Abstract
The peroxisome proliferator-activated receptor alpha (PPARα) is a ligand-activated transcription factor involved in the regulation of a variety of processes, ranging from inflammation and immunity to nutrient metabolism and energy homeostasis. PPARα serves as a molecular target for hypolipidemic fibrates drugs which bind the receptor with high affinity. Furthermore, PPARα binds and is activated by numerous fatty acids and fatty acid-derived compounds. PPARα governs biological processes by altering the expression of a large number of target genes. Accordingly, the specific role of PPARα is directly related to the biological function of its target genes. Here, we present an overview of the involvement of PPARα in lipid metabolism and other pathways through a detailed analysis of the different known or putative PPARα target genes. The emphasis is on gene regulation by PPARα in liver although many of the results likely apply to other organs and tissues as well.
1. Introduction
Nutrient metabolism and energy homeostasis are tightly controlled by numerous regulatory systems involving specific transcription factors. The peroxisome proliferator-activated receptors (PPARs) are ligand-activated transcription factors that belong to the superfamily of nuclear hormone receptors and play an important role in nutrient homeostasis [1–3]. Three different PPAR subtypes are known: PPARα, PPARβ/δ, and PPARγ. All PPARs share the same molecular mode of action via formation of heterodimers with the nuclear receptor RXR, followed by binding to specific DNA-response elements in target genes known as peroxisome proliferator response elements (PPREs). PPREs are characterized by a common core sequence consisting of a direct repeat of the consensus sequence AGGTCA interspaced by a single nucleotide [1, 4]. Expression of PPARα and PPARβ/δ is found ubiquitously, whereas PPARγ is mainly expressed in adipose tissue, macrophages, and colon [5, 6]. Activation of transcription by PPARs is dependent on a number of different steps including ligand binding to PPAR, binding of PPAR to the target gene, removal of corepressors and recruitment of coactivators, remodeling of the chromatin structure, and finally facilitation of gene transcription [7]. This paper will focus exclusively on PPARα.
PPARα was first discovered in the early 1990s and since then has been identified as the master regulator of hepatic lipid metabolism [8]. In addition, PPARα has been shown to govern glucose metabolism, lipoprotein metabolism, liver inflammation, amino acid metabolism, and hepatocyte proliferation (specifically in rodents). Synthetic agonists of PPARα lower plasma triglycerides and raise plasma high-density lipoprotein (HDL) levels and are thus used clinically in the treatment of dyslipidemia [2, 9–11].
In recent years, the advent of microarray technology has allowed the study of whole genome expression profiles. Accordingly, a wealth of new information has become available about the role of specific transcription factors in regulation of gene expression. Combined with data collected using more established methods, microarray has permitted the generation of a comprehensive picture of the impact of PPARα on gene expression, thereby providing key insight into the functional role of PPARα. The present paper is aimed at providing a detailed and updated overview of PPARα target genes in different biological processes and to highlight possible differences between mouse and human.
Although the presence of a functional PPRE is often used as a criteria for designating direct PPARα target genes, we did not apply this criteria very stringently in our analysis as the in vivo functionality of most of the identified PPREs remains uncertain. Recent studies indicate that the standard approach to screen for PPREs in the 1-2 kb region upstream of the transcriptional start site (TSS) is at odds with accumulating evidence that PPARs often bind quite distant from the TSS [12–14]. In those cases, contact with the basal transcription machinery is expected to be established via DNA looping. Thus, the absence of a PPRE in the 1-2 kb region upstream of the TSS cannot be used as a criterion to disqualify target genes. Other aspects that need to be taken into account include correspondence in gene function with better established PPAR targets and the timing of gene induction following PPARα activation.
2. PPARα Tissue Expression Profile in Mouse and Human
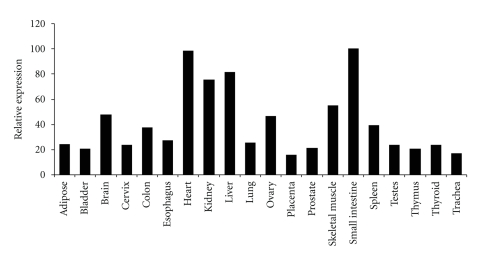
High expression levels of PPARα expression are found in liver and specifically in the parenchymal cell population. Expression of PPARα in nonparenchymal liver cells such as Kupffer cells is much lower [15, 16]. Other tissues with high PPARα mRNA levels are heart, kidney, intestine, and brown adipose tissue, all of which are characterized by an elevated rate of fatty acid catabolism [17]. PPARα expression has also been detected in immune cells such as the peripheral blood mononuclear cell population, and specifically in T-cells and macrophages [18–22]. Evidence suggests that mice and humans share similar PPARα tissue expression profiles [6, 17] (Figure 1). In the past, the importance of PPARα in human liver was questioned based on data showing approximately 10-fold lower PPARα mRNA levels in human liver compared with mouse liver [23]. A recent study using more advanced methodology revealed similar PPARα expression in mouse and human liver and in mouse and human hepatocytes, thus strongly arguing against this notion [24]. Given that PPARα has been most extensively studied in liver, most of the information on PPARα target genes presented here relates to hepatic gene regulation.
Figure 1.
Expression profile of P P A R α in human tissues. The FirstChoice Human Total RNA Survey Panel (Ambion) was reverse transcribed and used for qPCR using primers specific for human PPARα. Expression levels are expressed relative to small intestine, which showed the highest expression level (100%).
3. PPARα Structure in Mouse and Human
Analogous to other nuclear receptor superfamily members, PPARα has a domain structure consisting of an N-terminal activating function-1 (AF-1) domain, a central DNA-binding domain (DBD), and a C-terminal ligand-binding domain (LBD) [25, 26]. The N-terminal domain can be phosphorylated leading to changes in transcriptional activity and even ligand binding of the receptor [27]. The DBD is responsible for physical interaction with DNA and allows PPARα to bind to specific PPREs as a heterodimer with RXR [28]. The LBD harbors the ligand-binding pocket crucial for dimerization with RXR and contains the activating function-2 involved in physical interactions with coregulatory proteins [7, 29, 30]. Comparison of human and murine PPARα shows 85% identity at the nucleotide level and 91% identity at the amino acid level. Data have indicated that there is some genetic heterogeneity in the functional coding sequence of human PPARα that translates into functional differences in receptor activity. One identified variant of the human PPARα gene produces a protein that is mutated within the PPARα DNA-binding domain. This L162V gene variant exhibits greater ligand-induced activity compared to the wild-type receptor [31, 32]. While there is some evidence for a link between the L162V polymorphism and metabolic parameters such as plasma lipid levels, these correlations are not always found [32–37]. Interestingly, the effect of L162V polymorphism has been suggested to be modulated via gene-drug and gene-nutrient interactions [38–40]. The V227A polymorphism was found in Japanese population and has been associated with altered serum lipid levels and nonalcoholic fatty liver disease [41–44]. In addition to polymorphic variants, a truncated splice variant of human PPARα has been described that negatively interferes with wild-type PPARα activity [45].
4. PPARα Ligands
PPARα serves as receptor for a structurally diverse set of compounds. The most important class of synthetic PPARα ligands is the fibrates, including gemfibrozil, bezafibrate, clofibrate, fenofibrate, and Wy14643 [2, 9–11, 46]. This class of drugs is used in the treatment of dyslipidemia primarily associated with type 2 diabetes mellitus. In addition, PPARα is activated by plasticizers, insecticides, and other rodent hepatic carcinogens. Natural ligands of PPARα include a variety of fatty acids as well as numerous fatty acid derivatives and compounds showing structural resemblance to fatty acids, including acyl-CoAs, oxidized fatty acids, eicosanoids, endocannabinoids, and phytanic acid [47–53]. Endogenous ligand activation of PPARα in liver was initially suggested to occur primarily during fasting as large amounts of free fatty acids are released into the bloodstream and enter the liver [54, 55]. However, compelling evidence indicates that hepatic PPARα is not activated by plasma free fatty acids, whereas it can be activated by dietary fatty acids and fatty acids generated via de novo lipogenesis [56–60]. Recently, it was shown that the effects of dietary unsaturated fatty acids on hepatic gene expression are almost exclusively mediated by PPARα and mimic the effect of synthetic PPARα agonists [61].
5. PPARα and Hepatic Lipid Metabolism
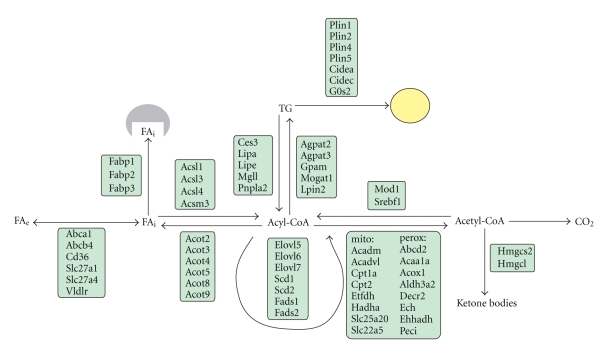
Regulation of lipid metabolism is mainly coordinated by liver, which actively metabolizes fatty acids as fuel and continuously produces very low-density lipoproteins (VLDLs) particles to provide a constant supply of fatty acids to peripheral tissues. Disturbances in these pathways are the basis for hepatic steatosis and alterations in plasma lipoprotein levels. Many aspects of hepatic lipid metabolism are under control of PPARα, including fatty acid uptake through membranes, fatty acid activation, intracellular fatty acid trafficking, fatty acid oxidation and ketogenesis, and triglyceride storage and lipolysis (Figure 2). It has been suggested that part of the effect of PPARα on hepatic ketogenesis may be mediated by induction of the PPARα target fibroblast growth factor 21 [90–92]. A detailed discussion of the specific genes within the various lipid metabolic pathways that are targeted by PPARα is provided below (Table 1).
Figure 2.
Schematic representation of PPARα target genes in different aspects of hepatic lipid metabolism.
Table 1.
List of P P A R α target genes in different biological processes in liver. Genes regulated by P P A R α in mouse are shown in lower case. Genes regulated in human and mouse are shown in CAPITAL BOLD. Genes regulated only in human are shown in CAPITAL, and genes with detected functional PPRE are shown in italic font.
| Lipid metabolism | Lipid/hormone transport | Adipor2 [24, 62], Cd36 [24, 62–64], LEPR [24, 62], Slc27a1 [62, 64–67], SLC27A2 [24, 62, 63, 68], SLC27A4 [24, 62] |
| Acyl-CoA formation/hydrolysis/ binding | Acot1 [24, 62, 69], Acot7 [62], ACOT12 [24, 62], ACSL1 [24, 62, 70–72], ACSL3 [24, 62], Acsl4 [24, 62, 63, 73], ACSL5 [24, 62, 63], ACSM3 [24, 62], Acss2 [62], FABP1 [24, 62, 74, 75], Fabp2 [24, 62, 63], FABP3 [24, 62], Fabp4 [62, 68], Fabp5 [62] | |
| Mitochondrial β-oxidation/oxidative phosphorylation | ACAA2 [24, 62, 68], Acadl [24, 62, 70, 74, 76], ACADM [24, 55, 62, 70], ACADS [24, 54, 62, 70], ACADVL [24, 62, 63, 70], Acad8 [62], Acad9[62], Acad10 [62], Acot2 [24, 62, 69], Acot9 [62], Acot10 [62], CPT1A [24, 54, 55, 62, 77–79], Cpt1b [24, 62], CPT2 [24, 62, 70, 80], Crat [24, 62, 74], Dci [24, 62, 65, 68, 76], Decr1 [24, 62, 76, 81], ETFA [24], Etfb [24, 62], ETFDH [24, 62], HADHA [24, 62, 63, 68, 82], HADHB [24, 62, 63, 68], Hadh [24, 62, 68, 74, 76, 83], Hadh2 [62], Hibch [24, 62], SLC25A20 [24, 62, 84], SLC22A5 [24, 62, 85, 86], TXNIP [24, 62], Ucp2 [24, 62, 87–89], Ucp3 [62] | |
| Ketogenesis/ketolysis | Acat1[24, 62, 68], Bdh [62], FGF21 [24, 62, 90–92], Hmgcl [62], HMGCS2[24, 62, 93–95] | |
| Peroxisomal β-oxidation | ABCD2 [24, 62, 96], ABCD3 [24, 62, 96], ACAA1A [24, 62, 68, 76], Acaa1b [24, 62], Acot3 [24, 62], Acot4 [24, 62], Acot5 [24, 62], Acot8 [62, 97], ACOX1 [24, 55, 62, 68, 70, 98, 99], Crot [24, 62, 74], Decr2 [24, 62, 68, 100] ECH1 [24, 62, 63, 68], Ehhadh [24, 62, 101, 102], HACL1 [24], HSD17B4 [24, 62, 103, 104], Peci [24, 62, 65, 100], Pex11a [24] | |
| Microsomal (ω-hydroxylation) | ALDH3A1[24], Aldh3a2 [24, 62, 105], ALDH9A1 [24], Cyp4a1 [74, 106–109], Cyp4a3 [55, 74, 108], Cyp4a10 [24, 62, 68, 84, 106, 110], Cyp4a12a [24, 62, 65], Cyp4a14 [24, 62, 68, 110, 111], Cyp4f15 [24], Cyp4x1 [24] | |
| Lipogenesis | Acaca [62], ACACB [62], AGPAT2 [62], Agpat3 [24, 62], Agpat5 [62], Agpat6 [62], Dgat1 [62, 112], ELOVL5 [62, 113, 114], ELOVL6 [24, 62, 113, 114], Elovl7 [62], FADS1 [24, 62, 113], Fads2 [62, 113, 115], Fasn [62, 112], GPAM [24, 62, 65], Hsd17b12 [62], Lpin2 [24, 56, 62], MLYCD [24, 62], Mogat1 [24, 62], MOD1 [24, 62, 70, 116], Scd1 [24, 62, 117, 118], Scd2 [62, 68], Slc25a10 [62, 100], Srebf1 [24, 62, 119] | |
| Lipases/lipid droplet proteins | ADFP[24, 62, 120], Ces1 [62], Ces3 [24, 62], Cidea [24, 62, 121], CIDEC [24, 62, 121], Gos2 [24, 62, 122, 123], Lipa [24, 62], Lipe [24, 62, 68], Mgll [24, 62, 63, 68], Oxpat/Lsdp5 [24, 62, 124, 125], Plin1 [24], PNPLA2 [24, 62], S3-12 [24, 62] | |
| Lipoprotein uptake and metabolism | ANGPTL4[24, 62, 126, 127], APOA1 [128–133], APOA2 [24, 134], APOA5 [24, 135, 136], APOCIII [137–139], LIPC [24, 62], Lipg [62], Lpl [62, 64, 65, 140], Lrp4 [24, 62], PCTP [24, 62], Pltp [62, 65, 141, 142], Mttp [24, 62, 143], VLDLR [24, 62] | |
| Cholesterol/Bile transport and metabolism | ABCA1 [24, 62, 144], ABCB4 [24, 62, 94, 144], Abcb11 [62], Abcg5 [62, 144], Abcg8 [62, 144], Cav1 [24], CYP7A1 [24, 62, 145–147], Cyp8b1 [62, 148], Cyp27a1 [145], FXR [62], LXR [144, 149], Npc1 [62], Rab9 [24, 62], Scarb2 [62], Slc10a1 [94], Slc10a2 [62, 150] | |
|
| ||
| Other pathways | Glucose/Glycerol transport and metabolism | AQP3 [24, 62, 151], Aqp7 [62], Aqp9 [62, 151], Fbp2 [24, 62], G6PC [24], Gpd1 [24, 62, 151], Gpd2 [62, 151], GYK [24, 62, 151], Gys-2 [152], Ldha [62], Pcx [62], PCK1 [24], Pdk1 [24], PDK4 [24, 62, 153, 154] |
| Biotransformation | AKR1B10 [24], AKR1C3 [24], CYP1A2 [24], Cyp2a5 [110], CYP2B6 [24], CYP2C8 [24], CYP2C9 [24], Cyp2c11 [155], Cyp2c12 [155], Cyp2c29 [110], CYP2J2 [24], CYP3A5 [24], CYP3A7 [24], CYP3A11 [24, 110], Cyp3a43 [24], EPHX2 [24, 156], Gsta3 [157], MGST3 [24], UGT1A9 [158] | |
| Amino Acid metabolism | ABAT [24, 159], Acmsd [159], AGXT2 [24, 160], Arg1 [160], ASL [24, 160], Ass1 [160], CBS [24, 159], CPS-1 [24, 160], Cth [159], Got1 [160], Got2 [62, 64, 160, 161], Gls [160], GLS2 [159], GPT [24, 159], Hal [159], Hpd [159], OAT [24, 159], ODC1 [24, 62, 159], OTC [24, 62, 160], PAH [24, 159], PSAT1 [24, 62, 159], Tat [159, 162] | |
| Inflammation | Apcs [163], Birc3 [163], Cebpb [164], Cd68 [24, 165], Crp [24, 163], Cxcl10/IP10 [165], FGB [24, 155, 166, 167], Emr1 (F4/80) [156], Icam-1 [24, 165], Ifi47 [24, 163], Igtp [163], Nfkbia [168–170], Il-1β [165], Il-1r1 [111], Il1rn [163], Il1rap [163], Il-6 [111, 171], Il-6ra [111, 163], Il18 [163], Lcn2 [163, 165], Lifr [163], Ccl2 [165], Ccl3 [165], Mt1 [24, 163], Mt2 [163, 165], Orm2 [163], Orm3 [24, 163], Nfkb1 [24, 164], Pla1a [24, 163], Saa2 [163], Saa4 [24, 163], Stat1 [165], Stat2 [163], Stat3 [163], Steap4 [24, 163], Stress induced protein [163], Tnfα [165], Traf2 [163], Vcam-1 [24, 165] | |
5.1. Peroxisomal Fatty Acid β-Oxidation
The first link between PPARα and fatty acid catabolism was established by the identification of the Acyl-CoA oxidase gene, encoding the rate-limiting enzyme in peroxisomal long-chain fatty acid oxidation, as a direct PPARα target gene [98, 172]. Peroxisomes are known to be involved in many aspects of lipid metabolism, including synthesis of bile acids and plasmalogens, synthesis of cholesterol and isoprenoids, alpha-oxidation, glyoxylate and H2O2 metabolism, and beta-oxidation of very-long-straight-chain or branched-chain acyl-CoAs. The beta-oxidation of straight-chain acyl-CoAs starts with a reaction catalyzed by acyl-CoA oxidase 1 (Acox1) followed by one of two enzymes carrying both enoyl-CoA-hydratase and 3-hydroxyacyl-CoA dehydrogenase activity (L-bifunctional enzyme, Ehhadh; D-bifunctional enzyme, Hsd17b4) and finally peroxisomal 3-ketoacyl-CoA thiolase (Acaa1a, Acaa1b). All genes mentioned above represent PPARα targets [24, 55, 62, 68, 70, 76, 98, 99, 101–104]. Additionally, genes involved in peroxisomal fatty acid uptake (Abcd2 and Abcd3), conversion of fatty acid to acyl-CoA (Crot), and numerous thioesterases (Acots) that convert acyl-CoAs back to fatty acids have been reported to be regulated by PPARα [24, 62, 74, 96, 97]. Activation of PPARα using synthetic agonists is known to cause massive proliferation of peroxisomes in rodents via induction of a large set of genes encoding peroxisomal fatty acid oxidation enzymes, as well as genes involved in peroxisomal biogenesis (Pex genes). Chronic exposure to these so-called peroxisome proliferators can also induce liver cancer in rodents [173]. In contrast, activation of PPARα in humans does not seem to induce hepatocellular carcinomas, suggesting a species specific response to PPARα activation. Initially, it was believed that the differential response was due to the lack activation of Acox1 and other peroxisomal genes by PPARα in humans [71, 174, 175]. However, recent data indicate that PPARα is able to induce a significant number of genes involved in peroxisomal fatty acid oxidation in human primary hepatocytes, including Acox1 [24]. Also, PPARα-mediated induction of the Pex11a gene involved in peroxisome proliferation is observed in both species [24].
