Abstract
Background
Lentils are oftentimes responsible for allergic reactions to legumes in Mediterranean children. Though the primary sequence of the major allergen, Len c 1 is known, the location of the IgE binding epitopes remains undefined.
Objective
We sought to identify IgE-binding epitopes of Len c 1 and relate epitope binding to clinical characteristics.
Methods
135 peptides corresponding to the primary sequence of Len c 1 were probed with sera from 33 lentil-allergic individuals and 15 non-atopic controls by means of microarray immunoassay. Lentil-specific IgE, Skin Prick Tests and clinical reactions to lentil were determined. Epitopes were defined as overlapping signal above inter- and intra-slide cut-offs and confirmed by inhibition assays using a peptide from the respective region. Hierarchical clustering of microarray data was used to correlate binding patterns with clinical findings.
Results
The lentil-allergic patients specifically recognized IgE-binding epitopes located in the C-terminal region, between peptide 107 and 135. Inhibition experiments confirmed the specificity of IgE binding in this region, identifying different epitopes. Linkage of cluster results with clinical data and lentil specific IgE levels displayed a positive correlation between lentil-specific IgE levels, epitope recognition and respiratory symptoms.
Modeling based on the three-dimensional structure of a homologous soy vicilin suggests that the Len c 1 epitopes identified are exposed on the surface of the molecule.
Conclusion
Several IgE-binding sequential epitopes of Len c 1 have been identified. Epitopes are located in the C-terminal region, and are predicted to be exposed on the surface of the protein. Epitope diversity is positively correlated with IgE levels, pointing to a more polyclonal IgE response.
Keywords: lentil allergy, peptide microarray, epitope recognition, epitope diversity, Len c 1
Key messages.
This study is the first to explore and locate IgE-binding B-cell epitopes of lentil allergens using a peptide microarray immunoassay.
Combining the findings of the microarray immunoassay with clinical characteristics and IgE levels shows a positive correlation.
Introduction
Lentils, together with chickpea, are important causes for IgE-mediated food hypersensitivity in the Mediterranean.1–4 Other legumes, such as peanut and soybean, are legumes most frequently involved in allergic reactions in the United States, the United Kingdom and Japan. The major allergens from soybean and especially peanut have been extensively studied,5–10 however, more interest has been shown recently in chickpea and lentil allergens.1
In Spain, allergy to lentils is the fifth most common cause of food allergy in the pediatric population.1 Ten percent of children with food allergy have a convincing clinical history of allergy to lentils. Lentils frequently induce systemic symptoms, such as wheeze, rhinorrhea and disseminated urticaria symptoms.2 Lentil has also been implicated in food-dependent exercise-induced anaphylaxis.4
Over 50% of individuals allergic to lentil are also allergic to chickpea and green pea.2 Inhibition experiments and oral challenge tests suggest a high degree of in vitro and in vivo cross-reactivity.2
Several allergens from lentil have been characterized to date, including Len c 1.01, 1.02 and 2. Len c 1.01 (Len c 1) is a protein of approximately 50 kDa, that has been identified as a mature vicilin chain.12 Three genetic isoforms of this allergen have been described: Len c 1.0101, Len c 1.0102, and Len c 1.0103. Len c 1.02 is a 12–16 kDa protein, the β-subunit of lentil vicilin that is probably produced by means of posttranslational proteolytic processing of the precursor Len c 1.01. Len c 2 is a distinct 66 kDa protein, corresponding to a seed-specific biotinylated protein.12,13 A clear structural relationship between Len c 1.01 and several allergens of the vicilin family, including Ara h 1, Jug r 2, Ana o 1, Ses i 3 and subunits of soybean conglycinin, has been described.14 Studies have shown IgE binding to a 50 kDa band in lentil extract in more than 65% of patients,15 later described as Len c 1.01. 77% of the lentil allergic patients recognized the purified Len c 1.12 However, the location of the IgE binding sequential epitopes of the major lentil protein Len c 1 remains unknown.
Here we report the mapping of IgE binding epitopes of Len c 1 using a peptide microarray based immunoassay (MIA). This sensitive technique allowed us to rapidly study several sets of the protein simultaneously, using only a minute quantity of sera.
Methods
Patients
Thirty-three lentil allergic patients were recruited from the Hospital Fundación Jimenez Diaz (Madrid, Spain; n=5), from the Hospital Niño Jesús (Madrid, Spain; n= 24) and from Mount Sinai Medical Center (New York, USA; n=4) from 2004 to 2006. Written informed consent was obtained from all subjects (or the legal guardian for children) before their inclusion in the study. The diagnosis of IgE-mediated lentil allergy was made by an allergist on the basis of a convincing history (objective symptoms) of an acute reaction (less than 30 minutes) following lentil ingestion together with evidence of specific IgE antibodies (positive skin prick test and/or lentil specific IgE >0.35 kUA/L).
