Abstract
Campylobacter jejuni is the most common zoonotic bacterium associated with human diarrhea, and chickens are considered to be one of the most important sources for human infection, with no effective prophylactic treatment available. We describe here a prophylactic strategy using chitosan-DNA intranasal immunization to induce specific immune responses. The chitosan used for intranasal administration is a natural mucus absorption enhancer, which results in transgenic DNA expression in chicken nasopharynx. Chickens immunized with chitosan-DNA nanoparticles, which carried a gene for the major structural protein FlaA, produced significantly increased levels of serum anti-Campylobacter jejuni IgG and intestinal mucosal antibody (IgA), compared to those treated with chitosan-DNA (pCAGGS). Chitosan-pCAGGS-flaA intranasal immunization induced reductions of bacterial expellation by 2-3 log10 and 2 log10 in large intestine and cecum of chickens, respectively, when administered with the isolated C. jejuni strain. This study demonstrated that intranasal delivery of chitosan-DNA vaccine successfully induced effective immune response and might be a promising vaccine candidate against C. jejuni infection.
1. Introduction
Gram-negative bacterium Campylobacter is responsible for an estimated 400 million human cases of enterocolitis worldwide each year, making it the leading cause of bacterial foodborne disease and a major causative agent of traveller's disease. In a limited number of cases, the enteric manifestations are followed by sequelae, such as reactive arthritis and the life-threatening neuropathy Guillain-Barré syndrome (GBS) [1]. Two thermophilic species, C. jejuni and C. coli, are responsible for the vast majority of human campylobacteriosis (~90% and ~10%, resp.) [1]. Animals such as chickens, cattle, pigs, sheep, and dogs may act as asymptomatic reservoirs [2], by shedding C. jejuni in their stools, which results in the contamination of animal food products and surface water during slaughter and carcass dressing [3]. Chickens, often heavily colonized with C. jejuni without signs of pathology, are considered to be one of the most important sources for human infection [4]. It is generally assumed that C. jejuni-contaminated poultry constitutes a major risk to human health.
A wide array of interventions has been developed to reducing the carriage of C. jejuni in livestock and poultry [5], including attempts to eliminate C. jejuni from farms by increasing biosecurity, separating contaminated flocks, and improving hygiene during slaughter. Although these measures undoubtedly help to control shedding of C. jejuni in infected animals, and may reduce the number of positive flocks and the contamination level of their products, vaccination of poultry against C. jejuni will probably be the most effective measure, and remains a major goal [6, 7]. Considering the physiology of Campylobacter and their intestinal ecological niche in poultry, conventional vaccines have a poor performance, including inactivated vaccines [8, 9].
Vaccines generally have a very good safety profile. The rapid advances in genomics, proteomics, and molecular design of vaccines provide hope that an effective vaccine for Campylobacter spp. can be developed in the near future [1, 10, 11]. In 1990, it was first reported that intramuscular injection of plasmid DNA in a simple saline solution could transfect muscle cells in vivo [12]. Genetic immunization is a novel vaccine strategy that conceptually combines some of the most desirable attributes of standard vaccine approaches. The immune responses comprise both cell-mediated and humoral (antibody) components. Most recent studies associated with the efficacy of DNA vaccine against C. jejuni focus on the potential vaccine antigens, plasmid vector, and DNA delivery (mucosal adjuvants). The gene flaA plays an important role in the pathogenicity of C. jejuni [13]. As a result of its biocompatibility, biodegradability, low cost, and ability to open intercellular tight junctions, chitosan can be used as a delivering vehicle for the mucosal vaccine and may have an adjuvant effect [14].
Here, we reported intranasal immunization of White Leghorn chickens using chitosan-DNA nanoparticles that carried the recombinant plasmid pCAGGS-flaA for the flaA gene encoding the major structural protein FlaA of C. jejuni, and demonstrated its efficacy in inducing specific immune responses and protection against homologous strain challenge.
2. Materials and Methods
2.1. Animals
White Leghorn chickens aged 1 day were obtained from the Comparative Medicine Center of Yangzhou University (Yangzhou University, Jiangsu, China). Animals were housed, handled, and immunized following approval by the institutional animal experimental committee.
2.2. Plasmids, Bacteria, and Antibodies
pCAGGS was a kind gift from Dr. Miyazaki (Faculty of Medicine, University of Tokyo, Japan). C. jejuni strain ALM-80 was isolated from chicken by the Jiangsu Key Laboratory of Zoonosis, Yangzhou University (Jiangsu, China). COS-7 cells were ordered from Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China), and grown in DMEM (GIBCO, Grand Island, NY, USA) supplemented with 10% (v/v) fetal bovine serum (FBS; GIBCO). Chitosan, FITC-labeled rabbit antichicken IgG, horseradish peroxidase- (HRP-) labeled goat antichicken IgA, HRP-labeled rabbit antichicken IgG and IgM were obtained from Sigma (St Louis, MO, USA). All other chemicals and reagents were of analytical grade, and obtained from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China).
