucose 1,6-bisphosphate
- Gl-PGM1 and Gl-PGM2
Giardia lamblia phosphoglucomutase genes
- GPI
glycophosphosphoinositol
- LGT
lateral gene transfer
- Man-1-P
mannose 1-phosphate
- MudPIT
multiple dimensional protein identification technology
- PGI
phosphoglucose isomerase
- PGM
phosphoglucomutase
- PMI
phosphomannose isomerase
- PMM
phosphomannomutase
- QC
quality control
Conflict of interest statement
None declared.
References
- Adam RD. Biology of Giardia lamblia. Clin Microbiol Rev. 2001;14:447–475. doi: 10.1128/CMR.14.3.447-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JO, Sjögren AM, Davis LA, Embley TM, Roger AJ. Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Curr Biol. 2003;13:94–104. doi: 10.1016/s0960-9822(03)00003-4. [DOI] [PubMed] [Google Scholar]
- Aurrecoechea C, Brestelli J, Brunk BP, Carlton JM, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, et al. GiardiaDB and TrichDB: Integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis. Nucleic Acids Res. 2008;37:D526–D530. doi: 10.1093/nar/gkn631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Cui J, Robbins PW, Samuelson J. Use of Giardia, which appears to have a single nucleotide-sugar transporter for UDP-GlcNAc, to identify the UDP-Glc transporter of Entamoeba. Mol Biochem Parasitol. 2008;159:44–53. doi: 10.1016/j.molbiopara.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Robbins PW, Samuelson J. Molecular characterization of nucleocytosolic O-GlcNAc transferases of Giardia lamblia and Cryptosporidium parvum. Glycobiology. 2009;19:331–336. doi: 10.1093/glycob/cwn107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Vishwanath P, Cui J, Kelleher DJ, Gilmore R, Robbins PW, Samuelson J. The evolution of N-glycan-dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc Natl Acad Sci USA. 2007;104:11676–11681. doi: 10.1073/pnas.0704862104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan P, Douglas HC. Genetic control of phosphoglucomutase variants in Saccharomyces cerevisiae. J Bacteriol. 1969;98:532–535. doi: 10.1128/jb.98.2.532-535.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromphout K, Vleugels W, Heykants L, Schollen E, Keldermans L, Sciot R, D’Hooge R, De Deyn PP, von Figura K, Hartmann D, et al. The normal phenotype of pmm1-deficient mice suggests that pmm1 is not essential for normal mouse development. Mol Cell Biol. 2006;26:5621–5635. doi: 10.1128/MCB.02357-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Smith T, Robbins PW, Samuelson J. Darwinian selection for sites of Asn-linked glycosylation in phylogenetically disparate eukaryotes and viruses. Proc Natl Acad Sci USA. 2009;106:13421–13426. doi: 10.1073/pnas.0905818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daran JM, Bell W, François J. Physiological and morphological effects of genetic alterations leading to a reduced synthesis of UDP-glucose in Saccharomyces cerevisiae. FEMS Microbiol Lett. 1997;153:89–96. doi: 10.1111/j.1574-6968.1997.tb10468.x. [DOI] [PubMed] [Google Scholar]
- Das S, Traynor-Kaplan A, Reiner DS, Meng TC, Gillin FD. A surface antigen of Giardia lamblia with a glycosylphosphatidylinositol anchor. J Biol Chem. 1991;266:21318–21325. [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzén O, Jerlström-Hultqvist J, Castro E, Sherwood E, Ankarklev J, Reiner DS, Palm D, Andersson JO, Andersson B, Svärd SG. Draft genome sequencing of Giardia intestinalis assemblage B isolate GS: Is human giardiasis caused by two different species? PLoS Pathog. 2009;5:e1000560. doi: 10.1371/journal.ppat.1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: Cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J Biol Chem. 2001;276:9838–9845. doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- Grabińska KA, Cui J, Chatterjee A, Guan Z, Raetz CR, Robbins PW, Samuelson J. Glycobiology. 2010. Molecular characterization of the cis-prenyltransferase of Giardia lamblia. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabińska KA, Ghosh SK, Guan Z, Cui J, Raetz CR, Robbins PW, Samuelson J. Dolichyl-phosphate-glucose is used to make O-glycans on glycoproteins of Trichomonas vaginalis. Eukaryot Cell. 2008;7:1344–1351. doi: 10.1128/EC.00061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Henze K, Horner DS, Suguri S, Moore DV, Sánchez LB, Müller M, Embley TM. Unique phylogenetic relationships of glucokinase and glucosephosphate isomerase of the amitochondriate eukaryotes Giardia intestinalis, Spironucleus barkhanus and Trichomonas vaginalis. Gene. 2001;281:123–131. doi: 10.1016/s0378-1119(01)00773-9. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Cherukuri PF, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, et al. CDD: A conserved domain database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- Nixon JEJ, Wang A, Field J, Morrison HG, McArthur AG, Sogin ML, Loftus B, Samuelson J. Evidence for lateral transfer of genes encoding ferredoxins, nitroreductases, NADH oxidase and alcohol dehydrogenase 3 from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica. Eukaryot Cell. 2002;1:181–190. doi: 10.1128/EC.1.2.181-190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlean P. Dolichol phosphate mannose synthase is required in vivo for glycosyl phosphatidylinositol membrane anchoring, O mannosylation, and N glycosylation of protein in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:5796–5805. doi: 10.1128/mcb.10.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal D, Banerjee S, Cui J, Schwartz A, Ghosh SK, Samuelson J. Giardia, Entamoeba, and Trichomonas enzymes activate metronidazole (nitroreductases) and inactivate metronidazole (NIMs) Antimicrob Agents Chemother. 2009;53:458–464. doi: 10.1128/AAC.00909-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirard M, Achouri Y, Collet JF, Schollen E, Matthijs G, Van Schaftingen E. Kinetic properties and tissular distribution of mammalian phosphomannomutase isozymes. Biochem J. 1999;339:201–207. [PMC free article] [PubMed] [Google Scholar]
- Quick CB, Fisher RA, Harris H. A kinetic study of the isozymes determined by the three human phosphoglucomutase loci PGM1, PGM2, and PGM3. Eur J Biochem. 1974;42:511–517. doi: 10.1111/j.1432-1033.1974.tb03366.x. [DOI] [PubMed] [Google Scholar]
- Ratner DM, Cui J, Steffen M, Moore LL, Robbins PW, Samuelson J. Changes in the N-glycome (glycoproteins with Asn-linked glycans) of Giardia lamblia with differentiation from trophozoites to cysts. Eukaryot Cell. 2008;7:1930–1940. doi: 10.1128/EC.00268-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regni C, Naught L, Tipton PA, Beamer LJ. Structural basis of diverse substrate recognition by the enzyme PMM/PGM from P. aeruginosa. Structure. 2004;12:55–63. doi: 10.1016/j.str.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Regni C, Schramm AM, Beamer LJ. The reaction of phosphohexomutase from Pseudomonas aeruginosa: Structural insights into a simple processive enzyme. J Biol Chem. 2006;281:15564–15571. doi: 10.1074/jbc.M600590200. [DOI] [PubMed] [Google Scholar]
- Samuelson J, Banerjee S, Magnelli P, Cui J, Kelleher DJ, Gilmore R, Robbins PW. The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc Natl Acad Sci USA. 2005;102:1548–1553. doi: 10.1073/pnas.0409460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Simpson AG, Inagaki Y, Roger AJ. Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of “primitive” eukaryotes. Mol Biol Evol. 2006;23:615–625. doi: 10.1093/molbev/msj068. [DOI] [PubMed] [Google Scholar]
- Steiner TS, Thielman NM, Guerrant RL. Protozoal agents: What are the dangers for the public water supply? Annu Rev Med. 1997;48:329–340. doi: 10.1146/annurev.med.48.1.329. [DOI] [PubMed] [Google Scholar]
- Wu G, Fiser A, ter Kuile B, Sali A, Müller M. Convergent evolution of Trichomonas vaginalis lactate dehydrogenase from malate dehydrogenase. Proc Natl Acad Sci USA. 1999;96:6285–6290. doi: 10.1073/pnas.96.11.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]