5.2. Mitochondrial Fatty Acid β-Oxidation
The crucial role of PPARα in mitochondrial fatty acid oxidation is illustrated by the phenotype of fasted PPARα −/− mice, which exhibit hypoketonemia, hepatic steatosis, and elevated plasma free fatty acid levels [54, 55, 176]. It is now evident that virtually every enzymatic step within the fatty acid oxidative pathway is under control of PPARα. Specifically, PPARα induces genes controlling fatty acid import into the mitochondria (Cpt1, Cpt2, Slc25a20, Slc22a5), as well as the major enzymes within the β-oxidation pathway, including various acyl-CoA dehydrogenases (Acad, step 1), mitochondrial trifunctional enzyme (Hadh, step 2–4), and genes involved in β-oxidation of unsaturated fatty acid (Dci, Decr) [24, 54, 55, 62, 63, 65, 68, 70, 74, 76–86].
Additionally, synthesis of ketone bodies via mitochondrial HMG-CoA synthase (Hmgcs2) and HMG-CoA lyase (Hmgcl) is governed by PPARα [24, 62, 93–95], as is the expression of genes encoding electron transferring flavoprotein and the corresponding dehydrogenase (Etfa, Etfb, Etfdh) [24, 62]. The latter proteins mediate the transfer of electrons from Acyl-CoA dehydrogenases to the membrane-bound electron transfer flavoprotein ubiquinone oxidoreductase, allowing further entry into the oxidative phosphorylation pathway [177, 178]. Finally, PPARα induces uncoupling proteins Ucp2 and Ucp3, which have been proposed to function as an outward transporter of nonesterified fatty acid anions from the mitochondrial matrix [24, 62, 87–89].
5.3. Microsomal Fatty Acid ω-Hydroxylation
Cyp4A enzymes are members of the cytochrome P450 monoxygenase superfamily and catalyze microsomal ω-hydroxylation of fatty acids [106, 179]. Studies using PPARα −/− mice have shown that hepatic expression of Cyp4a genes is almost completely dependent on PPARα (Cyp4a10, Cyp4a12, Cyp4a14 in mice, Cyp4a1, Cyp4a3 in rat, Cyp4a11 in human) [55, 62, 74, 84, 106–111]. Furthermore, expression is extremely sensitive to PPARα ligand activation, indicating that Cyp4a genes may serve as PPARα marker genes. Although previous studies performed in human primary hepatocytes could not show regulation of Cyp4a by human PPARα, our microarray data revealed significant induction of Cyp4a11 by Wy14643 in primary human hepatocytes [24, 68, 180, 181]. ω-hydroxylation of saturated and unsaturated fatty acids may lead to the generation of high-affinity PPARα ligands, including hydroxyeicosatetraenoic acids (HETEs), thus creating a positive feedback loop [182]. Alternatively, induction of ω-oxidation by PPARα has been suggested to promote the degradation of the PPARα agonist leukotriene B4 as part of a feedback mechanism aimed at controlling the duration of the inflammatory response [53].
5.4. Hepatic Lipogenesis
Whereas PPARα is mostly known for its ability to induce fatty acid oxidation, growing evidence points to a role of PPARα in regulation of lipogenesis. A functional PPRE was identified in the promoter of a limited number of lipogenic genes including Δ6 desaturase (Fads2), malic enzyme (Mod1), phosphatidate phosphatase (Lpin2), and Δ9 desaturase (Scd1) [56, 115–117]. Gene expression profiling showed that chronic in vivo treatment of mice with PPARα agonist causes the upregulation of a large set of lipid biosynthetic genes [62]. However, regulation is much less pronounced in primary hepatocytes, suggesting an indirect mechanism. Consistent with this notion, induction of lipogenic genes by chronic PPARα activation was completely abolished in SREBP1−/− mice [183]. The effect of PPARα agonists on SREBP targets has been attributed to increased activation of SREBP1c via enhanced proteolytic cleavage [112]. Such a mechanism may also lead to increased SREBP1 mRNA via an autoloop regulatory circuit [184]. Alternatively, it is possible that PPARα is recruited to promoters of SREBP targets and stimulates SREBP activity [12]. Interestingly, in rat FAO hepatoma cells, it was found that PPARα activation reduced expression of lipogenic genes, including Fasn, Gpam, and SREBP1c, while Insig1 expression was increased by PPARα [185]. The reason for the discrepancy is not clear.
In contrast to de novo fatty acid and cholesterol synthesis, synthesis of triglycerides may be directly targeted by PPARα. Several genes within these pathways are upregulated by PPARα activation, including Gpam, various Agpat genes, Mogat1, Dgat1, and Lpin2 [24, 62, 65, 112]. Induction of genes involved in triglyceride synthesis from fatty acids may reflect a broader role of PPARα in the hepatic response to fasting aimed at neutralizing large amounts of incoming adipose tissue-derived free fatty acids.
5.5. Fatty Acid Uptake and Binding
Before they can be metabolized in the liver, fatty acids have to be transferred across the cell membrane. Several proteins are involved in fatty acid transport across the plasma membrane, a number of which carry both fatty acid transporter and acyl-CoA synthetase activity. Studies have shown that the fatty acid transport proteins Slc27a1, Slc27a2, and Slc27a4 are upregulated by PPARα in liver [24, 62, 64–68, 101].
Slc27a1 is not expressed and not regulated by PPARα in isolated primary hepatocytes, suggesting regulation occurs in liver macrophages (Kupffer cells). So far, the only fatty acid transporter for which a PPAR response element has been identified is Slc27a1. PPARα agonists also markedly induce hepatic expression of the fatty acid transporter/scavenger receptor Cd36, which is expressed in various liver cell types [24, 62–64]. Additionally, expression of numerous acyl-CoA synthetases is induced by PPARα [24, 62, 63, 70–73]. Currently, limited information is available about the cellular localization and the structure/function relationship of acyl-CoA synthetase enzyme [186].
The Fabp gene family comprise a group of high-affinity intracellular fatty acid-binding proteins. Interestingly, Fabp1 was one of the first PPARα target genes identified [74, 75, 187, 188]. Recent studies indicate that Fabp1 may be involved in partitioning of FA to specific lipid metabolic pathways [189]. Other Fabp genes induced by PPARα activation in mouse liver include Fabp2, Fabp3, Fabp4, and Fabp5 [24, 62, 63]. Induction of Fabp4 (A-FABP, aP2) upon PPARα activation likely occurs via its expression in Kupffer cells. Fabp4 expression in hepatocytes is correlated with acquisition of a steatotic phenotype concurrent with upregulation of PPARγ mRNA [190].
5.6. Lipases and Lipid Droplet Proteins
PPARα −/− mice exhibit elevated hepatic TG accumulation, especially under fasting conditions [54, 191, 192]. Conversely, treatment with PPARα agonists lowers hepatic triglyceride levels in models of hepatic steatosis and can prevent the fasting-induced increase in liver TG [193, 194]. The antisteatotic effect of PPARα has mainly been attributed to stimulation of fatty acid oxidation, which would decrease the availability of fatty acids for TG storage.
Recently, hepatic lipid droplets were shown to be targeted by autophagy, which ultimately leads to TG hydrolysis via lysosomal acid hydrolase (Lipa). Which other lipases importantly contribute to intracellular lipolysis of hepatic TG stores remains unclear, but lipases active in adipocytes likely play a role, including Ces3, Lipe, Pnpla2, Mgll, and perhaps Pnpla3 [195–200]. With the exception of Pnpla3, all of the above genes are induced by short-term treatment with PPARα agonist in mouse hepatocytes. Regulation of Pnpla2 was also observed in human hepatocytes. Pnpla2 and Lipe were previously classified as direct target genes of PPARγ in adipose tissue, suggesting that they are direct target of PPARα as well [201, 202]. Thus, apart from induction of fatty acid oxidation, PPARα activation may also decrease hepatic TG storage by stimulating the TG hydrolysis pathway.
Lipid droplets are coated with one or more members of the perilipin family of proteins: perilipin (Plin1), Adrp/adipophilin (Plin2), Tip47 (Plin3), S3-12 (Plin4), and Oxpat/Lsdp5 (Plin5). Adrp and Lsdp5 have been identified as target genes of PPARα in liver [120, 124]. A recent study suggests that Adrp could serve as potential mediator of the effect of PPARα on VLDL production. Adrp induction by PPARα may diminish VLDL production by favoring fatty acids storage in cytosolic lipid droplets rather than directing through VLDL assembly [203]. Besides Adrp, expression of S3-12 and perilipin, which are known as PPARγ target genes in adipose tissue, is induced by PPARα agonist in human hepatocytes [24, 204]. Perilipin expression in human liver is correlated with development of steatotic liver [205].
Two recently identified lipid droplet-associated proteins that are not part of the perilipin family are Cidec (FSp27) and Cidea [206, 207]. Both proteins promote TG accumulation and are targets of PPARγ in adipocytes [208, 209]. In addition, they are regulated by PPARα in mouse liver, although the kinetics of induction of the two genes seems to be quite different [121]. Cidec but not Cidea upregulation by PPARα agonist could be confirmed in human primary hepatocytes [24].
Interestingly, the G(0)/G(1) switch gene 2 (G0s2) was recently identified as an inhibitor of Pnpla2 activity and located to lipid droplets in adipocytes stimulated with β-adrenergic receptor agonist [122]. Previously, G0s2 was shown to be a direct PPARα target gene in mouse liver and PPARγ target in adipocytes [123]. Whether G0s2 associates with lipid droplets in hepatocytes remains to be further investigated. Similar to the induction of triglyceride synthesis, regulation of numerous lipid droplet proteins by PPARα reflects a broader role of PPARα in the hepatic response to fasting aimed at deflecting large amounts of incoming adipose tissue-derived free fatty acids towards storage in lipid droplets.
6. PPARα and Lipoprotein Metabolism
Clinical studies in humans have provided ample evidence that fibrate drugs effectively lower fasting plasma triglycerides (TG) and raise plasma HDL [210–213]. At the molecular level, fibrates act as synthetic agonist for PPARα, indicating an important role of PPARα in the control of lipoprotein metabolism. PPARα lowers plasma TG in part by reducing very low-density lipoprotein (VLDL) production [194]. Traditionally, this effect of PPARα was ascribed to induction of genes involved in fatty acid oxidation and the concomitant reduction in lipid availability for VLDL production. However, this paper has made it evident that in addition to its role in fatty acid catabolism, PPARα influences multiple aspects of intracellular lipid trafficking and metabolism, some of which may oppose hepatic TG lowering. Furthermore, expression of Mttp, which is involved in the lipidation of apoB100 to form a nascent VLDL particle, is positively regulated by PPARα [214]. Thus, the precise target genes underlying the suppressive effect of PPARα agonist on hepatic VLDL production remain to be fully elucidated.
In addition to suppressing VLDL production, PPARα agonists are known to stimulate clearance of TG-rich lipoproteins [194]. Clearance of TG-rich lipoproteins VLDL and chylomicrons is mediated by the enzyme lipoprotein lipase (LPL), which is attached to the capillary endothelium of muscle and adipose tissue. Expression of Lpl in liver is restricted to Kupffer cells and upregulated by PPARα agonists [140, 215]. In contrast, no evidence is available indicating a stimulatory effect of PPARα on Lpl expression in heart and skeletal muscle, which account for the major share of plasma TG clearance [140, 216]. LPL activity is mostly regulated posttranslationally via altered secretion from liver of LPL-modulating factors, including apolipoprotein C-III (Apoc3), apolipoprotein A-V (Apoa5), Angiopoietin-like protein 3 (Angptl3), and Angiopoietin-like protein 4 (Angptl4). Firstly, PPARα agonists downregulate the expression of LPL inhibitor APOC3, supposedly via mechanisms involving the transcription factors REV-ERBα, HNF4α, or FOXO1 [137, 217–219]. Secondly, PPARα agonists increase hepatic expression and plasma levels of APOA5, which is a positive regulator of LPL [220]. A functional PPAR responsive element has been identified in the promoter of the human APOA5 gene, classifying APOA5 as a direct PPARα target gene [135, 136]. Thirdly, PPARα upregulates hepatic expression and plasma levels of Angptl4, which acts as inhibitor of LPL activity by converting active LPL dimers to inactive monomers [126]. The DNA response element conferring PPAR regulation was located to intron 3 of the Angptl4 gene [127]. Finally, PPARα stimulates hepatic expression of the VLDL receptor (Vldlr) [24, 62]. The functional significance of Vldlr regulation in liver is unclear, as Vldlr is most highly expressed in adipose tissue, heart, and skeletal muscle, where it plays an auxiliary role in plasma TG hydrolysis by LPL. Recently, Vldlr was shown to be under control of PPARγ in adipocytes [221]. Thus, it appears that both pro- and antilipolytic pathways are activated by PPARα. Under conditions of pharmacological PPARα activation, the prolipolytic actions of PPARα dominate, as illustrated by the stimulation of plasma TG clearance.