2.3. Construction of pCAGGS-flaA
A pair of primers were designed to amplify gene flaA of C. jejuni that contained XbaI and EcoRI sites, forward: 5′-CATCTAGAAGCATTTAACAAGTTCATGG-3′, reverse: 5′-TAGAATTCGTGTTTATCCTAAAACCCAT-3′. Amplification was conducted in a volume of 50 μL, containing 5 μL 10× buffer, 3 pM dNTP, 20 pM primers, 2 μL (10 pg) DNA, and 1 U Taq polymerase (Takara Diagnostics, Dalian, China). The reactions were performed with a thermal cycler (Bio-Rad Ltd, Hercules, CA, USA) using the following temperature-cycling parameters: initial denaturation at 95°C for 5 min, followed by 32 cycles of 1 min at 94°C, 1 min at 58°C, 1.5 min at 72°C, with a final extension step of 5 min at 72°C. Amplification generated 1935-bp DNA fragments. Eight microliters of PCR products were separated on a 1% (w/v) agarose gel following electrophoresis at 60 V for 1.5 h. Gels were stained with ethidium bromide (0.3 mg/l) and visualized under UV illumination. A 100-bp DNA ladder (Promega, Madison, WI, USA) was used as a molecular size standard.
Then, the PCR product was cloned into pGEM-T easy vector (Sangon Biotech, Shanghai, China) to give rise to pT-flaA. The EcoRI-XbaI-restricted insert of pT-flaA was cloned into eukaryotic expression vector pCAGGS, and then the recombinant vector pCAGGS-flaA was constructed and inserted in Escherichia coli (DH5α) by electrotransformation.
For immunization purposes, homogeneous stocks of DNA were purified from 2.5 L cultures using the Plasmid Giga Preparation kit (Qiagen, Hilden, Germany). The DNA preparations were quantified by optical density (OD) measurements at 260 nm and analyzed by 1% (w/v) agarose gel electrophoresis.
2.4. Preparation of Chitosan-DNA Nanoparticles
The chitosan/pCAGGS-flaA DNA nanoparticles were synthesized by Mao et al. [15]. In brief, a chitosan (Sigma, St Louis, MO, USA) solution (200 μg/mL in 5 mM sodium acetate buffer, pH 5.5), 400 kD, 85% deacetylated, and a pCAGGS-flaA solution (100 μg/mL in 5 mM sodium sulfate solution) were separately preheated to 50°C–52°C in parallel. Then, the pCAGGS-flaA solution was added into an equal volume of the chitosan solution. Uniform particles were obtained by coacervation between chitosan and DNA, and were left to stand for at least 1 h at room temperature.
2.5. Characterization of Chitosan-DNA Complexes
Freshly prepared chitosan-DNA complexes were characterized by morphological examination by transmission electron microscopy (TEM). The samples were placed on a cotton grid, allowed to sit for a few minutes and air-dried. Then, they were negatively stained with uranyl acetate for visualization under a transmission electron microscopy (TEM, accelerating voltage/120 kV, Philips Tecnai 12, Holland).
Naked DNA (1 μg in 20 μL of 25 mM Na2SO4 solution) or chitosan-DNA suspension (20 μL, equivalent to 1 μg of DNA) was incubated with 0.5 U DNase I (final concentration of 28 U/mL) for 15 min at 37°C. EDTA was added to stop the reaction to a final concentration of 50 mM. Then, chitosan-DNA was subjected to chitosanase and lysozyme (final concentrations of 0.15 and 15 U/mL) digestion at 37°C for 4 h. Integrity of DNA was analyzed by agarose electrophoresis.
2.6. Gene Expression In Vitro
Two micrograms of plasmid (Chitosan-pCAGGS-flaA) was transfected into 70% confluent COS-7 cells covering a 6 cm2 dish using a LipofectAMINE 2000 kit (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. After 2 days incubation at 37°C in 5% CO2, the cells were washed with phosphate-buffered saline (PBS) and fixed with 3% (v/v) paraformaldehyde in PBS for 20 min at room temperature. The cells were then washed three times and permeabilized with PBS containing 0.05% (v/v) Tween 20 for 30 min. After a further wash with PBS, the cells were incubated with a 1 : 50 dilution of mouse antiFlaA antiserum (from our laboratory) for 1 h followed by rinsing with PBS and incubation with fluorescein isothiocyanate- (FITC-) conjugated goat antimouse immunoglobulin G antibody (Sigma-Aldrich, St. Louis, MO, USA) for 45 min. The cells were washed again with PBS and visualized by a fluorescence microscope (Leica Microsystems GmbH,Wetzlar, Germany).
2.7. Association Efficiency of Chitosan-DNA Nanoparticles
To assay the association efficiency (AE) of pCAGGS-flaA, the nanoparticles suspension was first centrifuged (Heraeus Fresco 21; Thermo Scientific, Germany) at 30,000 g, at 4°C for 30 min. The total amount of the original pCAGGS-flaA and free (or unconjugated) pCAGGS-flaA were then analyzed using a microspectrophotometer (Biophotometer; Eppendorf, Germany). The association efficiency was calculated as follows:
| (1) |
2.8. Vaccination of Chickens and Challenge Experiment
One-day-old White Leghorn chickens, randomly divided into three groups (30 per group), were immunized intranasally with chitosan/pCAGGS-flaA nanoparticles, chitosan/pCAGGS, and saline control, respectively. These chickens were immunized three times on days 1, 15, and 29 at a dose of 150 μg in 200 μL (100 μL each nostril). On day 42, all chickens were challenged orally with 5×107 ALM-80 strain. Three chickens were sacrificed every 3 days from day 45 to day 63 to harvest the small intestine, large intestine, and cecum to calculate the number of C. jejuni ALM-80. The method of isolation and enumeration of Campylobacter can be found in our preliminary study [4].