Lentil (Lens esculenta) specific IgE was measured using the Immuno CAP System FEIA (Phadia, Uppsala, Sweden). The assay had a lower detection limit of 0.35 kUA/L and an upper limit of 100 kUA/L, with higher values reported as greater than 100 kUA/L (>100 kUA/L).
SPT with lentil (Lens culinaris) extract (Leti, Madrid, Spain) was performed by a standard technique using a needle (ALK-Abello, Madrid, Spain). The mean of the wheal’s largest diameter and its perpendicular in millimeters (mm) was measured after 15 minutes. A result was considered positive when the mean diameter of the wheal was ≥ 3 mm larger than the wheal of the negative control. Histamine (10 mg/ml) and saline solutions were used as positive and negative controls, respectively. When this commercial extract was not available, results of prick to prick skin tests performed with cooked lentils were reported as positive or negative.
Clinical information was gathered by means of a specific questionnaire. This information included a detailed clinical history, including the characteristics of reactions to lentil exposure, other plant-related food allergies, pollen allergy, and personal and family history of atopic diseases.
Preparation of Microarray: Peptides, Slides, Printing
Peptide libraries, slides and printing were prepared and performed as described elsewhere.16,17
Peptide Microarray Immunoassay
Immunolabelling was performed as previously described, with some modifications.8,10,16,18 In brief, an area around the arrays was demarcated using a hydrophobic pen (DakoCytomation Pen, DAKO, Glostrup, Denmark). The slides were first rinsed with phosphate-buffered saline containing 0.05% tween 20 (PBS-T) and non-specific binding sites were blocked with 400μl of 1% human serum albumin (HSA) in PBS-T (PBS-T/HSA) for 60 minutes at room temperature. After removing the PBS-T/HSA from the slide surface by aspiration, 50μl of patient serum diluted 1:5 in PBS-T/HSA was applied and allowed to incubate for 24 hours at 4° C. Slides were washed with PBS-T and incubated for 24 hours at 4° C with a cocktail of two monoclonal biotinylated anti-human IgE antibodies (Invitrogen, Carlsbad, CA, USA and BD Biosciences Pharmingen, San Jose, CA, USA) diluted 1:250 each.
Slides were washed with PBS-T, incubated for 4 minutes with ethylene diamine tetraacetic acid (EDTA) 1 mM in PBS-T, washed again with PBS-T, equilibrated for 1 minute with Dendrimer Buffer (Genisphere, Philadelphia, USA) followed by incubation for 3 hours at room temperature with Anti-Biotin-Dendrimer-Oyster 550 (Genisphere) in Dendrimer Buffer at 0.6 μg/ml with addition of 0.02μg/ml of salmon sperm DNA.
All incubations were performed in the dark in a humidity chamber (Binding Site, Birmingham, UK) on an orbital rotating platform with gentle agitation. To test for non-specific binding of secondary antibody or dendrimers to peptides, two immunolabelings were performed without patient’s serum. The immunoassay was performed as explained above except for substitution of serum by blocking solution.
The slides were then washed with PBS-T, 15 mM Tris buffer, centrifuge dried, followed by wash with 0.1 × PBS, centrifuge dried, washed again with 0.05 × PBS, centrifuge dried, and scanned using a ScanArray®Gx (PerkinElmer, Waltham, MA, USA). Images were saved as TIFF files.
Peptide Microarray Inhibition Experiment
A peptide inhibition assay was carried out in which the array was immunolabeled as described above except that the serum pool (diluted 1:50 in blocking buffer) was pre-incubated with 1μl of peptide at a concentration of 1 mM (1 peptide per array) for 2h at gentle agitation. The same serum pool without added peptides was processed in parallel and incubated under the same conditions as control.
Data Analysis
Lentil specific IgE, age, age of onset of lentil allergy and lentil SPT size were summarized as median with the interquartile range: median [Q1–Q3]. χ2 (or Fisher test if necessary), and Wilcoxon test were used for comparisons between groups. Correlation between variables was measured with the Pearson correlation coefficient.
Regarding peptide microarray data, fluorescence signal from one channel (for IgE binding) was digitized with the ScanArray Express Microarray Analysis System (PerkinElmer), exported as comma-delimited (CSV) files and analyzed using R analysis (R development Core Team.R: A language and environment for statistical computing. Version 2.5.0 Vienna, Austria: R foundation for Statistical Computing, 2007. http://r-project.org).