PPARα agonists raise plasma HDL levels in humans, which is most likely achieved via species specific mRNA induction of apolipoprotein A-I (Apoa1) and A-II (Apoa2) [128, 175, 222–224]. Apoa1 gene expression is not induced by PPARα in rodents due to the presence of disabling mutations within the PPAR-response element [129]. In fact, PPARα activation in mouse downregulates Apoa1 mRNA expression and plasma concentrations through an indirect pathway involving the PPARα-dependent induction of the nuclear receptor REV-ERBα, a negative regulator of transcription [129, 130, 225].
The impact of PPARα in HDL metabolism likely extends beyond regulation of apolipoproteins. Evidence suggests that both PPARα and PPARβ/δ stimulate expression of endothelial lipase (Lipg) in liver [62, 226]. Endothelial lipase mainly carries phospholipase activity and its overexpression was shown to significantly reduce plasma HDL cholesterol levels [227–229]. Since Lipg is expressed in endothelial cells, macrophages, and hepatocytes, regulation of hepatic Lipg by PPARα and PPARβ/δ may be mediated by different cell types. In as much as PPARα agonists raise plasma HDL levels, the physiological relevance of Lipg induction by PPARα remains to be established.
In our recent publication, the PPARα agonist Wy14643 modestly induced hepatic lipase (Lipc) expression in primary human hepatocytes [24]. Hepatic lipase exhibits both phospholipase and triglyceride hydrolase activity and hydrolyzes triglycerides and phospholipids of chylomicron remnants, IDL, and HDL [230]. Whether Lipc represents a direct target gene of PPARα in human remains unclear. Other genes involved in lipoprotein metabolism that are regulated by PPARα include phosphatidylcholine transfer protein (Pctp). Induction of Pctp mRNA by PPARα is conserved in primary human hepatocytes [24]. Pctp encodes a steroidogenic acute regulatory-related transfer domain protein that binds with high affinity to phosphatidylcholines. In a recent publication, a role for Pctp in the metabolic response to PPARα was proposed [231]. Overall, it is evident that PPARα governs multiple aspects of plasma lipoprotein metabolism.
7. PPARα and Glucose/Glycerol Metabolism
Although PPARα has mostly been linked to fatty acid metabolism, studies in mice have yielded considerable evidence for a role of PPARα in hepatic glucose metabolism. Indeed, fasted PPARα −/− mice display severe hypoglycemia [54, 55, 176]. Several mechanisms may account for the hypoglycemia, including decreased hepatic glucose production and increased peripheral glucose utilization. Genes involved in gluconeogenesis that have been identified as PPARα targets include phosphoenolpyruvate carboxykinase (Pck1), pyruvate carboxylase (Pcx), and lactate dehydrogenase A [62]. Interestingly, regulation of Pck1 by PPARα was only observed in human hepatocytes [24]. Pyruvate carboxylase was identified as direct target of PPARγ in adipocytes [232].
PPARα was shown to have a specific role in the metabolic conversion of glycerol in liver by directly upregulating expression of genes such as Gpd1, Gpd2, Gyk, Aqp3, and Aqp9 [151]. Besides governing glucose production, PPARα may also alter glucose utilization in numerous tissues via induction of pyruvate dehydrogenase kinase isoform 4 (Pdk4) [153, 154, 233–236]. Pdk4 phosphorylates and inactivates pyruvate dehydrogenase, thereby limiting carbon flux through glycolysis. Synthesis of glycogen is also affected in PPARα −/− mice, which may be mediated in part via defective regulation of Gys2 [152]. It is noteworthy that in contrast to studies in mice, human trials generally do not support an effect of PPARα activation on plasma glucose levels. Consistent with these data, it was found that upregulation of genes involved in the glycolysis/gluconeogenesis pathway by Wy14643 was uniquely observed in mouse hepatocytes and not human hepatocytes [24].
8. PPARα and Hepatic Cholesterol/Bile Metabolism
It has been demonstrated that PPARα activation increases efflux of cholesterol to HDL. Formation of nascent HDL is mediated by Abca1-dependent lipidation of newly-secreted Apoa1. Expression of Abca1 is upregulated by PPARα agonists in both human and mouse hepatocytes, as well as in mouse intestine [24, 237]. Presently, it is not clear if this effect of PPARα activation is mediated via LXRα, as was shown previously in macrophages [21]. Other genes involved in cholesterol uptake and transport that were shown to be under control of PPARα include Abcg5, Abcg8, Cav1, Npc1, and Rab9 [24, 62, 144].
While PPARα is known to govern specific genes involved in bile acid synthesis, the overall impact on bile acid homeostasis remains somewhat ambiguous. Expression of Cyp7a1, which represents the rate-limiting enzyme in bile acid synthesis, is markedly downregulated in PPARα −/− mice in fasting condition [62]. Paradoxically, synthetic PPARα agonists reduce Cyp7a1 expression in both mice and human [145, 238–240]. In agreement with the latter observation, fibrate treatment leads to decreased bile acid synthesis. To what extent the changes in Cyp7a1 expression reflect direct regulation by PPARα is unclear as PPARα also influences the expression of other nuclear hormone receptors involved in regulation of Cyp7a1 such as FXR and LXR. It has also been suggested that PPARα can antagonize LXR signaling and LXR-dependent activation of Cyp7a1 gene promoter [241–243].
Other genes involved in bile acid synthesis that are regulated by PPARα include Cyp27a1 which is downregulated by PPARα agonists in a PPARα-dependent manner [145], and Cyp8b1 which is upregulated by PPARα [62, 148]. Recently, CYP7b1 expression was shown to be suppressed by PPARα in a sex-specific manner, which was shown to occur via sumoylation of the LBD of PPARα [244]. Finally, PPARα stimulates expression of the hepatobiliary phospholipid transporter Abcb4 [24, 62, 94, 144].
9. PPARα and Amino Acid Metabolism
Accumulating evidence supports a role for PPARα in regulation of amino acid and urea metabolism [159, 160, 162, 245]. Studies in mice have shown that PPARα governs metabolism of amino acids by suppressing expression of genes involved in transamination (Aspartate amino transferase (Got1), Alanine amino transferase (Gpt), Alanine glyoxylate aminotransferase (Agtx2), and deamination (Glutaminase (Gls)), as well as numerous genes that are part of the urea cycle (Cps1, Otc, Ass1, and Asl) [160, 161, 245]. In agreement with these data, PPARα −/− mice exhibit increased plasma urea levels [160]. Several of the above genes were also downregulated by PPARα agonist in primary human hepatocytes, suggesting that regulation of nitrogen metabolism by PPARα is at least partially conserved between mice and human [24].
At the present time, the mechanism behind downregulation of nitrogen metabolism by PPARα remains elusive. It has been proposed that PPARα may modulate the activity of other transcription factors that are directly involved in amino acid homeostasis, including HNF4α and C/EBPα [160]. However, concrete evidence supporting such a mechanism is lacking.
Whereas PPARα activation decreases hepatic aminotransferase expression in mice, PPARα agonists were shown to increase expression of Gpt in human hepatocytes and HepG2 cells, which occurred via direct regulation of the gene promoter [161, 246]. The observed increase in plasma alanine amino transferase activity in patients treated with fibrates may thus be related to direct regulation of Gpt transcription, rather than drug-induced liver injury.
10. PPARα and Inflammation
Besides regulating numerous metabolic pathways, PPARα also governs inflammatory processes, which is mainly achieved by downregulating gene expression via a mechanism generally referred to as transrepression. The first clue towards anti-inflammatory effects of PPARα came from the observation that PPARα −/− mice exhibit a prolonged inflammatory response in the ear swelling test [53]. The anti-inflammatory effects of PPARα are likely explained by interference of PPARα with the activity of many proinflammatory transcription factors including signal transducer and activator of transcription (Stat), Activator protein-1 (AP-1), and NF-κB [247]. Specifically, it has been shown that activated PPARα binds to c-Jun and to the p65 subunit of NF-κB, thereby inhibiting AP-1- and NF-κB- mediated signaling [248]. Additionally, PPARα induces the inhibitory protein IκBα, which normally retains NF-κB in a nonactive form, leading to suppression of NF-κB DNA-binding activity [168]. Suppression of fibrinogen gene expression by PPARα activation is likely mediated by interference with the transcription factor CAATT/enhancer-binding protein (C/EBP) via sequestration of the coactivator glucocorticoid receptor-interacting protein 1/transcriptional intermediary factor 2 (GRIP1/TIF2) [166]. Finally, recent data indicate that activated PPARα may downregulate gene expression by causing the loss of STAT1 and STAT3 binding to DNA [12].
Specific genes downregulated by PPARα include a number of acute phase genes such as fibrinogen, serum amyloid P-component, lipocalin 2, metallothioneins, and serum amyloid A2, which were shown to be suppressed by the PPARα agonist Wy14643 in wild-type mice but not PPARα −/− mice [163]. Similarly, in humans fenofibrate treatment has been shown to decrease plasma levels of several acute phase proteins including C-reactive protein, fibrinogen-α and -β and interleukin 6 [166, 249, 250]. With the exception of the sIl-1 receptor antagonist and Vanin-1, to our knowledge no inflammatory genes have been identified as direct positive targets of PPARα [163].
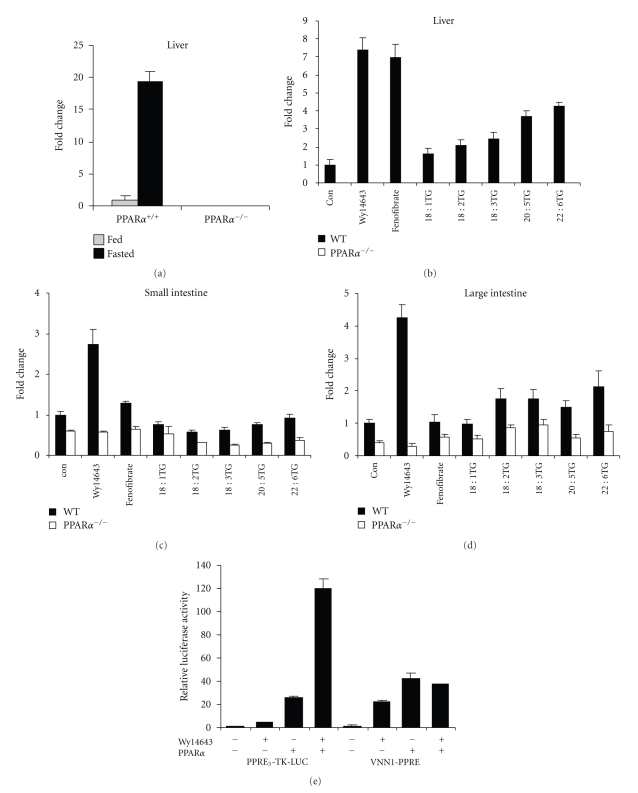
The Vanin-1 (Vnn1) gene encodes a glycosylphosphati-dylinositol-linked membrane-associated pantetheinase that generates cysteamine from pantothenic acid. Studies suggest that Vanin1 may promote inflammation. Mice lacking Vnn1 showed decreased NSAID- or Schistosoma-induced intestinal inflammation, which was associated with higher glutathione levels [251]. Other evidence indicates that Vanin-1 stimulates production of inflammatory mediators by intestinal epithelial cells and thereby controls the innate immune response, possibly by antagonizing PPARγ activity [252]. Epithelial Vanin-1 was also found to regulate inflammation-driven cancer development in a colitis-associated colon cancer model [253]. Evidence presented in Figure 3 demonstrates that Vnn1 likely represents a direct target gene of PPARα. Expression of Vnn1 in mouse liver was markedly increased by fasting in wildtype but not PPARα −/− mice (Figure 3(a)). Negligible Vnn1 expression was detected in PPARα −/− mouse liver. Moreover, hepatic Vnn1 expression was significantly induced by 6 h treatment with dietary fatty acids and by the synthetic PPARα agonists Wy14643 and fenofibrate (Figure 3(b)). Additional data lend strong support to the importance of PPARα in Vnn1 gene regulation in small and large intestine (Figures 3(c) and 3(d)), although the results are not quite as striking as in liver. Finally, it was shown that two adjacent and partially overlapping PPREs located around 4 kb downstream of the transcription start site of the mouse Vnn1 gene were functional in a luciferase reporter assay in HepG2 cells (Figure 3(e)). PPARα transfection and Wy14643 markedly increased luciferase activity, although for reasons that remain unclear, no synergism between the two treatments was observed. Overall, these data suggest that Vnn1 represents a direct PPARα target gene.
Figure 3.
Vanin-1 likely represents a direct P P A R α target gene. (a) Vnn1 expression in livers of ad libitum fed and 24 h fasted wildtype and PPARα −/− mice. (b) Vnn1 expression in liver, (c) small intestine, and (d) large intestine of wildtype and PPARα −/− mice 6 h after administration of a single oral dose of Wy14643 (4 mg), fenofibrate (4 mg), and synthetic triglycerides triolein, trilinolein, trilinolenin, trieicosapentaenoin, or tridocosahexaenoin (400 mL). (e) HepG2 cells were transiently transfected with reporters (PPRE)3-TK-LUC or PPRE-Vnn1-LUC (PPRE present in intron 3-4 of the Vnn1 gene cloned into pGL3-promoter) and PPARα expression plasmid (pSG5). After transfection, cells were treated with WY14643 (50 μM) for 24 hours followed by determination of luciferase and β-galactosidase activities in the cell lysates. Luciferase activities were normalized to β-galactosidase, and the relative luciferase activity of the cells treated with DMSO was set to 1. Error bars represent SEM.
The ability of PPARα to stimulate fatty acid oxidation and suppress hepatic inflammation has led to the exploration of PPARα agonists as a therapeutic option for nonalcohol fatty liver disease and specifically nonalcoholic steatohepatitis (NASH). Several studies in mice have shown that PPARα activation can reduce or even reverse the progression of steatohepatitis [193, 254–259]. The inhibitory effect of PPARα on progression of steatosis to steatohepatitis may be mediated in part by COX2 (Ptgs2), a candidate gene involved in steatohepatitis development that is suppressed by PPARα [260]. In the absence of PPARα, liver steatosis and inflammation are enhanced in mice chronically fed a HFD [165]. Whether the effects of PPARα on NASH are primarily related to changes in hepatic TG content or occur via direct suppression of inflammatory genes and markers remains unclear.
11. PPARα and Biotransformation
The detoxification of endogenous and exogenous molecules is generally divided into three distinct biotransformation phases. The phase I reaction involves the introduction of a polar group into the xenobiotic molecule and is catalyzed by members of the cytochrome P450 (CYP) superfamily [179, 180, 261]. Phase II enzymes are responsible for covalent linkage of the absorbed chemicals or products of the phase I reactions with compounds such as glutathione, glucuronic acid, or amino acids and are carried out by sulfotransferases, UDP-glucuronosyltransferases (UGTs), glutathione- S-transferases (GSTs), and N-acetyltransferases [261]. The third phase corresponds to elimination of the conjugated molecule from cells and their excretion into bile or urine via specific transporters, mainly members of the superfamily ATP-binding cassette transporter proteins [262, 263]. Studies have shown that peroxisome proliferators modulate exclusively the Cyp4a class of monooxygenases (involved in the metabolism of biologically important compounds such as fatty acids, see Section 5.3) in mouse while regulating various other Cyp genes in human hepatocytes, including members of the Cyp1a, Cyp2a, Cyp2c, and Cyp2e subfamilies [24]. Our recent microarray data confirmed the human specific regulation of Cyp genes belonging to classes 1-3 by PPARα in primary human hepatocytes. Interestingly, we also observed a significant induction of another subfamily member of Cyp4 enzymes, Cyp4x1, by PPARα in human primary hepatocytes which was not conserved in mouse [24]. Cyp4x1 has been shown to be involved in oxidation of anandamide, which represents one of the endocannabinoids. Besides upregulation of gene expression, a number of genes involved in phase I biotransformation are downregulated by PPARα in mice, including Cyp2a5, Cyp2c11, Cup2c12, and Cyp2c29 [110, 155].