2.9. Serum and Mucosal Antibody Titers to Campylobacter Measured by Enzyme-Linked Immunosorbent Assay (ELISA)
Blood was collected after 4 and 6 weeks of primary immunization and sera were stored at −70°C until they were tested by ELISA for antibodies. The intestinal mucosal secretion was collected at 2 and 4 weeks after booster immunization. The abdominal cavity was opened aseptically, and the entire small intestine, including the duodenum, jejunum, and ileum, was collected. Pancreas, connective tissue, and fat were removed into PBS, and the intestine was cut longitudinally and then cut into 1-cm-long sections. The intestinal antibodies were extracted using a PBS solution containing Tween 20 (0.05%, v/v), soybean trypsin inhibitor (0.1 mg/mL) (Sigma-Aldrich, St. Louis, MO, USA), EDTA (0.05 mg/mL), and phenylmethylsulfonyl fluoride (PMSF) (0.35 mg/mL) (Sigma-Aldrich, St. Louis, MO, USA). Intestinal lavage solutions were mixed with extraction solution and shaken for 2 h at 4°C. After centrifugation at 20,000 × g for 30 min at 4°C the supernatant was collected. Bovine serum albumin was added to a final concentration of 0.1% (w/v) and samples were preserved at −20°C. The titer of antibodies against Campylobacter in intestinal secretions was measured using an indirect enzyme-linked immunosorbent assay (ELISA) developed by laboratory procedure. Briefly, the ELISA involved the following steps: (1) 96-well microtitre plates were coated with the isolated strain ALM-80 (5 × 105 cfu/well) for 6 h at 42°C, and blocked with BSA solution (10%, w/v) for 1 h at 37°C, then washed with a 0.05% (v/v) solution of Tween 20 in PBS. (2) Samples were serially diluted and added. The plates were incubated for 2 h at 37°C and washed with PBS-Tween. (3) Goat antimouse IgG, IgM, or IgA was added. The plates were incubated for 1.5 h at 37°C and washed with PBS-Tween. (4) o-phenylenediamine dichloride (OPD; Sigma, USA) was added to the plates at room temperature. (5) H2SO4 (2.5 M) was added to stop the reaction 30 min after addition of the substrate. (6) The absorbance was recorded at 490 nm.
2.10. Ratio of CD4+/CD8+ T Cells in Cecal Tonsil and Spleen
To analyze the changes in T-cell composition upon immunization, spleen and cecal tonsil of each animal (four per group on investigation days 42 and 56) were crushed in ice-cold PBS. The homogenates were centrifuged at 300 rpm for 2 min at below 4°C before the insoluble matter settled. The isolated cells were incubated with FITC- (fluoresceinisothiocyanate- ) labeled CD4 and PE (phycoerythrin- ) labeled CD8α monoclonal antibodies (Southern Biotechnology Associates, Birmingham, AL, USA) for 30 min at 37°C in the dark. After the cells were washed, aliquots of 20,000 cells per sample were analyzed using a FACSAria flow cytometry system (Becton Dickinson, CA, USA).
2.11. Statistical Analysis
The data were presented as mean ± standard deviation. The Student t-test for comparison of two independent samples was used to evaluate differences between the groups (immunized group versus control). Viable counts of bacteria were converted into logarithmic form. For the purpose of statistical analysis, a log10 viable count of <1.47 (limit for direct plate detection), obtained from a sample that became positive only after enrichment, was rated as a log10 value of 1.0. A sample which yielded no C. jejuni growth after enrichment was rated as a log10 value of 0. Data were evaluated by multifactorial variance analysis. P-values less than .05 were considered to be significant, and P-values less than .01 were considered to be highly significant. The flow cytometry data were evaluated using BD Bioscience's FACS Diva software (Becton Dickinson, CA, USA).
3. Results
3.1. Identification of pCAGGS-flaA
flaA was amplified from C. jejuni ALM-80 by PCR, and sequence analysis showed that the sequence of amplified fragment was consistent with that of flaA published in GenBank. Recombinant plasmids pT-flaA and pCAGGS-flaA were confirmed by PCR and restriction enzyme digestion. It was demonstrated that recombinant plasmids contained the flaA gene.