For hierarchical cluster analysis the absolute signal intensity data (digital fluorescence units, dfu) of all patients on all array elements was used. Clusters were determined by using cluster software (http://rana.lbl.gov/EisenSoftware.htm) using the average linkage algorithm and uncentered measurements for features, array elements remained unchanged. Results are displayed using Java TreeView 1.60 (http://rana.lbl.gov/EisenSoftware.htm).
Results
Patient Description
Thirty-three lentil allergic patients were included in the study. Median age at inclusion was 8.0 [5.0–13.0] yrs. The majority of patients had family and personal history of atopy: 24 patients had allergic rhinitis, 21 had asthma and 14 had atopic dermatitis. Sixteen patients (48.5 %) were allergic to pollen. General characteristics of the patients are included in Table I.
Table I.
Patient characteristics
| ID | Age (yrs)/Sex | Pollen allergy | CAP to lentil (kUA/L) | Age of lentil allergy onset | Symptoms with lentil | SPT | Provocation with lentil | Allergy to tree nuts and sesame | Allergy to legumes (including peanut) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 14/M | No | ≥100 | 4 | U, OAS, DS, A, U, ANA | positive | ND | no | pea, bean, chickpea |
| 2 | 13/M | Yes | 24,20 | 3 | OAS | ND | Positive | no | pea, bean, chickpea, peanut |
| 3 | 13/M | Yes | 26,20 | 2 | OAS, U, DS, ANA | positive | Positive | no | pea, bean, chickpea |
| 4 | 12/M | Yes | 17,40 | 2 | OAS, R, A, U, ANA | positive | Positive | no | pea, bean, chickpea |
| 5 | 16/M | Yes | 22,00 | 1 | OAS | positive | Positive | no | pea |
| 6 | 11/M | Yes | 9,67 | 2 | OAS, R, A, U, ANA | positive | Positive | no | bean, pea |
| 7 | 12/M | Yes | 8,02 | 7 | R, A, U, ANA | positive | Positive | almond | chickpea, pea, peanut |
| 8 | 13/F | Yes | ≥100 | 2 | OAS, R, A, U, DS, ANA | positive | ND | no | chickpea, pea, bean |
| 9 | 10/F | No | 59,20 | 2 | R, A, U | positive | Positive | no | pea, bean, chickpea, peanut |
| 10 | 8/M | Yes | 21,00 | 1 | U, DS, ANA | positive | ND | almond | chickpea, pea, bean |
| 11 | 6/M | Yes | 65,40 | 1 | R, A, U, ANA | positive | Positive | no | chickpea, pea, bean |
| 12 | 5/F | ND | 30,10 | 3 | OAS, R, A, U | ND | Positive | almond | bean, pea, peanut |
| 13 | 7/M | No | ≥100 | 1,2 | OAS, R, A, U, DS, ANA | positive | ND | no | chickpea, pea, bean |
| 14 | 7/M | No | 11,30 | 3 | OAS, U | positive | Positive | no | pea, bean, chickpea, peanut |
| 15 | 5/M | Yes | 16,30 | 4 | OAS, R, A, U, ANA | positive | Positive | no | chickpea |
| 16 | 3/M | No | 14,50 | 2 | OAS, R, A, U, DS, ANA | positive | ND | no | pea, bean, chickpea, peanut |
| 17 | 6/M | No | 49,90 | 1,4 | systemic | positive | Positive | no | chickpea, pea, bean |
| 18 | 4/M | No | 3,80 | 2 | OAS, R, A, U, ANA | positive | Positive | no | pea, bean, chickpea, peanut |
| 19 | 32/M | Yes | 0,77 | 1 | OAS, R, A, DS, U, ANA | positive | ND | pistachiocashew, walnut, hazelnut, almond | pea, bean, chickpea, peanut |
| 20 | 5/F | No | 9,82 | 1 | OAS, R, A, U, ANA | positive | Positive | pistachio hazelnut, walnut, almond | chickpea, pea, bean |
| 21 | 4/M | Yes | 56,7 | 2 | OAS, R, A, U, ANA | positive | Positive | no | chickpea, pea, bean |
| 22 | 3/M | Yes | ≥100 | 2 | systemic | positive | Positive | no | chickpea, pea, bean, soy |
| 23 | 5/F | No | ≥100 | 1 | OAS, U | positive | ND | no | chickpea, pea, bean |
| 24 | 5/F | No | 19,00 | 2 | DS | positive | Positive | no | chickpea, pea |
| 25 | 2/M | ND | 16,00 | 1 | OAS | positive | Positive | no | chickpea, pea, bean |
| 26 | 49/F | No | 1,61 | 2 | OAS, U, R, A | positive | ND | no | chickpea, pea, bean, soy |
| 27 | 5/M | No | 3,60 | 1 | OAS, U, ANA | positive | ND | no | no |
| 28 | 5/F | No | 9,92 | 1 | U, DS, ANA | positive | ND | no | chickpea, pea, bean, soy |