With respect to phase II biotransformation, PPARα has been shown to downregulate Glutathione-S-transferase A [GSTA], possibly leading to decreased biliary excretion of glutathione conjugates [157, 264, 265]. In contrast, expression of UDP-glucuronosyltransferase 1A (Ugt1a9), which participates with other UGT enzymes in glucuronidation of bilirubin, arachidonic, and linoleic acid metabolites, is under direct stimulatory control of PPARα [158]. Overall, it is evident that PPARα is a major regulator of biotransformation enzymes and governs the expression of numerous cytochrome P-450 and conjugating enzymes. However, only a small portion of the regulation seems to be conserved between rodents and humans.
12. Conclusion
In 2010, we are celebrating the 20th anniversary of the discovery of PPARα by Isseman and Green. PPARα was initially isolated as a novel nuclear hormone receptor that serves as molecular target of a diverse class of rodent hepatocarcinogens. Since then it has become clear that PPARα can be activated by a large variety of endogenous and synthetic agonists including fibrate drugs. In fact, PPARα is nowadays considered as a crucial fatty acids sensor that mediates the effects of numerous fatty acids and fatty acid derivatives on gene expression. Furthermore, over the years PPARα has emerged as a crucial transcriptional regulator of numerous metabolic and inflammatory processes. Although PPARα has mostly been connected with stimulation of fatty acid oxidation, it is now evident that the effects of PPARα are much more widespread and cover numerous aspects of nutrient metabolism and energy homeostasis, including metabolism of lipoproteins, glucose/glycerol, cholesterol and bile acids, xenobiotics, and amino acids. Certainly, PPARα merits the classification as a master regulator of hepatic intermediary metabolism. Until recently, much confusion surrounded the effects of PPARα activation in human liver. Recent studies indicate that at least in terms of lipid metabolism, the function and specific target genes of PPARα are generally well conserved between mouse and human. One of the major challenges lying ahead is to gain better understanding of the molecular mechanism underlying downregulation of gene expression by PPARα, to improve insight into the specific mechanisms and pathways of endogenous PPARα activation, and to better link the functional consequences of PPARα activation to induction of specific PPARα target genes.
Acknowledgment
B. Knoch was funded by AgResearch within the Nutrigenomics New Zealand partnership.
References
- 1.Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocrine Reviews. 1999;20(5):649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 2.Kersten S, Desvergne B, Wahli W. Roles of PPARS in health and disease. Nature. 2000;405(6785):421–424. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- 3.Evans RM, Barish GD, Wang Y-X. PPARs and the complex journey to obesity. Nature Medicine. 2004;10(4):355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 4.IJpenberg A, Jeannin E, Wahli W, Desvergne B. Polarity and specific sequence requirements of peroxisome proliferator-activated receptor (PPAR)/retinoid X receptor heterodimer binding to DNA. A functional analysis of the malic enzyme gene PPAR response element. Journal of Biological Chemistry. 1997;272(32):20108–20117. doi: 10.1074/jbc.272.32.20108. [DOI] [PubMed] [Google Scholar]
- 5.Braissant O, Foufelle F, Scotto C, Dauça M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-α, -β, and -γ in the adult rat. Endocrinology. 1996;137(1):354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 6.Escher P, Braissant O, Basu-Modak S, Michalik L, Wahli W, Desvergne B. Rat PPARs: quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology. 2001;142(10):4195–4202. doi: 10.1210/endo.142.10.8458. [DOI] [PubMed] [Google Scholar]
- 7.Xu HE, Stanley TB, Montana VG, et al. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARα . Nature. 2002;415(6873):813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- 8.Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347(6294):645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 9.Thorp JM, Waring WS. Modification of metabolism and distribution of lipids by ethyl chlorophenoxyisobutyrate. Nature. 1962;194(4832):948–949. doi: 10.1038/194948a0. [DOI] [PubMed] [Google Scholar]
- 10.Willson TM, Brown PJ, Sternbach DD, Henke BR. The PPARs: from orphan receptors to drug discovery. Journal of Medicinal Chemistry. 2000;43(4):527–550. doi: 10.1021/jm990554g. [DOI] [PubMed] [Google Scholar]
- 11.Berger J, Moller DE. The mechanisms of action of PPARs. Annual Review of Medicine. 2002;53:409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- 12.van der Meer DLM, Degenhardt T, Väisänen S, et al. Profiling of promoter occupancy by PPARα in human hepatoma cells via ChIP-chip analysis. Nucleic Acids Research. 2010;38(9):2839–2850. doi: 10.1093/nar/gkq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lefterova MI, Zhang Y, Steger DJ, et al. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes and Development. 2008;22(21):2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen R, Pedersen TÅ, Hagenbeek D, et al. Genome-wide profiling of PPARγ:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes and Development. 2008;22(21):2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoekstra M, Kruijt JK, Van Eck M, Van Berkel TJC. Specific gene expression of ATP-binding cassette transporters and nuclear hormone receptors in rat liver parenchymal, endothelial, and Kupffer cells. Journal of Biological Chemistry. 2003;278(28):25448–25453. doi: 10.1074/jbc.M301189200. [DOI] [PubMed] [Google Scholar]
- 16.Peters JM, Rusyn I, Rose ML, Gonzalez FJ, Thurman RG. Peroxisome proliferator-activated receptor α is restricted to hepatic parenchymal cells, not Kupffer cells: implications for the mechanism of action of peroxisome proliferators in hepatocarcinogenesis. Carcinogenesis. 2000;21(4):823–826. doi: 10.1093/carcin/21.4.823. [DOI] [PubMed] [Google Scholar]
- 17.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126(4):789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marx N, Kehrle B, Kohlhammer K, et al. PPAR activators as antiinflammatory mediators in human T lymphocytes: implications for atherosclerosis and transplantation-associated arteriosclerosis. Circulation Research. 2002;90(6):703–710. doi: 10.1161/01.res.0000014225.20727.8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chinetti G, Griglio S, Antonucci M, et al. Activation of proliferator-activated receptors α and γ induces apoptosis of human monocyte-derived macrophages. Journal of Biological Chemistry. 1998;273(40):25573–25580. doi: 10.1074/jbc.273.40.25573. [DOI] [PubMed] [Google Scholar]
- 20.Marx N, Mackman N, Schönbeck U, et al. PPARα activators inhibit tissue factor expression and activity in human monocytes. Circulation. 2001;103(2):213–219. doi: 10.1161/01.cir.103.2.213. [DOI] [PubMed] [Google Scholar]
- 21.Chinetti G, Lestavel S, Bocher V, et al. PPAR-α and PPAR-γ activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. Nature Medicine. 2001;7(1):53–58. doi: 10.1038/83348. [DOI] [PubMed] [Google Scholar]
- 22.Gbaguidi FG, Chinetti G, Milosavljevic D, et al. Peroxisome proliferator-activated receptor (PPAR) agonists decrease lipoprotein lipase secretion and glycated LDL uptake by human macrophages. FEBS Letters. 2002;512(1–3):85–90. doi: 10.1016/s0014-5793(02)02223-8. [DOI] [PubMed] [Google Scholar]
- 23.Palmer CNA, Hsu M-H, Griffin KJ, Raucy JL, Johnson EF. Peroxisome proliferator activated receptor-α expression in human liver. Molecular Pharmacology. 1998;53(1):14–22. [PubMed] [Google Scholar]
- 24.Rakhshandehroo M, Hooiveld G, Müller M, Kersten S. Comparative analysis of gene regulation by the transcription factor PPARα between mouse and human. PLoS ONE. 2009;4(8) doi: 10.1371/journal.pone.0006796. Article ID e6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hi R, Osada S, Yumoto N, Osumi T. Characterization of the amino-terminal activation domain of peroxisome proliferator-activated receptor α. Importance of α-helical structure in the transactivating function. Journal of Biological Chemistry. 1999;274(49):35152–35158. doi: 10.1074/jbc.274.49.35152. [DOI] [PubMed] [Google Scholar]
- 26.Cronet P, Petersen JFW, Folmer R, et al. Structure of the PPARα and -γ ligand binding domain in complex with AZ 242; ligand selectivity and agonist activation in the PPAR family. Structure. 2001;9(8):699–706. doi: 10.1016/s0969-2126(01)00634-7. [DOI] [PubMed] [Google Scholar]
- 27.Diradourian C, Girard J, Pégorier J-P. Phosphorylation of PPARs: from molecular characterization to physiological relevance. Biochimie. 2005;87(1):33–38. doi: 10.1016/j.biochi.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 28.Wan Y-JY, Cai Y, Lungo W, et al. Peroxisome proliferator-activated receptor α-mediated pathways are altered in hepatocyte-specific retinoid x receptor α-deficient mice. Journal of Biological Chemistry. 2000;275(36):28285–28290. doi: 10.1074/jbc.M000934200. [DOI] [PubMed] [Google Scholar]
- 29.Xu HE, Lambert MH, Montana VG, et al. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(24):13919–13924. doi: 10.1073/pnas.241410198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu HE, Lambert MH, Montana VG, et al. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Molecular Cell. 1999;3(3):397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 31.Sapone A, Peters JM, Sakai S, et al. The human peroxisome proliferator-activated receptor α gene: identification and functional characterization of two natural allelic variants. Pharmacogenetics. 2000;10(4):321–333. doi: 10.1097/00008571-200006000-00006. [DOI] [PubMed] [Google Scholar]
- 32.Flavell DM, Pineda Torra I, Jamshidi Y, et al. Variation in the PPARα gene is associated with altered function in vitro and plasma lipid concentrations in type II diabetic subjects. Diabetologia. 2000;43(5):673–680. doi: 10.1007/s001250051357. [DOI] [PubMed] [Google Scholar]
- 33.Lacquemant C, Lepretre F, Pineda Torra I, et al. Mutation screening of the PPARα gene in type 2 diabetes associated with coronary heart disease. Diabetes and Metabolism. 2000;26(5):393–401. [PubMed] [Google Scholar]
- 34.Vohl M-C, Lepage P, Gaudet D, et al. Molecular scanning of the human PPARα gene: association of the L162V mutation with hyperapobetalipoproteinemia. Journal of Lipid Research. 2000;41(6):945–952. [PubMed] [Google Scholar]
- 35.Evans D, Aberle J, Wendt D, Wolf A, Beisiegel U, Mann WA. A polymorphism, L162V, in the peroxisome proliferator-activated receptor α (PPARα) gene is associated with lower body mass index in patients with non-insulin-dependent diabetes mellitus. Journal of Molecular Medicine. 2001;79(4):198–204. doi: 10.1007/s001090100189. [DOI] [PubMed] [Google Scholar]
- 36.Gouni-Berthold I, Giannakidou E, Müller-Wieland D, et al. Association between the PPARα L162V polymorphism, plasma lipoprotein levels, and atherosclerotic disease in patients with diabetes mellitus type 2 and in nondiabetic controls. American Heart Journal. 2004;147(6):1117–1124. doi: 10.1016/j.ahj.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 37.Flavell DM, Jamshidi Y, Hawe E, et al. Peroxisome proliferator-activated receptor α gene variants influence progression of coronary atherosclerosis and risk of coronary artery disease. Circulation. 2002;105(12):1440–1445. doi: 10.1161/01.cir.0000012145.80593.25. [DOI] [PubMed] [Google Scholar]
- 38.Tai ES, Corella D, Demissie S, et al. Polyunsaturated fatty acids interact with the PPARA-L162V polymorphism to affect plasma triglyceride and apolipoprotein C-III concentrations in the Framingham Heart Study. Journal of Nutrition. 2005;135(3):397–403. doi: 10.1093/jn/135.3.397. [DOI] [PubMed] [Google Scholar]
- 39.Brisson D, Ledoux K, Bosse Y, et al. Effect of apolipoprotein E, peroxisome proliferator-activated receptor alpha and lipoprotein lipase gene mutations on the ability of fenofibrate to improve lipid profiles and reach clinical guideline targets among hypertriglyceridemic patients. Pharmacogenetics. 2002;12(4):313–320. doi: 10.1097/00008571-200206000-00007. [DOI] [PubMed] [Google Scholar]
- 40.Bossé Y, Pascot A, Dumont M, et al. Influences of the PPARα-L162V polymorphism on plasma HDL 2-cholesterol response of abdominally obese men treated with gemfibrozil. Genetics in Medicine. 2002;4(4):311–315. doi: 10.1097/00125817-200207000-00010. [DOI] [PubMed] [Google Scholar]
- 41.Yamakawa-Kobayashi K, Ishiguro H, Arinami T, Miyazaki R, Hamaguchi H. A Val227Ala polymorphism in the peroxisome proliferator activated receptor alpha (PPARalpha) gene is associated with variations in serum lipid levels. Journal of Medical Genetics. 2002;39(3):189–191. doi: 10.1136/jmg.39.3.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naito H, Yamanoshita O, Kamijima M, et al. Association of V227A PPARα polymorphism with altered serum biochemistry and alcohol drinking in Japanese men. Pharmacogenetics and Genomics. 2006;16(8):569–577. doi: 10.1097/01.fpc.0000220565.90466.79. [DOI] [PubMed] [Google Scholar]
- 43.Naito H, Kamijima M, Yamanoshita O, et al. Differential effects of aging, drinking and exercise on serum cholesterol levels dependent on the PPARA-V227A polymorphism. Journal of Occupational Health. 2007;49(5):353–362. doi: 10.1539/joh.49.353. [DOI] [PubMed] [Google Scholar]
- 44.Chen S, Li Y, Li S, Yu C. A Val227Ala substitution in the peroxisome proliferator activated receptor alpha (PPAR alpha) gene associated with non-alcoholic fatty liver disease and decreased waist circumference and waist-to-hip ratio. Journal of Gastroenterology and Hepatology. 2008;23(9):1415–1418. doi: 10.1111/j.1440-1746.2008.05523.x. [DOI] [PubMed] [Google Scholar]
- 45.Gervois P, Pineda Torra I, Chinetti G, et al. A truncated human peroxisome proliferator-activated receptor α splice variant with dominant negative activity. Molecular Endocrinology. 1999;13(9):1535–1549. doi: 10.1210/mend.13.9.0341. [DOI] [PubMed] [Google Scholar]
- 46.Fruchart JC, Duriez P, Staels B. Molecular mechanism of action of the fibrates. Journal de la Societe de Biologie. 1999;193(1):67–75. [PubMed] [Google Scholar]
- 47.Schoonjans K, Staels B, Auwerx J. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. Journal of Lipid Research. 1996;37(5):907–925. [PubMed] [Google Scholar]
- 48.Khan SA, Vanden Heuvel JP. Role of nuclear receptors in the regulation of gene expression by dietary fatty acids (review) Journal of Nutritional Biochemistry. 2003;14(10):554–567. doi: 10.1016/s0955-2863(03)00098-6. [DOI] [PubMed] [Google Scholar]