3.2. Synthesis and Characterization of Chitosan-DNA
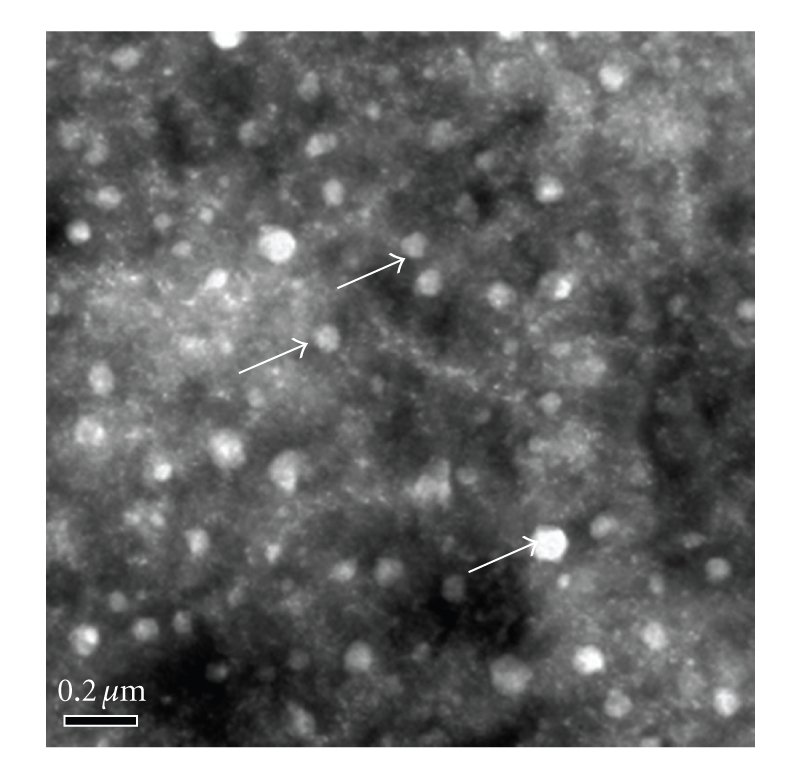
Chitosan-DNA complexes were synthesized when DNA (pCAGGS-flaA) solution (100 μg/mL in 5 mM Na2SO4) was added to 0.02% (w/v) chitosan (pH 5.5) at 55°C during high-speed vortexing. Uniform particles were obtained by coacervation between chitosan and DNA. TEM observation confirmed that freshly synthesized chitosan-DNA complexes were 80–100 nm in size and nearly spherical (Figure 1), with an association efficiency of 91.9%.
Figure 1.
TEM image of chitosan-DNA. Freshly prepared chitosan-DNA solution was negatively stained with uranyl acetate. Scale bars represent 200 nm. The freshly synthesized chitosan-DNA complexes were approximately 80–100 nm in size and nearly spherical under TEM.
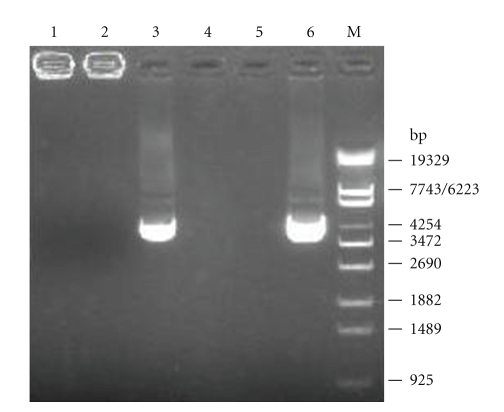
The ability of the chitosan/pCAGGS-flaA nanoparticles to protect pCAGGS-flaA from nuclease degradation was evaluated by enzymatic digestion and agarose gel electrophoresis. As shown in Figure 2, 1 μg naked DNA was completely degraded (lane 5) under 28 U/mL DNAse I digestion. However, even at such a high DNAse I concentration, chitosan-DNA complex remained intact (lane 2). As a control, 2 μg chitosan showed entire degradation after chitosanase and lysozyme digestion releasing most of the DNA inside the complex (lane 3). These results suggest that chitosan could protect DNA from markedly high concentration of DNAse I digestion. At the physiological condition such as mucosal membrane surfaces with large quantities but lower concentration of nucleases, this protection of DNA by chitosan would be meaningful for the maintenance of integration and function of DNA vaccine.
Figure 2.
Protection of DNA from DNAse I digestion by chitosan. Naked DNA and chitosan-DNA complex containing 1 μg DNA were both incubated with DNAse I for 15 min at 37°C. Then the reaction was stopped by adding with 0.5 M EDTA. chitosan-DNA was then subjected to chitosanase and lysozyme digestion. Finally, all samples were run on 0.8% (w/v) agarose gel and stained with ethidium bromide. Lane 1: chitosan-DNA; 2: chitosan-DNA + DNAse I (28 U/mL); 3: chitosan-DNA + chitosanase (0.15 U/mL) + lysozyme (15 U/mL); 4: 3 + DNAse I (28 U/mL); 5: DNA+ DNAse I (28 U/mL); 6: DNA (pCAGGS-flaA, 4.1 kb).