| 29 | 19/F | No | 1,78 | R, U, A, DS, angioedema | positive | ND | pistachio hazelnut, walnut, almond | chickpea, peanut | |
| 30 | 10/M | Yes | 14,50 | 6 | R, A, U, DS, ANA | Positive (PP) | ND | no | bean, pea, peanut |
| 31 | 37/F | No | 10,40 | 5 | OAS, R, A, U, DS, ANA | ND | ND | pistachio cashew, hazelnut, sesame | chickpea, pea, peanut, soy |
| 32 | 9/F | Yes | 35,50 | 4 | OAS, R, A, U, DS, ANA | Positive (PP) | ND | sesame | bean, pea, peanut, soy |
| 33 | 12/F | Yes | 73,70 | 1 | OAS, R, A, U, ANA | Positive (PP) | ND | no | no |
A, Asthma; ANA, Anaphylaxis; DS, Digestive Symptoms; F, female; M, male; ND, Not Done or Not Determined; OAS, oral allergy syndrome; PP, prick-prick with cooked lentils; SPT, Skin Prick Test; R, Rhinoconjunctivitis; U, Urticaria
The median age of onset for lentil allergy was 2 yrs [1.0–3.0]. Lentil specific IgE at the time of enrollment in the study was 19.0 [9.9–58.0] kUA/L and lentil SPT was 5.0 [3.4–6.9] mm. Most subjects reported systemic symptoms following lentil ingestion (87.9%), mostly generalized urticaria, and upper and/or lower respiratory symptoms (wheeze, cough, shortness of breath, rhino-conjunctivitis).
A significant number of patients reported symptoms following ingestion of other legumes: 29 of 33 patients reacted to green pea, 26/28 reacted to chickpea (28 patients had eaten chickpea previously and therefore were able to answer), 25/32 to bean, 12/27 to peanut, and 5/19 to soy. With respect to reactions to tree nuts, 6/22 reacted with almond, 4/19 with pistachio, 3/20 with walnut, 4/19 with hazelnut and 2/16 with cashew.
Peptide Microarray Immunoassay
Epitope Recognition
Commercially synthesized peptides, of 15 AA in length with an overlap of 12, corresponding to the primary sequence of Len c 1 were robotically arrayed in two sets of triplicate and attached to derivatized glass slides. The peptide arrays were assayed in parallel with sera from the 33 lentil allergic patients and 15 healthy volunteers.
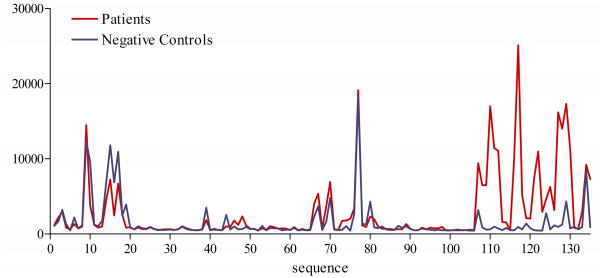
Based on previous experiments 8–10,18, we expected to see a non-random distribution of signal intensity corresponding to antibody recognition of overlapping peptides, even though the peptides were printed on different locations on the array. Figure 1 (intensity of patients and negative controls) displays the median signal (dfu) by peptide ordered according to the primary sequence of Len c 1 from N- to C-terminus. As expected, signal intensity is present in peaks in distinct regions of the protein, represented by sets of overlapping peptides indicating recognition of specific sequences.
FIG 1.
Average signal intensity (in dfu) plotted from N-terminal to C-terminal position of the Len c 1 sequence for patients (red) and negative controls (blue).
The region at the C-terminal side (peptides 106–135) of the molecule appears to be specifically recognized by 84.8% of the lentil allergic patients above inter- and intra-slide cut-off values. The intensity of the IgE signal was significantly higher in the allergic group (p<0.001).
Some signal associated with peptides in the N-terminal side of the molecule and a region in the center of the molecule (peptides 70–89) can be seen in both allergic individuals and negative controls. Non-specific artifact was suspected. The non-specificity of these signals was confirmed by two immunolabellings substituting patient’s sera with blocking solution. Without serum incubation, fluorescence was absent in the C-terminal region of the molecule, but was still present in the N-terminus and central part (peptides 70–89). Inhibition experiments were not performed for these regions. We focused our attention on the C-terminal portion of the molecule.