- 49.Kliewer SA, Sundseth SS, Jones SA, et al. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ . Proceedings of the National Academy of Sciences of the United States of America. 1997;94(9):4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ . Proceedings of the National Academy of Sciences of the United States of America. 1997;94(9):4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keller H, Dreyer C, Medin J, Mahfoudi A, Ozato K, Wahli W. Fatty acids and retinoids control lipid metabolism through activation of peroxisome proliferator-activated receptor-retinoid X receptor heterodimers. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(6):2160–2164. doi: 10.1073/pnas.90.6.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu K, Bayona W, Kallen CB, et al. Differential activation of peroxisome proliferator-activated receptors by eicosanoids. Journal of Biological Chemistry. 1995;270(41):23975–23983. doi: 10.1074/jbc.270.41.23975. [DOI] [PubMed] [Google Scholar]
- 53.Devchand PR, Keller H, Peters JM, Vazquez M, Gonzalez FJ, Wahli W. The PPARα-leukotriene B4 pathway to inflammation control. Nature. 1996;384(6604):39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
- 54.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor α mediates the adaptive response to fasting. Journal of Clinical Investigation. 1999;103(11):1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor α (PPARα) in the cellular fasting response: the PPARα-null mouse as a model of fatty acid oxidation disorders. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(13):7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanderson LM, Degenhardt T, Koppen A, et al. Peroxisome proliferator-activated receptor β/δ (PPARβ/δ) but not PPARα serves as a plasma free fatty acid sensor in liver. Molecular and Cellular Biology. 2009;29(23):6257–6267. doi: 10.1128/MCB.00370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chakravarthy MV, Pan Z, Zhu Y, et al. “New” hepatic fat activates PPARα to maintain glucose, lipid, and cholesterol homeostasis. Cell Metabolism. 2005;1(5):309–322. doi: 10.1016/j.cmet.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 58.Patsouris D, Reddy JK, Müller M, Kersten S. Peroxisome proliferator-activated receptor α mediates the effects of high-fat diet on hepatic gene expression. Endocrinology. 2006;147(3):1508–1516. doi: 10.1210/en.2005-1132. [DOI] [PubMed] [Google Scholar]
- 59.Martin PGP, Guillou H, Lasserre F, et al. Novel aspects of PPARα-mediated regulation of lipid and xenobiotic metabolism revealed through a nutrigenomic study. Hepatology. 2007;45(3):767–777. doi: 10.1002/hep.21510. [DOI] [PubMed] [Google Scholar]
- 60.Ren B, Thelen AP, Peters JM, Gonzalez FJ, Jump DB. Polyunsaturated fatty acid suppression of hepatic fatty acid synthase and S14 gene expression does not require peroxisome proliferator-activated receptor α . Journal of Biological Chemistry. 1997;272(43):26827–26832. doi: 10.1074/jbc.272.43.26827. [DOI] [PubMed] [Google Scholar]
- 61.Sanderson LM, de Groot PJ, Hooiveld GJEJ, et al. Effect of synthetic dietary triglycerides: a novel research paradigm for nutrigenomics. PLoS ONE. 2008;3(2) doi: 10.1371/journal.pone.0001681. Article ID e1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rakhshandehroo M, Sanderson LM, Matilainen M, et al. Comprehensive analysis of PPARα-dependent regulation of hepatic lipid metabolism by expression profiling. PPAR Research. 2007;2007:13 pages. doi: 10.1155/2007/26839. Article ID 26839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo L, Fang H, Collins J, et al. Differential gene expression in mouse primary hepatocytes exposed to the peroxisome proliferator-activated receptor α agonists. BMC Bioinformatics. 2006;7(supplement 2):p. S18. doi: 10.1186/1471-2105-7-S2-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Motojima K, Passilly P, Peters JM, Gonzalez FJ, Latruffe N. Expression of putative fatty acid transporter genes are regulated by peroxisome proliferator-activated receptor α and γ activators in a tissue- and inducer-specific manner. Journal of Biological Chemistry. 1998;273(27):16710–16714. doi: 10.1074/jbc.273.27.16710. [DOI] [PubMed] [Google Scholar]
- 65.Yamazaki K, Kuromitsu J, Tanaka I. Microarray analysis of gene expression changes in mouse liver induced by peroxisome proliferator-activated receptor α agonists. Biochemical and Biophysical Research Communications. 2002;290(3):1114–1122. doi: 10.1006/bbrc.2001.6319. [DOI] [PubMed] [Google Scholar]
- 66.Martin G, Schoonjans K, Lefebvre A-M, Staels B, Auwerx J. Coordinate regulation of the expression of the fatty acid transport protein and acyl-CoA synthetase genes by PPARα and PPARγ activators. Journal of Biological Chemistry. 1997;272(45):28210–28217. doi: 10.1074/jbc.272.45.28210. [DOI] [PubMed] [Google Scholar]
- 67.Frohnert BI, Hui TY, Bernlohr DA. Identification of a functional peroxisome proliferator-responsive element in the murine fatty acid transport protein gene. Journal of Biological Chemistry. 1999;274(7):3970–3977. doi: 10.1074/jbc.274.7.3970. [DOI] [PubMed] [Google Scholar]
- 68.Richert L, Lamboley C, Viollon-Abadie C, et al. Effects of clofibric acid on mRNA expression profiles in primary cultures of rat, mouse and human hepatocytes. Toxicology and Applied Pharmacology. 2003;191(2):130–146. doi: 10.1016/s0041-008x(03)00231-x. [DOI] [PubMed] [Google Scholar]
- 69.Hunt MC, Lindquist PJG, Peters JM, Gonzalez FJ, Diczfalusy U, Alexson SEH. Involvement of the peroxisome proliferator-activated receptor α in regulating long-chain acyl-CoA thioesterases. Journal of Lipid Research. 2000;41(5):814–823. [PubMed] [Google Scholar]
- 70.Aoyama T, Peters JM, Iritani N, et al. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor α (PPARα) Journal of Biological Chemistry. 1998;273(10):5678–5684. doi: 10.1074/jbc.273.10.5678. [DOI] [PubMed] [Google Scholar]
- 71.Hsu M-H, Savas Ü, Griffin KJ, Johnson EF. Identification of peroxisome proliferator-responsive human genes by elevated expression of the peroxisome proliferator-activated receptor alpha in HepG2 cells. Journal of Biological Chemistry. 2001;276(30):27950–27958. doi: 10.1074/jbc.M100258200. [DOI] [PubMed] [Google Scholar]
- 72.Schoonjans K, Watanabe M, Suzuki H, et al. Induction of the acyl-coenzyme A synthetase gene by fibrates and fatty acids is mediated by a peroxisome proliferator response element in the C promoter. Journal of Biological Chemistry. 1995;270(33):19269–19276. doi: 10.1074/jbc.270.33.19269. [DOI] [PubMed] [Google Scholar]
- 73.Lewin TM, Van Horn CG, Krisans SK, Coleman RA. Rat liver acyl-CoA synthetase 4 is a peripheral-membrane protein located in two distinct subcellular organelles, peroxisomes, and mitochondrial-associated membrane. Archives of Biochemistry and Biophysics. 2002;404(2):263–270. doi: 10.1016/s0003-9861(02)00247-3. [DOI] [PubMed] [Google Scholar]
- 74.Hashimoto T, Fujita T, Usuda N, et al. Peroxisomal and mitochondrial fatty acid β-oxidation in mice nullizygous for both peroxisome proliferator-activated receptor and peroxisomal fatty acyl-CoA oxidase: genotype correlation with fatty liver phenotype. Journal of Biological Chemistry. 1999;274(27):19228–19236. doi: 10.1074/jbc.274.27.19228. [DOI] [PubMed] [Google Scholar]
- 75.Poirier H, Niot I, Monnot M-C, et al. Differential involvement of peroxisome-proliferator-activated receptors α and δ in fibrate and fatty-acid-mediated inductions of the gene encoding liver fatty-acid-binding protein in the liver and the small intestine. Biochemical Journal. 2001;355(2):481–488. doi: 10.1042/0264-6021:3550481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vanden Heuvel JP, Kreder D, Belda B, et al. Comprehensive analysis of gene expression in rat and human hepatoma cells exposed to the peroxisome proliferator WY14,643. Toxicology and Applied Pharmacology. 2003;188(3):185–198. doi: 10.1016/s0041-008x(03)00015-2. [DOI] [PubMed] [Google Scholar]
- 77.Brandt JM, Djouadi F, Kelly DP. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor α . Journal of Biological Chemistry. 1998;273(37):23786–23792. doi: 10.1074/jbc.273.37.23786. [DOI] [PubMed] [Google Scholar]
- 78.Mascaró C, Acosta E, Ortiz JA, Marrero PF, Hegardt FG, Haro D. Control of human muscle-type carnitine palmitoyltransferase I gene transcription by peroxisome proliferator-activated receptor. Journal of Biological Chemistry. 1998;273(15):8560–8563. doi: 10.1074/jbc.273.15.8560. [DOI] [PubMed] [Google Scholar]
- 79.Napal L, Marrero PF, Haro D. An intronic peroxisome proliferator-activated receptor-binding sequence mediates fatty acid induction of the human carnitine palmitoyltransferase 1A. Journal of Molecular Biology. 2005;354(4):751–759. doi: 10.1016/j.jmb.2005.09.097. [DOI] [PubMed] [Google Scholar]
- 80.Barrero MJ, Camarero N, Marrero PF, Haro D. Control of human carnitine palmitoyltransferase II gene transcription by peroxisome proliferator-activated receptor through a partially conserved peroxisome proliferator-responsive element. Biochemical Journal. 2003;369(3):721–729. doi: 10.1042/BJ20020851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hakkola EH, Hiltunen JK, Autio-Harmainen HI. Mitochondrial 2,4-dienoyl-CoA reductases in the rat: differential responses to clofibrate treatment. Journal of Lipid Research. 1994;35(10):1820–1828. [PubMed] [Google Scholar]
- 82.Tachibana K, Kobayashi Y, Tanaka T, et al. Gene expression profiling of potential peroxisome proliferator-activated receptor (PPAR) target genes in human hepatoblastoma cell lines inducibly expressing PPAR isoforms. Nuclear Receptor. 2005;3 doi: 10.1186/1478-1336-3-3. Article ID 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang B, Marcus SL, Saijadi FG, et al. Identification of a peroxisome proliferator-responsive element upstream of the gene encoding rat peroxisomal enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(16):7541–7545. doi: 10.1073/pnas.89.16.7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gutgesell A, Wen G, König B, et al. Mouse carnitine-acylcarnitine translocase (CACT) is transcriptionally regulated by PPARα and PPARδ in liver cells. Biochimica et Biophysica Acta. 2009;1790(10):1206–1216. doi: 10.1016/j.bbagen.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 85.van Vlies N, Ferdinandusse S, Turkenburg M, Wanders RJA, Vaz FM. PPARα-activation results in enhanced carnitine biosynthesis and OCTN2-mediated hepatic carnitine accumulation. Biochimica et Biophysica Acta. 2007;1767(9):1134–1142. doi: 10.1016/j.bbabio.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 86.Luci S, Geissler S, König B, et al. PPARα agonists up-regulate organic cation transporters in rat liver cells. Biochemical and Biophysical Research Communications. 2006;350(3):704–708. doi: 10.1016/j.bbrc.2006.09.099. [DOI] [PubMed] [Google Scholar]
- 87.Kelly LJ, Vicario PP, Thompson GM, et al. Peroxisome proliferator-activated receptors γ and α mediate in vivo regulation of uncoupling protein (UCP-1, UCP-2, UCP-3) gene expression. Endocrinology. 1998;139(12):4920–4927. doi: 10.1210/endo.139.12.6384. [DOI] [PubMed] [Google Scholar]
- 88.Armstrong MB, Towle HC. Polyunsaturated fatty acids stimulate hepatic UCP-2 expression via a PPARα-mediated pathway. American Journal of Physiology. 2001;281(6):E1197–E1204. doi: 10.1152/ajpendo.2001.281.6.E1197. [DOI] [PubMed] [Google Scholar]
- 89.Tsuboyama-Kasaoka N, Takahashi M, Kim H, Ezaki O. Up-regulation of liver uncoupling protein-2 mRNA by either fish oil feeding or fibrate administration in mice. Biochemical and Biophysical Research Communications. 1999;257(3):879–885. doi: 10.1006/bbrc.1999.0555. [DOI] [PubMed] [Google Scholar]
- 90.Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos-Flier E. Hepatic fibroblast growth factor 21 is regulated by PPARα and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metabolism. 2007;5(6):426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 91.Lundåsen T, Hunt MC, Nilsson L-M, et al. PPARα is a key regulator of hepatic FGF21. Biochemical and Biophysical Research Communications. 2007;360(2):437–440. doi: 10.1016/j.bbrc.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 92.Inagaki T, Dutchak P, Zhao G, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metabolism. 2007;5(6):415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 93.Le May C, Pineau T, Bigot K, Kohl C, Girard J, Pégorier J-P. Reduced hepatic fatty acid oxidation in fasting PPARα null mice is due to impaired mitochondrial hydroxymethylglutaryl-CoA synthase gene expression. FEBS Letters. 2000;475(3):163–166. doi: 10.1016/s0014-5793(00)01648-3. [DOI] [PubMed] [Google Scholar]
- 94.Kok T, Bloks VW, Wolters H, et al. Peroxisome proliferator-activated receptor α (PPARα)-mediated regulation of multidrug resistance 2 (Mdr2) expression and function in mice. Biochemical Journal. 2003;369(3):539–547. doi: 10.1042/BJ20020981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rodríguez JC, Gil-Gómez G, Hegardt FG, Haro D. Peroxisome proliferator-activated receptor mediates induction of the mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase gene by fatty acids. Journal of Biological Chemistry. 1994;269(29):18767–18772. [PubMed] [Google Scholar]
- 96.Fourcade S, Savary S, Albet S, et al. Fibrate induction of the adrenoleukodystrophy-related gene (ABCD2): promoter analysis and role of the peroxisome proliferator-activated receptor PPARα . European Journal of Biochemistry. 2001;268(12):3490–3500. doi: 10.1046/j.1432-1327.2001.02249.x. [DOI] [PubMed] [Google Scholar]
- 97.Hunt MC, Solaas K, Kase BF, Alexson SEH. Characterization of an acyl-CoA thioesterase that functions as a major regulator of peroxisomal lipid metabolism. Journal of Biological Chemistry. 2002;277(2):1128–1138. doi: 10.1074/jbc.M106458200. [DOI] [PubMed] [Google Scholar]
- 98.Goldberg C. The pursuit of the fictional self. American Journal of Psychotherapy. 2004;58(2):209–219. doi: 10.1176/appi.psychotherapy.2004.58.2.209. [DOI] [PubMed] [Google Scholar]
- 99.Corton JC, Bocos C, Moreno ES, Merritt A, Cattley RC, Gustafsson J-Å. Peroxisome proliferators alter the expression of estrogen-metabolizing enzymes. Biochimie. 1997;79(2-3):151–162. doi: 10.1016/s0300-9084(97)81508-8. [DOI] [PubMed] [Google Scholar]
- 100.De Souza AT, Dai X, Spencer AG, et al. Transcriptional and phenotypic comparisons of Ppara knockout and siRNA knockdown mice. Nucleic Acids Research. 2006;34(16):4486–4494. doi: 10.1093/nar/gkl609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alvares K, Fan C, Dadras SS, et al. An upstream region of the enoyl-coenzyme A hydratase/3-hydroxyacyl- coenzyme A dehydrogenase gene directs luciferase expression in liver in response to peroxisome proliferators in transgenic mice. Cancer Research. 1994;54(9):2303–2306. [PubMed] [Google Scholar]
- 102.Guo Y, Jolly RA, Halstead BW, et al. Underlying mechanisms of pharmacology and toxicity of a novel PPAR agonist revealed using rodent and canine hepatocytes. Toxicological Sciences. 2007;96(2):294–309. doi: 10.1093/toxsci/kfm009. [DOI] [PubMed] [Google Scholar]