3.3. Specific Serum and Intestinal Antibody Responses
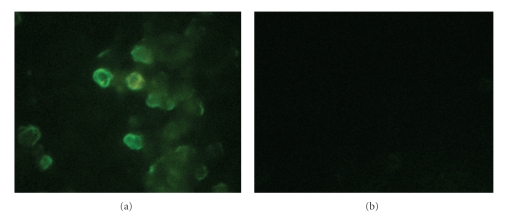
The ability of constructed chitosan-DNA vaccine complex to express flaA in eukaryotic cells (COS-7 cell line) transfected with pCAGGS-flaA, and the negative control pCAGGS was elucidated. At 48 h post transfection, only pCAGGS-flaA-transfected cells expressed protein FlaA, which was detected by immunofluorescent assay (Figure 3).
Figure 3.
In vitro gene expression of chitosan-encapsulated pCAGGS-flaA in COS-7 cell line by immunofluorescent assay. Ability of constructed chitosan-DNA vaccine complex to express flaA in eukaryotic cells (COS-7 cell line) transfected with pCAGGS-flaA (a), and the negative control pCAGGS (b) was evaluated. After 48 h transfection, only pCAGGS-flaA-transfected cells expressed protein flaA, which was detected by fluorescent microscopy.
In order to evaluate the potential utility of chitosan-DNA immunization in inducing specific responses, 1-day-old commercial White Leghorn chickens were vaccinated orally with chitosan-plasmid nanoparticles at 2-week intervals. Negative control groups included blank and CS-pCAGGS controls. Low titers of specific antibodies were observed in all immunized groups at day 28. However, the titers of specific antibodies in the CS-pCAGGS-flaA group were elevated significantly at day 42 (P < .01) (Table 1).
Table 1.
Immunogenicity assay of C. jejuni chitosan-DNA vaccine in chickens.
| Group | Titers of antibodies in serum | Titers of antibodies in intestinal mucosa | ||
|---|---|---|---|---|
| Second immunization | Third immunization | Second immunization | Third immunization | |
| CS-pCAGGS-flaA | 96.0 ± 55.4a | 426.7 ± 147.8a | 65.3 ± 62.0b | 170.7 ± 73.9a |
| CS-pCAGGS | 26.7 ± 9.2 | 23.3 ± 18.5 | 34.7 ± 28.1 | 13.3 ± 4.6 |
| PBS | 26.7 ± 9.2 | 23.3 ± 18.5 | 34.7 ± 28.1 | 29.3 ± 30.3 |
aHigh significantly different (P < .01) among vaccine groups as calculated by Student's t-test.
bSignificantly different (P < .05) among vaccine groups as calculated by Student's t-test.
“CS” is the short form for chitosan.
3.4. Ratio of CD4+/CD8+ T Cells in Spleen and Cecal Tonsil
The number of CD4+/CD8+ cells in cecal tonsil and spleen was detected by flow cytometry. Experimental data showed that the ratio of CD4+/CD8+ T cells in the spleen of the CS-pCAGGS-flaA group was significantly increased at day 28 and day 42, and the number of CD4+ cells was greater than the number of CD8+ cells. In the cecal tonsils, the ratio of CD4+/CD8+ cells significantly increased after the third immunization compared to that in the control group. The results indicated that the ratio of CD4+/CD8+ T cells in the cecal tonsil and spleen of the CS-pCAGGS-flaA groups was increased after the third immunization, and the number of CD4+ cells was greater than the number of CD8+ cells (Table 2).
Table 2.
Ratio of CD4+/CD8+ T cells in spleen and cecal tonsil of immunized chickens.
| Group | Ratioa of CD4+/CD8+ T cells in spleen | Ratioa of CD4+/CD8+ T cells in cecal tonsil | ||
|---|---|---|---|---|
| Second immunization | Third immunization | Second immunization | Third immunization | |
| CS-pCAGGS-flaA | 1.34 ± 0.53b | 1.54 ± 0.23b | 0.98 ± 0.17 | 1.94 ± 0.21b |
| CS-pCAGGS | 0.63 ± 0.22 | 0.94 ± 0.22 | 0.92 ± 0.26 | 1.29 ± 0.18 |
| PBS | 0.62 ± 0.05 | 1.00 ± 0.08 | 1.17 ± 0.30 | 1.27 ± 0.30 |
aValues are means ± standard deviations (four samples per group) for ratio of CD4+/CD8+ T cells treated with CS-pCAGGS-flaA and CS-pCAGGS or PBS.
bSignificantly different (P < .05) among vaccine groups as calculated by Student's t-test.
3.5. Shedding of C. jejuni in Immunized Chickens after Challenge
After oral challenge with 5 × 107 bacteria of strain ALM-80, the percentage of chickens with C. jejuni-positive cloaca showed a downward trend in the vaccine group, and bacteria were not detected at day 63. However, the negative control groups retained a high C. jejuni-positive ratio (Table 3).
Table 3.
Average positive rate of swab specimens of White Leghorn chickens challenged with strain ALM-80.
| Group | Average positive rate of swab specimens after oral challenge | ||
|---|---|---|---|
| 7 days | 14 days | 21 days | |
| CS-pCAGGS-flaA | 47.62% (10/21) | 30.77% (4/13)a | 0.00% (0/9)a |
| CS-pCAGGS | 47.62% (10/21) | 53.33% (8/15) | 44.44% (4/9) |
| PBS | 42.86% (9/21) | 53.33% (8/15) | 55.56% (5/9) |
aSignificantly different (P < .05) among vaccine groups as calculated by Student's t-test.