As previously described, dominant antigenic regions can be identified using this approach, although there is a high inter-patient heterogeneity. Some individuals’ IgE antibodies appear to recognize slightly different peptides; others recognize more or less regions than the majority. Figure 2 (Cluster image) displays individual heterogeneity in recognition patterns for the 33 patients. Red pixels represent intensity of individual IgE binding by pixel intensity. Patients are displayed in rows, subsequent peptides in columns. Negative controls are not included in this analysis as no binding to the described region was detected. The region from peptide 106 to 135 is recognized by the majority of patients. Differences, however, exist in the number of peptides recognized and intensity of binding. Patients cluster in two main groups: node 25 (r: 0.729) and node 27 (r: 0.6768) (Figure 2, Cluster Image). The lentil-specific IgE level is different between those two groups. Patients clustering in node 25 have a significantly higher lentil specific IgE level than the rest of patients (p<0.001): (median [Q1–Q3]): 62.3 [17.1–100.0] kUA/L vs. 11.3 [8.0–22.0] kUA/L. The patients clustered in node 25 also have more respiratory symptoms including rhino-conjunctivitis and wheezing after exposure to lentil (p = 0.024). Patients clustering in node 27 have a significantly lower median lentil specific IgE level than the rest of patients (p<0.005) (median [Q1–Q3]): 12.9 [6.5–21.3] kUA/L vs. 49.9 [14.5–100.0] kUA/L.
FIG 2.
Treeview display of hierarchical cluster analysis of IgE signal (positive in increasing brightness of red, negative in black). Peptides are in columns, individual patients (arrays) in rows. The arrows indicate peptide clusters (nodes) that were chosen for additional analysis of patient characteristics. See Methods and Results for details.
Lentil specific IgE levels positively correlated with both the number of positive peptides (r=0.514, p<0.005) and the signal intensity (r=0.799, p<0.0001) for the region of interest (peptide 106–135).
Inhibition Experiments
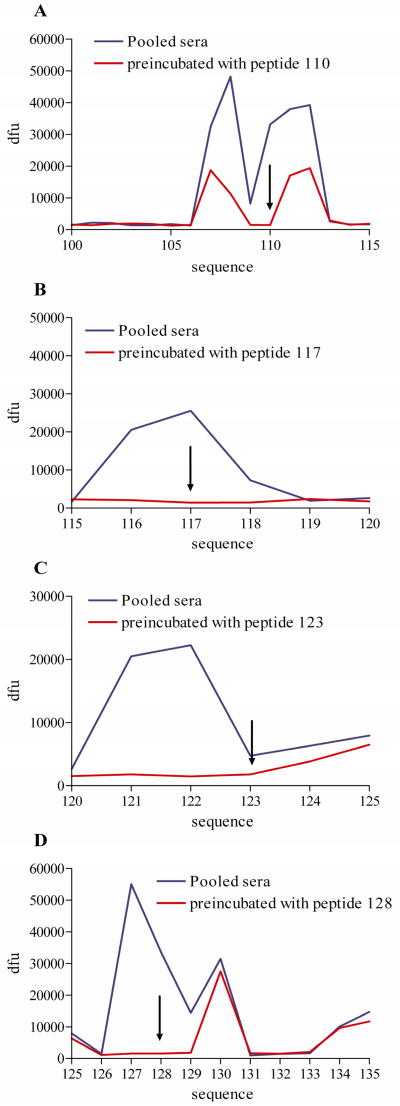
To confirm the sequence-specificity of IgE signal, pooled sera were incubated for 2 hours with individual peptides prior to immunolabeling, as described above. Four peptides from the region of interest were chosen for inhibition studies (peptides 110, 117, 123, 128).
Signal from overlapping peptides was attenuated using single peptides as inhibitors. For example, peptide 110 blocked the epitope 106–113. Peptide 117 blocked epitope 115–119. Peptide 123 blocked the epitope 120–123, and peptide 128 blocked the epitope 126–129 (Figure 3). Therefore, four distinct epitopes could be identified in the C-terminal region of the Len c 1 protein. The location of these epitopes on the Len c 1 sequence is shown in the online repository as figure E1.
FIG 3.
Average signal intensity (in dfu) plotted from N-terminal to C-terminal position of the Len c 1 peptide for pooled sera preincubated with a single peptide for 2 hours (red) and pooled sera without preincubation (blue). Four separate experiments with different peptides respectively were conducted (A–D). The arrows indicate the localization of the peptide used for the inhibition.