- 103.Corton JC, Bocos C, Moreno ES, et al. Rat 17β-hydroxysteroid dehydrogenase type IV is a novel peroxisome proliferator-inducible gene. Molecular Pharmacology. 1996;50(5):1157–1166. [PubMed] [Google Scholar]
- 104.Fan L-Q, Cattley RC, Corton JC. Tissue-specific induction of 17β-hydroxysteroid dehydrogenase type IV by peroxisome proliferator chemicals is dependent on the peroxisome proliferator-activated receptor α . Journal of Endocrinology. 1998;158(2):237–246. doi: 10.1677/joe.0.1580237. [DOI] [PubMed] [Google Scholar]
- 105.Wang T, Shah YM, Matsubara T, et al. Control of steroid 21-oic acid synthesis by peroxisome proliferator-activated receptor α and role of the hypothalamic-pituitary-adrenal axis. Journal of Biological Chemistry. 2010;285(10):7670–7685. doi: 10.1074/jbc.M109.090175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Johnson EF, Palmer CNA, Griffin KJ, Hsu M-H. Role of the peroxisome proliferator-activated receptor in cytochrome P450 4A gene regulation. FASEB Journal. 1996;10(11):1241–1248. doi: 10.1096/fasebj.10.11.8836037. [DOI] [PubMed] [Google Scholar]
- 107.Aldridge TC, Tugwood JD, Green S. Identification and characterization of DNA elements implicated in the regulation of CYP4A1 transcription. Biochemical Journal. 1995;306(2):473–479. doi: 10.1042/bj3060473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee SS-T, Pineau T, Drago J, et al. Targeted disruption of the α isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Molecular and Cellular Biology. 1995;15(6):3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kroetz DL, Yook P, Costet P, Bianchi P, Pineau T. Peroxisome proliferator-activated receptor controls the hepatic CYP4A induction adaptive response to starvation and diabetes. Journal of Biological Chemistry. 1998;273(47):31581–31589. doi: 10.1074/jbc.273.47.31581. [DOI] [PubMed] [Google Scholar]
- 110.Barclay TB, Peters JM, Sewer MB, Ferrari L, Gonzalez FJ, Morgan ET. Modulation of cytochrome P-450 gene expression in endotoxemic mice is tissue specific and peroxisome proliferator-activated receptor-α dependent. Journal of Pharmacology and Experimental Therapeutics. 1999;290(3):1250–1257. [PubMed] [Google Scholar]
- 111.Anderson SP, Yoon L, Richard EB, Dunn CS, Cattley RC, Corton JC. Delayed liver regeneration in peroxisome proliferator-activated receptor-α-null mice. Hepatology. 2002;36(3):544–554. doi: 10.1053/jhep.2002.35276. [DOI] [PubMed] [Google Scholar]
- 112.Knight BL, Hebbach A, Hauton D, et al. A role for PPARα in the control of SREBP activity and lipid synthesis in the liver. Biochemical Journal. 2005;389(2):413–421. doi: 10.1042/BJ20041896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang Y, Botolin D, Xu J, et al. Regulation of hepatic fatty acid elongase and desaturase expression in diabetes and obesity. Journal of Lipid Research. 2006;47(9):2028–2041. doi: 10.1194/jlr.M600177-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang Y, Botolin D, Christian B, Busik J, Xu J, Jump DB. Tissue-specific, nutritional, and developmental regulation of rat fatty acid elongases. Journal of Lipid Research. 2005;46(4):706–715. doi: 10.1194/jlr.M400335-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tang C, Cho HP, Nakamura MT, Clarke SD. Regulation of human Δ-6 desaturase gene transcription: identification of a functional direct repeat-1 element. Journal of Lipid Research. 2003;44(4):686–695. doi: 10.1194/jlr.M200195-JLR200. [DOI] [PubMed] [Google Scholar]
- 116.Castelein H, Gulick T, Declercq PE, Mannaerts GP, Moore DD, Baes MI. The peroxisome proliferator activated receptor regulates malic enzyme gene expression. Journal of Biological Chemistry. 1994;269(43):26754–26758. [PubMed] [Google Scholar]
- 117.Miller CW, Ntambi JM. Peroxisome proliferators induce mouse liver stearoyl-CoA desaturase 1 gene expression. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18):9443–9448. doi: 10.1073/pnas.93.18.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guillou H, Martin P, Jan S, et al. Comparative effect of fenofibrate on hepatic desaturases in wild-type and peroxisome proliferator-activated receptor α-deficient mice. Lipids. 2002;37(10):981–989. doi: 10.1007/s11745-006-0990-3. [DOI] [PubMed] [Google Scholar]
- 119.Gibbons GF, Patel D, Wiggins D, Knight BL. The functional efficiency of lipogenic and cholesterogenic gene expression in normal mice and in mice lacking the peroxisomal proliferator-activated receptor-alpha (PPAR-α) Advances in Enzyme Regulation. 2002;42:227–247. doi: 10.1016/s0065-2571(01)00033-4. [DOI] [PubMed] [Google Scholar]
- 120.Dalen KT, Ulven SM, Arntsen BM, Solaas K, Nebb HI. PPARα activators and fasting induce the expression of adipose differentiation-related protein in liver. Journal of Lipid Research. 2006;47(5):931–943. doi: 10.1194/jlr.M500459-JLR200. [DOI] [PubMed] [Google Scholar]
- 121.Viswakarma N, Yu S, Naik S, et al. Transcriptional regulation of Cidea, mitochondrial cell death-inducing DNA fragmentation factor α-like effector A, in mouse liver by peroxisome proliferator-activated receptor α and γ . Journal of Biological Chemistry. 2007;282(25):18613–18624. doi: 10.1074/jbc.M701983200. [DOI] [PubMed] [Google Scholar]
- 122.Yang X, Lu X, Lombès M, et al. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metabolism. 2010;11(3):194–205. doi: 10.1016/j.cmet.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zandbergen F, Mandard S, Escher P, et al. The G0/G1 switch gene 2 is a novel PPAR target gene. Biochemical Journal. 2005;392(2):313–324. doi: 10.1042/BJ20050636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yamaguchi T, Matsushita S, Motojima K, Hirose F, Osumi T. MLDP, a novel PAT family protein localized to lipid droplets and enriched in the heart, is regulated by peroxisome proliferator-activated receptorα . Journal of Biological Chemistry. 2006;281(20):14232–14240. doi: 10.1074/jbc.M601682200. [DOI] [PubMed] [Google Scholar]
- 125.Dalen KT, Dahl T, Holter E, et al. LSDP5 is a PAT protein specifically expressed in fatty acid oxidizing tissues. Biochimica et Biophysica Acta. 2007;1771(2):210–227. doi: 10.1016/j.bbalip.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 126.Kersten S, Mandard S, Tan NS, et al. Characterization of the fasting-induced adipose factor FIAF, a novel peroxisome proliferator-activated receptor target gene. Journal of Biological Chemistry. 2000;275(37):28488–28493. doi: 10.1074/jbc.M004029200. [DOI] [PubMed] [Google Scholar]
- 127.Mandard S, Zandbergen F, Tan NS, et al. The direct peroxisome proliferator-activated receptor target fasting-induced adipose factor (FIAF/PGAR/ANGPTL4) is present in blood plasma as a truncated protein that is increased by fenofibrate treatment. Journal of Biological Chemistry. 2004;279(33):34411–34420. doi: 10.1074/jbc.M403058200. [DOI] [PubMed] [Google Scholar]
- 128.Berthou L, Duverger N, Emmanuel F, et al. Opposite regulation of human versus mouse apolipoprotein A-I by fibrates in human apolipoprotein A-I transgenic mice. Journal of Clinical Investigation. 1996;97(11):2408–2416. doi: 10.1172/JCI118687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vu-Dac N, Chopin-Delannoy S, Gervois P, et al. The nuclear receptors peroxisome proliferator-activated receptor α and rev-erbα mediate the species-specific regulation of apolipoprotein A-I expression by fibrates. Journal of Biological Chemistry. 1998;273(40):25713–25720. doi: 10.1074/jbc.273.40.25713. [DOI] [PubMed] [Google Scholar]
- 130.Peters JM, Hennuyer N, Staels B, et al. Alterations in lipoprotein metabolism in peroxisome proliferator-activated receptor α-deficient mice. Journal of Biological Chemistry. 1997;272(43):27307–27312. doi: 10.1074/jbc.272.43.27307. [DOI] [PubMed] [Google Scholar]
- 131.Vu-Dac N, Schoonjans K, Laine B, Fruchart J-C, Auwerx J, Staels B. Negative regulation of the human apolipoprotein A-I promoter by fibrates can be attenuated by the interaction of the peroxisome proliferator-activated receptor with its response element. Journal of Biological Chemistry. 1994;269(49):31012–31018. [PubMed] [Google Scholar]
- 132.Raspé E, Madsen L, Lefebvre A-M, et al. Modulation of rat liver apolipoprotein gene expression and serum lipid levels by tetradecylthioacetic acid (TTA) via PPARα activation. Journal of Lipid Research. 1999;40(11):2099–2110. [PubMed] [Google Scholar]
- 133.Berthou L, Saladin R, Yaqoob P, et al. Regulation of rat liver apolipoprotein A-I, apolipoprotein A-II and acyl-coenzyme A oxidase gene expression by fibrates and dietary fatty acids. European Journal of Biochemistry. 1995;232(1):179–187. doi: 10.1111/j.1432-1033.1995.tb20797.x. [DOI] [PubMed] [Google Scholar]
- 134.Vu-Dac N, Schoonjans K, Kosykh V, et al. Fibrates increase human apolipoprotein A-II expression through activation of the peroxisome proliferator-activated receptor. Journal of Clinical Investigation. 1995;96(2):741–750. doi: 10.1172/JCI118118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Prieur X, Coste H, Rodríguez JC. The human apolipoprotein AV gene is regulated by peroxisome proliferator-activated receptor-α and contains a novel farnesoid X-activated receptor response element. Journal of Biological Chemistry. 2003;278(28):25468–25480. doi: 10.1074/jbc.M301302200. [DOI] [PubMed] [Google Scholar]
- 136.Vu-Dac N, Gervois P, Jakel H, et al. Apolipoprotein A5, a crucial determinant of plasma triglyceride levels, is highly responsive to peroxisome proliferator-activated receptor α activators. Journal of Biological Chemistry. 2003;278(20):17982–17985. doi: 10.1074/jbc.M212191200. [DOI] [PubMed] [Google Scholar]
- 137.Hertz R, Bishara-Shieban J, Bar-Tana J. Mode of action of peroxisome proliferators as hypolipidemic drugs. Suppression of apolipoprotein C-III. Journal of Biological Chemistry. 1995;270(22):13470–13475. doi: 10.1074/jbc.270.22.13470. [DOI] [PubMed] [Google Scholar]
- 138.Staels B, Vu-Dac N, Kosykh VA, et al. Fibrates downregulate apolipoprotein C-III expression independent of induction of peroxisomal acyl coenzyme A oxidase. A potential mechanism for the hypolipidemic action of fibrates. Journal of Clinical Investigation. 1995;95(2):705–712. doi: 10.1172/JCI117717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Haubenwallner S, Essenburg AD, Barnett BC, et al. Hypolipidemic activity of select fibrates correlates to changes in hepatic apolipoprotein C-III expression: a potential physiologic basis for their mode of action. Journal of Lipid Research. 1995;36(12):2541–2551. [PubMed] [Google Scholar]
- 140.Schoonjans K, Peinado-Onsurbe J, Lefebvre A-M, et al. PPARα and PPARγ activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO Journal. 1996;15(19):5336–5348. [PMC free article] [PubMed] [Google Scholar]
- 141.Bouly M, Masson D, Gross B, et al. Induction of the phospholipid transfer protein gene accounts for the high density lipoprotein enlargement in mice treated with fenofibrate. Journal of Biological Chemistry. 2001;276(28):25841–25847. doi: 10.1074/jbc.M101160200. [DOI] [PubMed] [Google Scholar]
- 142.Tu A-Y, Albers JJ. Functional analysis of the transcriptional activity of the mouse phospholipid transfer protein gene. Biochemical and Biophysical Research Communications. 2001;287(4):921–926. doi: 10.1006/bbrc.2001.5687. [DOI] [PubMed] [Google Scholar]
- 143.Spann NJ, Kang S, Li AC, et al. Coordinate transcriptional repression of liver fatty acid-binding protein and microsomal triglyceride transfer protein blocks hepatic very low density lipoprotein secretion without hepatosteatosis. Journal of Biological Chemistry. 2006;281(44):33066–33077. doi: 10.1074/jbc.M607148200. [DOI] [PubMed] [Google Scholar]
- 144.Kok T, Wolters H, Bloks VW, et al. Induction of hepatic ABC transporter expression is part of the PPARα-mediated fasting response in the mouse. Gastroenterology. 2003;124(1):160–171. doi: 10.1053/gast.2003.50007. [DOI] [PubMed] [Google Scholar]
- 145.Post SM, Duez H, Gervois PP, Staels B, Kuipers F, Princen HMG. Fibrates suppress bile acid synthesis via peroxisome proliferator-activated receptor-α-mediated downregulation of cholesterol 7α-hydroxylase and sterol 27-hydroxylase expression. Arteriosclerosis, Thrombosis, and Vascular Biology. 2001;21(11):1840–1845. doi: 10.1161/hq1101.098228. [DOI] [PubMed] [Google Scholar]
- 146.Patel DD, Knight BL, Soutar AK, Gibbons GF, Wade DP. The effect of peroxisome-proliferator-activated receptor-α on the activity of the cholesterol 7α-hydroxylase gene. Biochemical Journal. 2000;351(3):747–753. [PMC free article] [PubMed] [Google Scholar]
- 147.Cheema SK, Agellon LB. The murine and human cholesterol 7α-hydroxylase gene promoters are differentially responsive to regulation by fatty acids mediated via peroxisome proliferator-activated receptor α . Journal of Biological Chemistry. 2000;275(17):12530–12536. doi: 10.1074/jbc.275.17.12530. [DOI] [PubMed] [Google Scholar]
- 148.Hunt MC, Yang Y-Z, Eggertsen G, et al. The peroxisome proliferator-activated receptor α (PPARα) regulates bile acid biosynthesis. Journal of Biological Chemistry. 2000;275(37):28947–28953. doi: 10.1074/jbc.M002782200. [DOI] [PubMed] [Google Scholar]
- 149.Tobin KAR, Steineger HH, Albert S, et al. Cross-talk between fatty acid and cholesterol metabolism mediated by liver X receptor-α . Molecular Endocrinology. 2000;14(5):741–752. doi: 10.1210/mend.14.5.0459. [DOI] [PubMed] [Google Scholar]
- 150.Jung D, Fried M, Kullak-Ublick GA. Human apical sodium-dependent bile salt transporter gene (SLC10A2) is regulated by the peroxisome proliferator-activated receptor α . Journal of Biological Chemistry. 2002;277(34):30559–30566. doi: 10.1074/jbc.M203511200. [DOI] [PubMed] [Google Scholar]
- 151.Patsouris D, Mandard S, Voshol PJ, et al. PPARα governs glycerol metabolism. Journal of Clinical Investigation. 2004;114(1):94–103. doi: 10.1172/JCI20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mandard S, Stienstra R, Escher P, et al. Glycogen synthase 2 is a novel target gene of peroxisome proliferator-activated receptors. Cellular and Molecular Life Sciences. 2007;64(9):1145–1157. doi: 10.1007/s00018-007-7006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sugden MC, Bulmer K, Gibbons GF, Knight BL, Holness MJ. Peroxisome-proliferator-activated receptor-α (PPARα) deficiency leads to dysregulation of hepatic lipid and carbohydrate metabolism by fatty acids and insulin. Biochemical Journal. 2002;364(2):361–368. doi: 10.1042/BJ20011699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wu P, Peters JM, Harris RA. Adaptive increase in pyruvate dehydrogenase kinase 4 during starvation is mediated by peroxisome proliferator-activated receptor α . Biochemical and Biophysical Research Communications. 2001;287(2):391–396. doi: 10.1006/bbrc.2001.5608. [DOI] [PubMed] [Google Scholar]