3.6. Protection Effects of Immunized Chickens after Challenge with C. jejuni
After challenge, in vaccine group, the number of strain ALM-80 bacteria in the small intestine, large intestine, and cecum reached a peak from day 51 to day 57, and then decreased. The number of bacteria was significantly lower than control, which showed an upward trend of bacterial number in small intestine, large intestine and cecum. In the CS-pCAGGS-flaA group, ALM-80 was cleared from the small intestine on day 60 (Table 4). Compared with the negative control groups, the number of C. jejuni was reduced by 2-3 log10 in the large intestine and by 2 log10 in the cecum when chickens were challenged with the homologous strain (Table 4). Infection tests induced expression of that chitosan-DNA nanoparticle vaccine that delivered flaA of C. jejuni could reduce the number of C. jejuni in small intestine, large intestine, and cecum effectively.
Table 4.
The number of C. jejuni in small intestine, large intestine, and cecum after oral challenge.
| Organ | Group | CFU (CFU/g) | ||||||
|---|---|---|---|---|---|---|---|---|
| 3 days | 6 days | 9 days | 12 days | 15 days | 18 days | 21 days | ||
| Small intestine | 1 | 9.2 × 103 | 3.4 × 103 | 6.0 × 102 | 8.2 × 103 | 1.9 × 103 | 0 | 0 |
| 2 | 3.5 × 102 | 7.6 × 102 | 1.2 × 102 | 1.4 × 104 | 5.9 × 104 | 6.3 × 103 | 7.6 × 104 | |
| 3 | 2.0 × 103 | 6.3 × 102 | 7.3 × 102 | 1.7 × 103 | 1.2 × 105 | 2.9 × 104 | 3.8 × 104 | |
| Large intestine | 1 | 4.6 × 105 | 1.1 × 106 | 1.5 × 107 | 4.8 × 106 | 2.9 × 106 | 3.5 × 106 | 6.5 × 105 |
| 2 | 5.4 × 107 | 3.2 × 107 | 2.2 × 107 | 1.1 × 107 | 1.8 × 108 | 4.2 × 107 | 1.5 × 108 | |
| 3 | 4.7 × 107 | 2.1 × 107 | 2.2 × 107 | 9.3 × 107 | 3.5 × 108 | 3.2 × 107 | 1.8 × 107 | |
| Cecum | 1 | 2.2 × 106 | 4.5 × 106 | 2.8 × 107 | 2.9 × 107 | 3.7 × 107 | 3.0 × 107 | 1.2 × 107 |
| 2 | 2.7 × 108 | 1.8 × 108 | 7.4 × 108 | 2.4 × 108 | 1.9 × 108 | 2.6 × 109 | 2.3 × 109 | |
| 3 | 4.3 × 108 | 2.2 × 108 | 4.0 × 108 | 4.0 × 108 | 2.6 × 108 | 4.7 × 109 | 2.7 × 109 | |
Group 1: CS-pCAGGS-flaA group.
Group 2: CS-pCAGGS group.
Group 3: PBS group.
4. Discussion
Vaccination has played an important role in the protection of chickens against C. jejuni infections. However, the conventional killed vaccines against C. jejuni do not completely prevent infection and bacterial shedding [1]. Thus, an improved vaccine and/or vaccination schedule is needed to induce immunity in chickens. In the current study, we have developed chitosan/pCAGGS-flaA nanoparticles to inhibit Campylobacter jejuni in a White Leghorn model, which is a common layer chicken breed in China.
DNA vaccine can provide effective immunostimulation in terms of humoral and cell-mediated responses, therefore overcoming the influence of maternal antibody. Meanwhile, DNA vaccine has broad prospects in the prevention and treatment of bacterial, viral, parasitic, and other infections [12]. The strong immunogenicity and cross-protection of antigen genes are the prerequisites for constructing DNA vaccines, which is a good strategy to improve their immunological effects. For C. jejuni, adhesion to, invasion, and colonization of host intestinal cells require the participation of flagellin, which is essential for C. jejuni to adhere to host cells. Therefore, it is a key step in the process of host infection [16]. In our study, based on the analysis of virulence genes mainly carried by isolated strains, we selected flagellin gene flaA, which is closely involved in the infection process, as a protective antigen gene to construct a DNA vaccine. The flagella gene plasmid DNA constructed by Wyszyńska et al. can reduce the number of Campylobacter by 2 log2 [17]. Widders et al. found that titers of IgG and IgM in serum and IgG in intestinal mucosa were high after primary immunization with intraperitoneal injection and secondary immunization with intraperitoneal injection or oral administration of purified flagellin protein, and results indicated that immunization could reduce the level of intestinal infection with C. jejuni by 1-2 log2 [18].