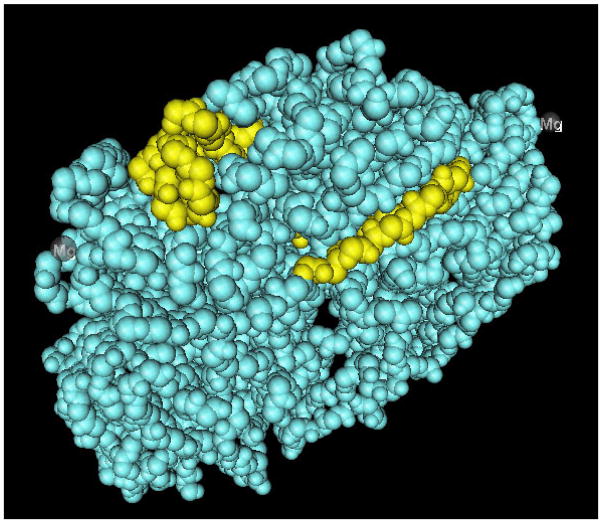
Modeling using the known three-dimensional structure of another vicilin (soy beta-conglycin; PDB 1UIK), predicts that these epitopes of Len c 1 are exposed on the surface of the molecule (Figure 4, 3D image).
FIG 4.
Epitope 1 and epitope 4 as defined in this study superimposed on the 3D structure of the vicilin soy beta-conglycin. Epitopes are located on the outside of the molecule. On top epitope 1, below epitope 2.
Discussion
This study focused on the specificity and diversity of the IgE response to the major lentil allergen, Len c 1. We have identified 4 putative epitopes in the C-terminus. No epitopes in Len c 1 have been previously identified. We could also demonstrate that IgE levels and clinical characteristics correlated with features of epitope recognition. This is consistent with previous studies of IgE recognition patterns to sequential (linear) epitopes, which have been associated with the natural history of food allergy in individual patients 18,19,20 and the severity of allergic reactions.8–10
Epitope detection and definition have been a main focus of several studies in allergy, all of which are confronted with the problem of definition criteria 8–10,18,21. Epitopes may be defined by a set of overlapping peptides that are identified by a certain percentage of patients. An important uncertainty, however, lies in the definition of the start and end points of the epitope. For example, in the case of Len c 1, there are four major signal peaks in the region from peptide 106–135 (Figure 1). However, no clear distinction can be made between single epitopes in the region by taking into account only the percentage of patients with positive binding. Cluster analysis of binding patterns of multiple peptides comprised of different amino acid numbers covering the allergen sequence, as described previously,9 offers one possible approach to address this vagueness. The specific design of our experiment did not include peptide sequences of different lengths covering this sequence of the protein. Therefore, the cluster analysis performed here was solely used to link binding patterns to patient characteristics.
Another approach is the implementation of inhibition experiments. In our study, pre-incubation of patients’ sera with a peptide selected from the area of interest showed a clear inhibition of IgE binding to that peptide, and due to sequence overlap, inhibition of the adjacent peptides could also be seen.
All four of the epitopes that we have defined in this assay are predicted to be located on the surface of the molecule by three dimensional modeling based on the sequence of the homologous beta-conglycinin of soy. Highlighting the Len c 1 epitopes on a related food protein with a known 3D structure (beta-conglycinin alpha subunit of soy bean)14 indicates that all four epitopes are exposed on the surface of the protein. Vicilins, such as those of soy bean and lentil, are very stable proteins,22 largely resistant to heat and digestion. A recent publication emphasizes the extreme heat resistance of some lentil allergens.1 Therefore, we can assume that Len c 1 reactive patients probably react to the whole protein as a trimer. The epitope location on the outer surface of the molecule makes them readily accessible for IgE antibodies.
As mentioned above, a positive correlation between the number of peptides recognized by an individual and the level of peanut specific IgE (correlation factor 0.579) has been studied and published previously.8 Our study shows that peptide recognition in numbers of positive peptides and binding intensity to epitopes is positively correlated to lentil specific IgE (Number = r 0.514, p<0.005; Intensity= r: 0.799, p<0.0001)
However, not all lentil allergic patients had detectable Len c 1 epitope binding using the microarray immunoassay. In our study 84.8% of lentil allergic individuals recognized peptide epitopes of Len c 1. The remaining patients may have antibody to conformational epitopes only, which are not detected with this experimental setup, or to other lentil proteins. Other studies have confirmed that Len c 1-specific antibodies are not present in all lentil allergic individuals; approximately 65% of lentil allergic patients recognize a 50 kDa band in lentil extract,5 later described as Len c 1, and Lopez-Torrejon et al. 12 reported a positive recognition by 77% of lentil-allergic individuals studied. These results indicate that Len c 1 is a major lentil allergen but not the only one responsible for symptoms.