- 155.Corton JC, Fan L-Q, Brown S, et al. Down-regulation of cytochrome P450 2C family members and positive acute-phase response gene expression by peroxisome proliferator chemicals. Molecular Pharmacology. 1998;54(3):463–473. doi: 10.1124/mol.54.3.463. [DOI] [PubMed] [Google Scholar]
- 156.Arand M, Coughtrie MWH, Burchell B, Oesch F, Robertson LW. Selective induction of bilirubin UDP-glucuronosyl-transferase by perfluorodecanoic acid. Chemico-Biological Interactions. 1991;77(1):97–105. doi: 10.1016/0009-2797(91)90008-u. [DOI] [PubMed] [Google Scholar]
- 157.Foliot A, Beaune P. Effects of microsomal enzyme inducers on glutathione S-transferase isoenzymes in livers of rats and hamsters. Biochemical Pharmacology. 1994;48(2):293–300. doi: 10.1016/0006-2952(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 158.Barbier O, Villeneuve L, Bocher V, et al. The UDP-glucuronosyltransferase 1A9 enzyme is a peroxisome proliferator-activated receptor α and γ target gene. Journal of Biological Chemistry. 2003;278(16):13975–13983. doi: 10.1074/jbc.M300749200. [DOI] [PubMed] [Google Scholar]
- 159.Sheikh K, Camejo G, Lanne B, Halvarsson T, Landergren MR, Oakes ND. Beyond lipids, pharmacological PPARα activation has important effects on amino acid metabolism as studied in the rat. American Journal of Physiology. 2007;292(4):E1157–E1165. doi: 10.1152/ajpendo.00254.2006. [DOI] [PubMed] [Google Scholar]
- 160.Kersten S, Mandard S, Escher P, et al. The peroxisome proliferator-activated receptor α regulates amino acid metabolism. FASEB Journal. 2001;15(11):1971–1978. doi: 10.1096/fj.01-0147com. [DOI] [PubMed] [Google Scholar]
- 161.Edgar AD, Tomkiewicz C, Costet P, et al. Fenofibrate modifies transaminase gene expression via a peroxisome proliferator activated receptor α-dependent pathway. Toxicology Letters. 1998;98(1-2):13–23. doi: 10.1016/s0378-4274(98)00042-3. [DOI] [PubMed] [Google Scholar]
- 162.Makowski L, Noland RC, Koves TR, et al. Metabolic profiling of PPARα-/- mice reveals defects in carnitine and amino acid homeostasis that are partially reversed by oral carnitine supplementation. FASEB Journal. 2009;23(2):586–604. doi: 10.1096/fj.08-119420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Stienstra R, Mandard S, Tan NS, et al. The Interleukin-1 receptor antagonist is a direct target gene of PPARα in liver. Journal of Hepatology. 2007;46(5):869–877. doi: 10.1016/j.jhep.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 164.Kleemann R, Gervois PP, Verschuren L, Staels B, Princen HMG, Kooistra T. Fibrates down-regulate IL-1-stimulated C-reactive protein gene expression in hepatocytes by reducing nuclear p50-NFκB-C/EBP-β complex formation. Blood. 2003;101(2):545–551. doi: 10.1182/blood-2002-06-1762. [DOI] [PubMed] [Google Scholar]
- 165.Stienstra R, Mandard S, Patsouris D, Maass C, Kersten S, Müller M. Peroxisome proliferator-activated receptor α protects against obesity-induced hepatic inflammation. Endocrinology. 2007;148(6):2753–2763. doi: 10.1210/en.2007-0014. [DOI] [PubMed] [Google Scholar]
- 166.Gervois P, Vu-Dac N, Kleemann R, et al. Negative regulation of human fibrinogen gene expression by peroxisome proliferator-activated receptor alpha agonists via inhibition of CCAAT box/enhancer-binding protein beta. Journal of Biological Chemistry. 2001;276(36):33471–33477. doi: 10.1074/jbc.M102839200. [DOI] [PubMed] [Google Scholar]
- 167.Kockx M, Gervois PP, Poulain P, et al. Fibrates suppress fibrinogen gene expression in rodents via activation of the peroxisome proliferator-activated receptor-α . Blood. 1999;93(9):2991–2998. [PubMed] [Google Scholar]
- 168.Delerive P, De Bosscher K, Berghe WV, Fruchart J-C, Haegeman G, Staels B. DNA binding-independent induction of IκBα gene transcription by PPARα . Molecular Endocrinology. 2002;16(5):1029–1039. doi: 10.1210/mend.16.5.0826. [DOI] [PubMed] [Google Scholar]
- 169.Delerive P, Gervois P, Fruchart J-C, Staels B. Induction of IκBα expression as a mechanism contributing to the anti-inflammatory activities of peroxisome proliferator-activated receptor-α activators. Journal of Biological Chemistry. 2000;275(47):36703–36707. doi: 10.1074/jbc.M004045200. [DOI] [PubMed] [Google Scholar]
- 170.Vanden Berghe W, Vermeulen L, Delerive P, De Bosscher K, Staels B, Haegeman G. A paradigm for gene regulation: inflammation, NF-κB and PPAR. Advances in Experimental Medicine and Biology. 2003;544:181–196. doi: 10.1007/978-1-4419-9072-3_22. [DOI] [PubMed] [Google Scholar]
- 171.Gervois P, Kleemann R, Pilon A, et al. Global suppression of IL-6-induced acute phase response gene expression after chronic in vivo treatment with the peroxisome proliferator-activated receptor-alpha activator fenofibrate. Journal of Biological Chemistry. 2004;279(16):16154–16160. doi: 10.1074/jbc.M400346200. [DOI] [PubMed] [Google Scholar]
- 172.Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W. Control of the peroxisomal β-oxidation pathway by a novel family of nuclear hormone receptors. Cell. 1992;68(5):879–887. doi: 10.1016/0092-8674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- 173.Klaunig JE, Babich MA, Baetcke KP, et al. PPARα agonist-induced rodent tumors: modes of action and human relevance. Critical Reviews in Toxicology. 2003;33(6):655–780. doi: 10.1080/713608372. [DOI] [PubMed] [Google Scholar]
- 174.Lawrence JW, Li Y, Chen S, et al. Differential gene regulation in human versus rodent hepatocytes by peroxisome proliferator-activated receptor (PPAR) α. PPARα fails to induce peroxisome proliferation-associated genes in human cells independently of the level of receptor expression. Journal of Biological Chemistry. 2001;276(34):31521–31527. doi: 10.1074/jbc.M103306200. [DOI] [PubMed] [Google Scholar]
- 175.Roglans N, Bellido A, Rodríguez C, et al. Fibrate treatment does not modify the expression of acyl coenzyme A oxidase in human liver. Clinical Pharmacology and Therapeutics. 2002;72(6):692–701. doi: 10.1067/mcp.2002.128605. [DOI] [PubMed] [Google Scholar]
- 176.Hashimoto T, Cook WS, Qi C, Yeldandi AV, Reddy JK, Rao MS. Defect in peroxisome proliferator-activated receptor α-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. Journal of Biological Chemistry. 2000;275(37):28918–28928. doi: 10.1074/jbc.M910350199. [DOI] [PubMed] [Google Scholar]
- 177.Frerman FE. Acyl-CoA dehydrogenases, electron transfer flavoprotein and electron transfer flavoprotein dehydrogenase. Biochemical Society Transactions. 1988;16(3):416–418. doi: 10.1042/bst0160416. [DOI] [PubMed] [Google Scholar]
- 178.Beckmann JD, Frerman FE. Electron-transfer flavoprotein-ubiquinone oxidoreductase from pig liver: purification and molecular, redox, and catalytic properties. Biochemistry. 1985;24(15):3913–3921. doi: 10.1021/bi00336a016. [DOI] [PubMed] [Google Scholar]
- 179.Honkakoski P, Negishi M. Regulation of cytochrome P450 (CYP) genes by nuclear receptors. Biochemical Journal. 2000;347(2):321–337. doi: 10.1042/0264-6021:3470321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Johnson EF, Hsu M-H, Savas U, Griffin KJ. Regulation of P450 4A expression by peroxisome proliferator activated receptors. Toxicology. 2002;181-182:203–206. doi: 10.1016/s0300-483x(02)00282-2. [DOI] [PubMed] [Google Scholar]
- 181.Cattley RC, DeLuca J, Elcombe C, et al. Do peroxisome proliferating compounds pose a hepatocarcinogenic hazard to humans? Regulatory Toxicology and Pharmacology. 1998;27(1, part 1):47–60. doi: 10.1006/rtph.1997.1163. [DOI] [PubMed] [Google Scholar]
- 182.Cowart LA, Wei S, Hsu M-H, et al. The CYP4A isoforms hydroxylate epoxyeicosatrienoic acids to form high affinity peroxisome proliferator-activated receptor ligands. Journal of Biological Chemistry. 2002;277(38):35105–35112. doi: 10.1074/jbc.M201575200. [DOI] [PubMed] [Google Scholar]
- 183.Oosterveer MH, Grefhorst A, van Dijk TH, et al. Fenofibrate simultaneously induces hepatic fatty acid oxidation, synthesis, and elongation in mice. Journal of Biological Chemistry. 2009;284(49):34036–34044. doi: 10.1074/jbc.M109.051052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Takeuchi Y, Yahagi N, Izumida Y, et al. Polyunsaturated fatty acids selectively suppress sterol regulatory element-binding protein-1 through proteolytic processing and autoloop regulatory circuit. Journal of Biological Chemistry. 2010;285(15):11681–11691. doi: 10.1074/jbc.M109.096107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.König B, Koch A, Spielmann J, et al. Activation of PPARα and PPARγ reduces triacylglycerol synthesis in rat hepatoma cells by reduction of nuclear SREBP-1. European Journal of Pharmacology. 2009;605(1–3):23–30. doi: 10.1016/j.ejphar.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 186.Soupene E, Kuypers FA. Mammalian long-chain acyl-CoA synthetases. Experimental Biology and Medicine. 2008;233(5):507–521. doi: 10.3181/0710-MR-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Issemann I, Prince R, Tugwood J, Green S. A role for fatty acids and liver fatty acid binding protein in peroxisome proliferation? Biochemical Society Transactions. 1992;20(4):824–827. doi: 10.1042/bst0200824. [DOI] [PubMed] [Google Scholar]
- 188.Simon TC, Roth KA, Gordon JI. Use of transgenic mice to map cis-acting elements in the liver fatty acid- binding protein gene (Fabpl) that regulate its cell lineage-specific, differentiation-dependent, and spatial patterns of expression in the gut epithelium and in the liver acinus. Journal of Biological Chemistry. 1993;268(24):18345–18358. [PubMed] [Google Scholar]
- 189.Storch J, McDermott L. Structural and functional analysis of fatty acid-binding proteins. Journal of Lipid Research. 2009;50:S126–S131. doi: 10.1194/jlr.R800084-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yu S, Matsusue K, Kashireddy P, et al. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor γ1 (PPARγ1) overexpression. Journal of Biological Chemistry. 2003;278(1):498–505. doi: 10.1074/jbc.M210062200. [DOI] [PubMed] [Google Scholar]
- 191.Costet P, Legendre C, Moré J, Edgar A, Galtier P, Pineau T. Peroxisome proliferator-activated receptor α-isoform deficiency leads to progressive dyslipidemia with sexually dimorphic obesity and steatosis. Journal of Biological Chemistry. 1998;273(45):29577–29585. doi: 10.1074/jbc.273.45.29577. [DOI] [PubMed] [Google Scholar]
- 192.Reddy JK. Nonalcoholic steatosis and steatohepatitis III. Peroxisomal β-oxidation, PPARα, and steatohepatitis. American Journal of Physiology. 2001;281(6):G1333–G1339. doi: 10.1152/ajpgi.2001.281.6.G1333. [DOI] [PubMed] [Google Scholar]
- 193.Fernández-Miranda C, Pérez-Carreras M, Colina F, López-Alonso G, Vargas C, Solís-Herruzo JA. A pilot trial of fenofibrate for the treatment of non-alcoholic fatty liver disease. Digestive and Liver Disease. 2008;40(3):200–205. doi: 10.1016/j.dld.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 194.Kersten S. Peroxisome proliferator activated receptors and lipoprotein metabolism. PPAR Research. 2008;2008:11 pages. doi: 10.1155/2008/132960. Article ID 132960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Reid BN, Ables GP, Otlivanchik OA, et al. Hepatic overexpression of hormone-sensitive lipase and adipose triglyceride lipase promotes fatty acid oxidation, stimulates direct release of free fatty acids, and ameliorates steatosis. Journal of Biological Chemistry. 2008;283(19):13087–13099. doi: 10.1074/jbc.M800533200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zimmermann R, Strauss JG, Haemmerle G, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306(5700):1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 197.Haemmerle G, Zimmermann R, Hayn M, et al. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. Journal of Biological Chemistry. 2002;277(7):4806–4815. doi: 10.1074/jbc.M110355200. [DOI] [PubMed] [Google Scholar]
- 198.Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. Journal of Biological Chemistry. 2004;279(47):48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 199.He S, McPhaul C, Li JZ, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. Journal of Biological Chemistry. 2010;285(9):6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Huang Y, He S, Li JZ, et al. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(17):7892–7897. doi: 10.1073/pnas.1003585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Deng T, Shan S, Li P-P, et al. Peroxisome proliferator-activated receptor-γ transcriptionally up-regulates hormone-sensitive lipase via the involvement of specificity protein-1. Endocrinology. 2006;147(2):875–884. doi: 10.1210/en.2005-0623. [DOI] [PubMed] [Google Scholar]
- 202.Kim JY, Tillison K, Lee J-H, Rearick DA, Smas CM. The adipose tissue triglyceride lipase ATGL/PNPLA2 is downregulated by insulin and TNF-α in 3T3-L1 adipocytes and is a target for transactivation by PPARγ . American Journal of Physiology. 2006;291(1):E115–E127. doi: 10.1152/ajpendo.00317.2005. [DOI] [PubMed] [Google Scholar]
- 203.Magnusson B, Asp L, Boström P, et al. Adipocyte differentiation-related protein promotes fatty acid storage in cytosolic triglycerides and inhibits secretion of very low-density lipoproteins. Arteriosclerosis, Thrombosis, and Vascular Biology. 2006;26(7):1566–1571. doi: 10.1161/01.ATV.0000223345.11820.da. [DOI] [PubMed] [Google Scholar]
- 204.Dalen KT, Schoonjans K, Ulven SM, et al. Adipose tissue expression of the lipid droplet-associating proteins S3-12 and perilipin is controlled by peroxisome proliferator-activated receptor-γ . Diabetes. 2004;53(5):1243–1252. doi: 10.2337/diabetes.53.5.1243. [DOI] [PubMed] [Google Scholar]
- 205.Straub BK, Stoeffel P, Heid H, Zimbelmann R, Schirmacher P. Differential pattern of lipid droplet-associated proteins and de novo perilipin expression in hepatocyte steatogenesis. Hepatology. 2008;47(6):1936–1946. doi: 10.1002/hep.22268. [DOI] [PubMed] [Google Scholar]
- 206.Puri V, Konda S, Ranjit S, et al. Fat-specific protein 27, a novel lipid droplet protein that enhances triglyceride storage. Journal of Biological Chemistry. 2007;282(47):34213–34218. doi: 10.1074/jbc.M707404200. [DOI] [PubMed] [Google Scholar]
- 207.Puri V, Ranjit S, Konda S, et al. Cidea is associated with lipid droplets and insulin sensitivity in humans. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(22):7833–7838. doi: 10.1073/pnas.0802063105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kim Y-J, Cho SY, Yun CH, Moon YS, Lee TR, Kim SH. Transcriptional activation of Cidec by PPARγ2 in adipocyte. Biochemical and Biophysical Research Communications. 2008;377(1):297–302. doi: 10.1016/j.bbrc.2008.09.129. [DOI] [PubMed] [Google Scholar]