DNA vaccine expression vectors with different promoters have significant impacts on expression of immunogenic genes [19]. The plasmid vector pCAGGS contains chicken β-actin promoter and cytomegalovirus enhancer of eukaryotic expression plasmid, therefore, the level of pCAGGS will be significantly high after transient transfection of most cell lines [20, 21]. Many studies have suggested that the expression efficiency of pCAGGS is high, therefore, it is presently being applied widely [22]. On the other hand, using chitosan, a natural bioadhesive material, as a gene carrier turned out to be an ideal way to stimulate the immune system. The aminoglucose and DNA can form complexes (polyplexes) via their electrostatic interaction. Chitosan can condense DNA into relatively compact nanoparticles and these can overcome physiological barriers when drugs are absorbed by the mucosa and arrive at the submucosal lymphoid tissue by endocytotic channels. The advantages of chitosan nanoparticles as gene vectors are as follows. (i) They can protect DNA from degradation in vivo [15]. (ii) Positively charged nanoparticles together with negatively charged mucosal cells and sialic acid can generate stronger electrostatic interactions and prolong the reaction time between vaccines and the mucosa, and increase vaccine uptake [23]. The preparation conditions of chitosan-DNA nanoparticles are mild, without using organic solvents, which solves the problem of DNA destruction by solvent residues. Chitosan is regarded as a potential nonviral gene carrier. Mao et al. were the first to combine DNA vaccine and chitosan to create a chitosan-DNA nanovaccine, which successfully generated immunological protection in a murine model of peanut allergy, and proved to be an effective oral gene delivery system [15]. In our study, high encapsulation efficiency was obtained (>90%), as demonstrated by electrophoresis and electron microscopy. Protection tests showed that chitosan-DNA nanoparticle vaccine that contained flaA of C. jejuni reduced effectively colonization of C. jejuni in the small intestine, large intestine, and cecum.
At present, investigation of the protective immune responses against C. jejuni is at the initial stage [24]. Campylobacter vaccine can induce significant immune protection against experimental infection of mice, ferrets, rabbits, or primates, and might relieve or eliminate clinical symptoms in these animals [25]. Nevertheless, vaccines have different effects on mammals and chickens, which needs to be considered when research is conducted using chicken models of colonization of the intestinal tract.
In addition, since flaA of C. jejuni is highly variable [26], it is interesting to assess the protective efficacy of the chitosan/pCAGGS-flaA nanoparticles against challenge with other C. jejuni strains in future studies. What's more, they need to replicate more efficiently in broilers by optimizing the immunization strategy.
5. Conclusions
This study demonstrated that intranasal delivery of chitosan-DNA vaccine successfully induced effective immune response and might be a promising vaccine candidate against C. jejuni infection.
Acknowledgments
This work was supported by the Grant 2007AA02Z419 and 2009BADB9B01 from the Ministry of Science and Technology of China, and Grants BE2008655 and BK2008011 from Jiangsu Provincial Government.
References
- 1.de Zoete MR, van Putten JPM, Wagenaar JA. Vaccination of chickens against Campylobacter. Vaccine. 2007;25(30):5548–5557. doi: 10.1016/j.vaccine.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Newell DG. Animal models of Campylobacter jejuni colonization and disease and the lessons to be learned from similar Helicobacter pylori models. Journal of Applied Microbiology. 2001;90(30):57S–67S. doi: 10.1046/j.1365-2672.2001.01354.x. [DOI] [PubMed] [Google Scholar]
- 3.Verwoerd DJ. Ostrich diseases. Revue Scientifique et Technique. 2000;19(2):638–661. doi: 10.20506/rst.19.2.1235. [DOI] [PubMed] [Google Scholar]
- 4.Huang JL, Xu HY, Bao GY, et al. Epidemiological surveillance of Campylobacter jejuni in chicken, dairy cattle and diarrhoea patients. Epidemiology and Infection. 2009;137(8):1111–1120. doi: 10.1017/S0950268809002039. [DOI] [PubMed] [Google Scholar]
- 5.Wagenaar JA, Mevius DJ, Havelaar AH. Campylobacter in primary animal production and control strategies to reduce the burden of human campylobacteriosis. Revue Scientifique et Technique. 2006;25(2):581–594. [PubMed] [Google Scholar]
- 6.Lee MD, Newell DG. Campylobacter in poultry: filling an ecological niche. Avian Diseases. 2006;50(1):1–9. doi: 10.1637/7474-111605R.1. [DOI] [PubMed] [Google Scholar]
- 7.Zhao C, Ge B, De Villena J, et al. Prevalence of Campylobacter spp., Escherichia coli, and Salmonella serovars in Retail Chicken, Turkey, Pork, and Beef from the Greater Washington, D.C., Area. Applied and Environmental Microbiology. 2001;67(12):5431–5436. doi: 10.1128/AEM.67.12.5431-5436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ringoir DD, Korolik V. Colonisation phenotype and colonisation potential differences in Campylobacter jejuni strains in chickens before and after passage in vivo. Veterinary Microbiology. 2003;92(3):225–235. doi: 10.1016/s0378-1135(02)00378-4. [DOI] [PubMed] [Google Scholar]