Panallergens, or common protein families (e.g., vicilins), may result in cross-reactivity among foods. The 2 clones of the lentil allergen Len c 1.02 were shown to have greater than 50% identity with other vicilins, including the major peanut allergen Ara h 1, and with soybean conglutinin subunits.12 In a study of 18 Spanish patients with pea allergy, many also reacted to lentil. Vicilin and convicilin were shown to be potential major allergens from pea and these cross-reacted with the major lentil allergen Len c 1.13 The 3-dimensional models of vicilins allergens (Ara h 1 from peanut, Len c 1 from lentil, and Pis s 1 from pea) built by homology-based modeling from the soybean beta-conglycinin were shown to be very similar, helping to account for their IgE-binding cross-reactivity.14
Many of the patients included in this study had symptoms when exposed to other legumes, mainly chickpea (92.9%) and pea (87.9%). Many of the patients (12/27) reacted to peanut. In the group of patients studied here, the lentil specific IgE levels appeared to correlate with the number of epitopes bound and the intensity of binding, rather than presence of tolerance/reaction to other legumes. Further analysis of IgE binding to vicilins with a larger number of patients is needed to fully understand the above described legume in vivo and in vitro cross reactivity 2 at an epitope level.
Performing the same epitope identifying experiments in Len c 1 using sera from patients allergic to peanut or chickpea, but tolerant to lentil; and analysis of Ara h 1 epitope binding in a larger cohort of lentil allergic patients, including a comparison of peanut tolerant and allergic individuals (with similar specific IgE levels), might further clarify mechanism of legume cross-reactivity. We believe that understanding the molecular basis of clinical reactivity will help us in counseling our patients more accurately. Peptide microarray immunoassays offer a versatile platform to address this issue. Our results provide a first step in helping to resolve the above described questions.
In conclusion, we have identified the IgE binding epitopes of a major lentil allergen, Len c 1, using a peptide microarray. Len c 1 epitopes detected by this assay appear to be located on the surface of
Supplementary Material
Acknowledgments
We wish to acknowledge Michelle Mishoe for the lentil specific IgE determination, and Galina Grishina and Rosalia Ayuso for their helpful discussion of the data.
Andrea Vereda thanks the World Allergy Organization for their support.
Declaration of all sources of funding: Andrea Vereda was funded by the World Allergy Organization; H. A. Sampson and W. G. Shreffler have received research support from the National Institutes of Health/National Institute of Allergy and Infectious Diseases and the Food Allergy and Anaphylaxis Network.
Abbreviations used
- AA
amino acid
- dfu
digital fluorescence units
- HSA
Human serum albumin
- MIA
microarray based immunoassay
- PBS-T
Phosphate Buffered Saline with Tween
- SPT
skin prick test
- kUA/L
Kilounits of antibody per liter
- yrs
years
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cuadrado C, Cabanillas B, Pedrosa MM, Varela A, Guillamon E, Muzquiz M, et al. Influence of thermal processing on IgE reactivity to lentil and chickpea proteins. Mol Nutr Food Res. 2009;53(11):1462–8. doi: 10.1002/mnfr.200800485. [DOI] [PubMed] [Google Scholar]
- 2.Martínez San Ireneo M, Ibáñez MD, Sánchez JJ, Carnés J, Fernández-Caldas E. Clinical features of legume allergy in children from a Mediterranean area. Ann Allergy Asthma Immunol. 2008;101(2):179–84. doi: 10.1016/s1081-1206(10)60207-4. [DOI] [PubMed] [Google Scholar]
- 3.Kalogeromitros D, Armenaka M, Galatas I, Capellou O, Katsarou A. Anaphylaxis induced by lentils. Ann Allergy Asthma Immunol. 1996;77(6):480–2. doi: 10.1016/S1081-1206(10)63354-6. [DOI] [PubMed] [Google Scholar]
- 4.Orhan F, Karakas T. Food-dependent exercise-induced anaphylaxis to lentil and anaphylaxis to chickpea in a 17-year-old boy. J Investig Allergol Clin Immunol. 2008;18(6):465–8. [PubMed] [Google Scholar]
- 5.Krishnan HB, Kim WS, Jang S, Kerley MS. All three subunits of soybean beta-conglycinin are potential food allergens. J Agric Food Chem. 2009;57(3):938–43. doi: 10.1021/jf802451g. [DOI] [PubMed] [Google Scholar]