- 9.Walker RI. Considerations for development of whole cell bacterial vaccines to prevent diarrheal diseases in children in developing countries. Vaccine. 2005;23(26):3369–3385. doi: 10.1016/j.vaccine.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 10.Huang S, Sahin O, Zhang Q. Infection-induced antibodies against the major outer membrane protein of Campylobacter jejuni mainly recognize conformational epitopes. FEMS Microbiology Letters. 2007;272(2):137–143. doi: 10.1111/j.1574-6968.2007.00752.x. [DOI] [PubMed] [Google Scholar]
- 11.Scott NE, Cordwell SJ. Campylobacter proteomics: guidelines, challenges and future perspectives. Expert Review of Proteomics. 2009;6(1):61–74. doi: 10.1586/14789450.6.1.61. [DOI] [PubMed] [Google Scholar]
- 12.Wolff JA, Malone RW, Williams P, et al. Direct gene transfer into mouse muscle in vivo. Science. 1990;247(4949, part 1):1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 13.Wassenaar TM, Bleumink-Pluym NMC, Newell DG, Nuijten PJM, Van der Zeijst BAM. Differential flagellin expression in a flaA−/ flaB+ mutant of Campylobacter jejuni. Infection and Immunity. 1994;62(9):3901–3906. doi: 10.1128/iai.62.9.3901-3906.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNeela EA, O’Connor D, Jabbal-Gill I, et al. A mucosal vaccine against diphtheria: formulation of cross reacting material (CRM197) of diphtheria toxin with chitosan enhances local and systemic antibody and Th2 responses following nasal delivery. Vaccine. 2000;19(9-10):1188–1198. doi: 10.1016/s0264-410x(00)00309-1. [DOI] [PubMed] [Google Scholar]
- 15.Mao H-Q, Roy K, Troung-Le VL, et al. Chitosan-DNA nanoparticles as gene carriers: synthesis, characterization and transfection efficiency. Journal of Controlled Release. 2001;70(3):399–421. doi: 10.1016/s0168-3659(00)00361-8. [DOI] [PubMed] [Google Scholar]
- 16.Müller J, Schulze F, Müller W, Hänel I. PCR detection of virulence-associated genes in Campylobacter jejuni strains with differential ability to invade Caco-2 cells and to colonize the chick gut. Veterinary Microbiology. 2006;113(1-2):123–129. doi: 10.1016/j.vetmic.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 17.Wyszyńska A, Raczko A, Lis M, Jagusztyn-Krynicka EK. Oral immunization of chickens with avirulent Salmonella vaccine strain carrying C. jejuni 72Dz/92 cjaA gene elicits specific humoral immune response associated with protection against challenge with wild-type Campylobacter. Vaccine. 2004;22(11-12):1379–1389. doi: 10.1016/j.vaccine.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Widders PR, Thomas LM, Long KA, Tokhi MA, Panaccio M, Apos E. The specificity of antibody in chickens immunised to reduce intestinal colonisation with Campylobacter jejuni. Veterinary Microbiology. 1998;64(1):39–50. doi: 10.1016/s0378-1135(98)00251-x. [DOI] [PubMed] [Google Scholar]
- 19.Suarez DL, Schultz-Cherry S. The effect of eukaryotic expression vectors and adjuvants on DNA vaccines in chickens using an avian influenza model. Avian Diseases. 2000;44(4):861–868. [PubMed] [Google Scholar]
- 20.Bu Z, Ye L, Compans RW, Yang C. Enhanced cellular immune response against SIV Gag induced by immunization with DNA vaccines expressing assembly and release-defective SIV Gag proteins. Virology. 2003;309(2):272–281. doi: 10.1016/s0042-6822(03)00066-7. [DOI] [PubMed] [Google Scholar]
- 21.Bu Z, Ye L, Skeen MJ, Ziegler HK, Compans RW, Yang C. Enhancement of immune responses to an HIV env DNA vaccine by a C-terminal segment of listeriolysin O. AIDS Research and Human Retroviruses. 2003;19(5):409–420. doi: 10.1089/088922203765551755. [DOI] [PubMed] [Google Scholar]
- 22.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 23.Soane RJ, Frier M, Perkins AC, Jones NS, Davis SS, Illum L. Evaluation of the clearance characteristics of bioadhesive systems in humans. International Journal of Pharmaceutics. 1999;178(1):55–65. doi: 10.1016/s0378-5173(98)00367-6. [DOI] [PubMed] [Google Scholar]
- 24.Sahin O, Luo N, Huang S, Zhang Q. Effect of Campylobacter-specific maternal antibodies on Campylobacter jejuni colonization in young chickens. Applied and Environmental Microbiology. 2003;69(9):5372–5379. doi: 10.1128/AEM.69.9.5372-5379.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baqar S, Bourgeois AL, Applebee LA, et al. Murine intranasal challenge model for the study of Campylobacter pathogenesis and immunity. Infection and Immunity. 1996;64(12):4933–4939. doi: 10.1128/iai.64.12.4933-4939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owen RJ, Fayos A, Hernandez J, Lastovica A. PCR-based restriction fragment length polymorphism analysis of DNA sequence diversity of flagellin genes of Campylobacter jejuni and allied species. Molecular and Cellular Probes. 1993;7(6):471–480. doi: 10.1006/mcpr.1993.1070. [DOI] [PubMed] [Google Scholar]