- 6.Holzhauser T, Wackermann O, Ballmer-Weber BK, Bindslev-Jensen C, Scibilia J, Perono-Garoffo L, et al. Soybean (Glycine max) allergy in Europe: Gly m 5 (beta-conglycinin) and Gly m 6 (glycinin) are potential diagnostic markers for severe allergic reactions to soy. J Allergy Clin Immunol. 2009;123(2):452–8. doi: 10.1016/j.jaci.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 7.Jin T, Guo F, Chen YW, Howard A, Zhang YZ. Crystal structure of Ara h 3, a major allergen in peanut. Mol Immunol. 2009;46(8–9):1796–804. doi: 10.1016/j.molimm.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 8.Flinterman AE, Knol EF, Lencer DA, Bardina L, den Hartog Jager CF, Lin J, et al. Peanut epitopes for IgE and IgG4 in peanut-sensitized children in relation to severity of peanut allergy. J Allergy Clin Immunol. 2008;121(3):737–43. doi: 10.1016/j.jaci.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 9.Shreffler WG, Beyer K, Chu TH, Burks AW, Sampson HA. Microarray immunoassay: association of clinical history, in vitro IgE function, and heterogeneity of allergenic peanut epitopes. J Allergy Clin Immunol. 2004;113(4):776–82. doi: 10.1016/j.jaci.2003.12.588. [DOI] [PubMed] [Google Scholar]
- 10.Shreffler WG, Lencer DA, Bardina L, Sampson HA. IgE and IgG4 epitope mapping by microarray immunoassay reveals the diversity of immune response to the peanut allergen, Ara h 2. J Allergy Clin Immunol. 2005;116(4):893–9. doi: 10.1016/j.jaci.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 11.Ibáñez MD, Martínez M, Sánchez JJ, Fernández-Caldas E. Legume cross-reactivity. Allergol Immunopathol (Madr ) 2003;31(3):151–61. [PubMed] [Google Scholar]
- 12.López-Torrejón G, Salcedo G, Martín-Esteban M, Díaz-Perales A, Pascual CY, Sanchez-Monge R. Len c 1, a major allergen and vicilin from lentil seeds: protein isolation and cDNA cloning. J Allergy Clin Immunol. 2003;112(6):1208–15. doi: 10.1016/j.jaci.2003.08.035. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez-Monge R, Pascual CY, Díaz-Perales A, Fernandez-Crespo J, Martín-Esteban M, Salcedo G. Isolation and characterization of relevant allergens from boiled lentils. J Allergy Clin Immunol. 2000;106(5):955–61. doi: 10.1067/mai.2000.109912. [DOI] [PubMed] [Google Scholar]
- 14.Barre A, Borges JP, Rouge P. Molecular modelling of the major peanut allergen Ara h 1 and other homotrimeric allergens of the cupin superfamily: a structural basis for their IgE-binding cross-reactivity. Biochimie. 2005;87(6):499–506. doi: 10.1016/j.biochi.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 15.Ibáñez SD, Martínez SI, Maranón LF, Fernández-Caldas E, Alonso LE, Laso BT. Specific IgE determinations to crude and boiled lentil (Lens culinaris) extracts in lentil-sensitive children and controls. Allergy. 1999;54(11):1209–14. doi: 10.1034/j.1398-9995.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- 16.Lin J, Bardina L, Shreffler WG, Andreae DA, Ge Y, Wang J, et al. Development of a novel peptide microarray for large-scale epitope mapping of food allergens. J Allergy Clin Immunol. 2009;124(2):315–22. doi: 10.1016/j.jaci.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J, Bardina L, Shreffler WG. Microarrayed allergen molecules for diagnostics of allergy. Methods Mol Biol. 2009;524:259–72. doi: 10.1007/978-1-59745-450-6_19. [DOI] [PubMed] [Google Scholar]
- 18.Cerecedo I, Zamora J, Shreffler WG, Lin J, Bardina L, Dieguez MC, et al. Mapping of the IgE and IgG4 sequential epitopes of milk allergens with a peptide microarray-based immunoassay. J Allergy Clin Immunol. 2008;122(3):589–94. doi: 10.1016/j.jaci.2008.06.040. [DOI] [PubMed] [Google Scholar]
- 19.Jarvinen KM, Beyer K, Vila L, Bardina L, Mishoe M, Sampson HA. Specificity of IgE antibodies to sequential epitopes of hen’s egg ovomucoid as a marker for persistence of egg allergy. Allergy. 2007;62(7):758–65. doi: 10.1111/j.1398-9995.2007.01332.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Lin J, Bardina L, Goldis M, Nowak-Wegrzyn A, Shreffler WG, et al. Correlation of IgE/IgG4 milk epitopes and affinity of milk-specific IgE antibodies with different phenotypes of clinical milk allergy. J Allergy Clin Immunol. 2010;125(3):695–702. doi: 10.1016/j.jaci.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuo H, Kohno K, Niihara H, Morita E. Specific IgE determination to epitope peptides of omega-5 gliadin and high molecular weight glutenin subunit is a useful tool for diagnosis of wheat-dependent exercise-induced anaphylaxis. J Immunol. 2005;175(12):8116–22. doi: 10.4049/jimmunol.175.12.8116. [DOI] [PubMed] [Google Scholar]
- 22.Breiteneder H, Radauer C. A classification of plant food allergens. J Allergy Clin Immunol. 2004;113(5):821–30. doi: 10.1016/j.jaci.2004.01.779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.