- 209.Matsusue K, Kusakabe T, Noguchi T, et al. Hepatic steatosis in leptin-deficient mice is promoted by the PPARgamma target gene Fsp27. Cell Metabolism. 2008;7(4):302–311. doi: 10.1016/j.cmet.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Frick MH, Elo O, Haapa K. Helsinki Heart Study: primary-prevention trial with gemfibrozil in middle-aged men with dyslipidemia. Safety of treatment, changes in risk factors, and incidence of coronary heart disease. The New England Journal of Medicine. 1987;317(20):1237–1245. doi: 10.1056/NEJM198711123172001. [DOI] [PubMed] [Google Scholar]
- 211.Rubins HB, Robins SJ, Collins D, et al. Gemfibrozil for the secondary prevention of coronary heart disease in men with low levels of high-density lipoprotein cholesterol. The New England Journal of Medicine. 1999;341(6):410–418. doi: 10.1056/NEJM199908053410604. [DOI] [PubMed] [Google Scholar]
- 212.Behar S, Brunner D, Kaplinsky E, Mandelzweig L, Benderly M. Secondary prevention by raising HDL cholesterol and reducing triglycerides in patients with coronary artery disease: the bezafibrate infarction prevention (BIP) study. Circulation. 2000;102(1):21–27. doi: 10.1161/01.cir.102.1.21. [DOI] [PubMed] [Google Scholar]
- 213.Steiner G, Hamsten A, Hosking J, et al. Effect of fenofibrate on progression of coronary-artery disease in type 2 diabetes: the Diabetes Atherosclerosis Intervention Study, a randomised study. The Lancet. 2001;357(9260):905–910. [PubMed] [Google Scholar]
- 214.Améen C, Edvardsson U, Ljungberg A, et al. Activation of peroxisome proliferator-activated receptor α increases the expression and activity of microsomal triglyceride transfer protein in the liver. Journal of Biological Chemistry. 2005;280(2):1224–1229. doi: 10.1074/jbc.M412107200. [DOI] [PubMed] [Google Scholar]
- 215.Camps L, Reina M, Llobera M, Bengtsson-Olivecrona G, Olivecrona T, Vilaro S. Lipoprotein lipase in lungs, spleen, and liver: synthesis and distribution. Journal of Lipid Research. 1991;32(12):1877–1888. [PubMed] [Google Scholar]
- 216.Carroll R, Severson DL. Peroxisome proliferator-activated receptor-α ligands inhibit cardiac lipoprotein lipase activity. American Journal of Physiology. 2001;281(2):H888–H894. doi: 10.1152/ajpheart.2001.281.2.H888. [DOI] [PubMed] [Google Scholar]
- 217.Raspé E, Duez H, Mansén A, et al. Identification of Rev-erbα as a physiological repressor of apoC-III gene transcription. Journal of Lipid Research. 2002;43(12):2172–2179. doi: 10.1194/jlr.m200386-jlr200. [DOI] [PubMed] [Google Scholar]
- 218.Gervois P, Chopin-Delannoy S, Fadel A, et al. Fibrates increase human REV-ERBα expression in liver via a novel peroxisome proliferator-activated receptor response element. Molecular Endocrinology. 1999;13(3):400–409. doi: 10.1210/mend.13.3.0248. [DOI] [PubMed] [Google Scholar]
- 219.Qu S, Su D, Altomonte J, et al. PPARα mediates the hypolipidemic action of fibrates by antagonizing FoxO1. American Journal of Physiology. 2007;292(2):E421–E434. doi: 10.1152/ajpendo.00157.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Schultze AE, Alborn WE, Newton RK, Konrad RJ. Administration of a PPARα agonist increases serum apolipoprotein A-V levels and the apolipoprotein A-V/apolipoprotein C-III ratio. Journal of Lipid Research. 2005;46(8):1591–1595. doi: 10.1194/jlr.C500010-JLR200. [DOI] [PubMed] [Google Scholar]
- 221.Tao H, Aakula S, Abumrad NN, Hajri T. Peroxisome proliferator-activated receptor-γ regulates the expression and function of very-low-density lipoprotein receptor. American Journal of Physiology. 2010;298(1):E68–E79. doi: 10.1152/ajpendo.00367.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bard J-M, Parra H-J, Camare R, et al. A multicenter comparison of the effects of simvastatin and fenofibrate therapy in severe primary hypercholesterolemia, with particular emphasis on lipoproteins defined by their apolipoprotein composition. Metabolism. 1992;41(5):498–503. doi: 10.1016/0026-0495(92)90208-r. [DOI] [PubMed] [Google Scholar]
- 223.Lussier-Cacan S, Bard J-M, Boulet L, et al. Lipoprotein composition changes induced by fenofibrate in dysbetalipoproteinemia type III. Atherosclerosis. 1989;78(2-3):167–182. doi: 10.1016/0021-9150(89)90221-9. [DOI] [PubMed] [Google Scholar]
- 224.Duezt H, Chao Y-S, Hernandez M, et al. Reduction of atherosclerosis by the peroxisome proliferator-activated receptor α agonist fenofibrate in mice. Journal of Biological Chemistry. 2002;277(50):48051–48057. doi: 10.1074/jbc.M206966200. [DOI] [PubMed] [Google Scholar]
- 225.Staels B, Van Tol A, Andreu T, Auwerx J. Fibrates influence the expression of genes involved in lipoprotein metabolism in a tissue-selective manner in the rat. Arteriosclerosis and Thrombosis. 1992;12(3):286–294. doi: 10.1161/01.atv.12.3.286. [DOI] [PubMed] [Google Scholar]
- 226.Sanderson LM, Boekschoten MV, Desvergne B, Müller M, Kersten S. Transcriptional profiling reveals divergent roles of PPARα and PPARβ/δ in regulation of gene expression in mouse liver. Physiological Genomics. 2010;41(1):42–52. doi: 10.1152/physiolgenomics.00127.2009. [DOI] [PubMed] [Google Scholar]
- 227.Ishida T, Choi S, Kundu RK, et al. Endothelial lipase is a major determinant of HDL level. Journal of Clinical Investigation. 2003;111(3):347–355. doi: 10.1172/JCI16306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Jin W, Millar JS, Broedl U, Glick JM, Rader DJ. Inhibition of endothelial lipase causes increased HDL cholesterol levels in vivo. Journal of Clinical Investigation. 2003;111(3):357–362. doi: 10.1172/JCI16146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Broedl UC, Maugeais C, Marchadier D, Glick JM, Rader DJ. Effects of nonlipolytic ligand function of endothelial lipase on high density lipoprotein metabolism in vivo. Journal of Biological Chemistry. 2003;278(42):40688–40693. doi: 10.1074/jbc.M304367200. [DOI] [PubMed] [Google Scholar]
- 230.Desager J-P, Horsmans Y, Vandenplas C, Harvengt C. Pharmacodynamic activity of lipoprotein lipase and hepatic lipase, and pharmacokinetic parameters measured in normolipidaemic subjects receiving ciprofibrate (100 or 200 mg/day) or micronised fenofibrate (200 mg/day) therapy for 23 days. Atherosclerosis. 1996;124(supplement):S65–S73. doi: 10.1016/0021-9150(96)05859-5. [DOI] [PubMed] [Google Scholar]
- 231.Kang HW, Kanno K, Scapa EF, Cohen DE. Regulatory role for phosphatidylcholine transfer protein/StarD2 in the metabolic response to peroxisome proliferator activated receptor alpha (PPARα) Biochimica et Biophysica Acta. 2010;1801(4):496–502. doi: 10.1016/j.bbalip.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Jitrapakdee S, Slawik M, Medina-Gomez G, et al. The peroxisome proliferator-activated receptor-γ regulates murine pyruvate carboxylase gene expression in vivo and in vitro. Journal of Biological Chemistry. 2005;280(29):27466–27476. doi: 10.1074/jbc.M503836200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sugden MC, Bulmer K, Gibbons GF, Holness MJ. Role of peroxisome proliferator-activated receptor-α in the mechanism underlying changes in renal pyruvate dehydrogenase kinase isoform 4 protein expression in starvation and after refeeding. Archives of Biochemistry and Biophysics. 2001;395(2):246–252. doi: 10.1006/abbi.2001.2586. [DOI] [PubMed] [Google Scholar]
- 234.Wu P, Inskeep K, Bowker-Kinley MM, Popov KM, Harris RA. Mechanism responsible for inactivation of skeletal muscle pyruvate dehydrogenase complex in starvation and diabetes. Diabetes. 1999;48(8):1593–1599. doi: 10.2337/diabetes.48.8.1593. [DOI] [PubMed] [Google Scholar]
- 235.Holness MJ, Bulmer K, Gibbons GF, Sugden MC. Up-regulation of pyruvate dehydrogenase kinase isoform 4 (PDK4) protein expression in oxidative skeletal muscle does not require the obligatory participation of peroxisome-proliferator-activated receptor α (PPARα) Biochemical Journal. 2002;366(3):839–846. doi: 10.1042/BJ20020754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Holness MJ, Smith ND, Bulmer K, Hopkins T, Gibbons GF, Sugden MC. Evaluation of the role of peroxisome-proliferator-activated receptor α in the regulation of cardiac pyruvate dehydrogenase kinase 4 protein expression in response to starvation, high-fat feeding and hyperthyroidism. Biochemical Journal. 2002;364(3):687–694. doi: 10.1042/BJ20011841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Knight BL, Patel DD, Humphreys SM, Wiggins D, Gibbons GF. Inhibition of cholesterol absorption associated with a PPARα-dependent increase in ABC binding cassette transporter A1 in mice. Journal of Lipid Research. 2003;44(11):2049–2058. doi: 10.1194/jlr.M300042-JLR200. [DOI] [PubMed] [Google Scholar]
- 238.Stahlberg D, Angelin B, Einarsson K. Effects of treatment with clofibrate, bezafibrate, and ciprofibrate on the metabolism of cholesterol in rat liver microsomes. Journal of Lipid Research. 1989;30(7):953–958. [PubMed] [Google Scholar]
- 239.Ståhlberg D, Reihnér E, Rudling M, Berglund L, Einarsson K, Angelin BO. Influence of bezafibrate on hepatic cholesterol metabolism in gallstone patients: reduced activity of cholesterol 7α-hydroxylase. Hepatology. 1995;21(4):1025–1030. doi: 10.1002/hep.1840210421. [DOI] [PubMed] [Google Scholar]
- 240.Bertolotti M, Concari M, Loria P, et al. Effects of different phenotypes of hyperlipoproteinemia and of treatment with fibric acid derivatives on the rates of cholesterol 7α-hydroxylation in humans. Arteriosclerosis, Thrombosis, and Vascular Biology. 1995;15(8):1064–1069. doi: 10.1161/01.atv.15.8.1064. [DOI] [PubMed] [Google Scholar]
- 241.Miyata KS, McCaw SE, Patel HV, Rachubinski RA, Capone JP. The orphan nuclear hormone receptor LXRα interacts with the peroxisome proliferator-activated receptor and inhibits peroxisome proliferator signaling. Journal of Biological Chemistry. 1996;271(16):9189–9192. doi: 10.1074/jbc.271.16.9189. [DOI] [PubMed] [Google Scholar]
- 242.Yoshikawa T, Ide T, Shimano H, et al. Cross-talk between peroxisome proliferator-activated receptor (PPAR) α and liver X receptor (LXR) in nutritional regulation of fatty acid metabolism. I. PPARS suppress sterol regulatory element binding protein-1c promoter through inhibition of LXR signaling. Molecular Endocrinology. 2003;17(7):1240–1254. doi: 10.1210/me.2002-0190. [DOI] [PubMed] [Google Scholar]
- 243.Gbaguidi GF, Agellon LB. The atypical interaction of peroxisome proliferator-activated receptor α with liver X receptor α antagonizes the stimulatory effect of their respective ligands on the murine cholesterol 7α-hydroxylase gene promoter. Biochimica et Biophysica Acta. 2002;1583(2):229–236. doi: 10.1016/s1388-1981(02)00217-2. [DOI] [PubMed] [Google Scholar]
- 244.Leuenberger N, Pradervand S, Wahli W. Sumoylated PPARα mediates sex-specific gene repression and protects the liver from estrogen-induced toxicity in mice. Journal of Clinical Investigation. 2009;119(10):3138–3148. doi: 10.1172/JCI39019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Walters MW, Wallace KB. Urea cycle gene expression is suppressed by PFOA treatment in rats. Toxicology Letters. 2010;197(1):46–50. doi: 10.1016/j.toxlet.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 246.Thulin P, Rafter I, Stockling K, et al. PPARα regulates the hepatotoxic biomarker alanine aminotransferase (ALT1) gene expression in human hepatocytes. Toxicology and Applied Pharmacology. 2008;231(1):1–9. doi: 10.1016/j.taap.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 247.Delerive P, De Bosscher K, Besnard S, et al. Peroxisome proliferator-activated receptor α negatively regulates the vascular inflammatory gene response by negative cross-talk with transcription factors NF-κB and AP-1. Journal of Biological Chemistry. 1999;274(45):32048–32054. doi: 10.1074/jbc.274.45.32048. [DOI] [PubMed] [Google Scholar]
- 248.Delerive P, Fruchart J-C, Staels B. Peroxisome proliferator-activated receptors in inflammation control. Journal of Endocrinology. 2001;169(3):453–459. doi: 10.1677/joe.0.1690453. [DOI] [PubMed] [Google Scholar]
- 249.Staels B, Koenig W, Habib A, et al. Activation of human aortic smooth-muscle cells is inhibited by PPARα but not by PPARγ activators. Nature. 1998;393(6687):790–793. doi: 10.1038/31701. [DOI] [PubMed] [Google Scholar]
- 250.Madej A, Okopien B, Kowalski J, et al. Effects of fenofibrate on plasma cytokine concentrations in patients with atherosclerosis and hyperlipoproteinemia IIb. International Journal of Clinical Pharmacology and Therapeutics. 1998;36(6):345–349. [PubMed] [Google Scholar]
- 251.Martin F, Penet M-F, Malergue F, et al. Vanin-1-/- mice show decreased NSAID- and Schistosoma-induced intestinal inflammation associated with higher gluthathione stores. Journal of Clinical Investigation. 2004;113(4):591–597. doi: 10.1172/JCI19557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Berruyer C, Pouyet L, Millet V, et al. Vanin-1 licenses inflammatory mediator production by gut epithelial cells and controls colitis by antagonizing peroxisome proliferator-activated receptor γ activity. Journal of Experimental Medicine. 2006;203(13):2817–2827. doi: 10.1084/jem.20061640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Pouyet L, Roisin-Bouffay C, Clément A, et al. Epithelial vanin-1 controls inflammation-driven carcinogenesis in the colitis-associated colon cancer model. Inflammatory Bowel Diseases. 2010;16(1):96–104. doi: 10.1002/ibd.21031. [DOI] [PubMed] [Google Scholar]
- 254.Ip E, Farrell GC, Robertson G, Hall P, Kirsch R, Leclercq I. Central role of PPARα-dependent hepatic lipid turnover in dietary steatohepatitis in mice. Hepatology. 2003;38(1):123–132. doi: 10.1053/jhep.2003.50307. [DOI] [PubMed] [Google Scholar]
- 255.Ip E, Farrell G, Hall P, Robertson G, Leclercq I. Administration of the potent PPARα agonist, Wy-14,643, reverses nutritional fibrosis and steatohepatitis in mice. Hepatology. 2004;39(5):1286–1296. doi: 10.1002/hep.20170. [DOI] [PubMed] [Google Scholar]
- 256.Kashireddy PV, Rao MS. Lack of peroxisome proliferator-activated receptor α in mice enhances methionine and choline deficient diet-induced steatohepatitis. Hepatology Research. 2004;30(2):104–110. doi: 10.1016/j.hepres.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 257.Shiri-Sverdlov R, Wouters K, Gorp PJV, et al. Early diet-induced non-alcoholic steatohepatitis in APOE2 knock-in mice and its prevention by fibrates. Journal of Hepatology. 2006;44(4):732–741. doi: 10.1016/j.jhep.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 258.Nakajima T, Kamijo Y, Tanaka N, et al. Peroxisome proliferator-activated receptor α protects against alcohol-induced liver damage. Hepatology. 2004;40(4):972–980. doi: 10.1002/hep.20399. [DOI] [PubMed] [Google Scholar]
- 259.Romics L, Jr., Kodys K, Dolganiuc A, et al. Diverse regulation of NF-κB and peroxisome proliferator-activated receptors in murine nonalcoholic fatty liver. Hepatology. 2004;40(2):376–385. doi: 10.1002/hep.20304. [DOI] [PubMed] [Google Scholar]
- 260.Yu J, Ip E, Dela Peña A, et al. COX-2 induction in mice with experimental nutritional steatohepatitis: role as pro-inflammatory mediator. Hepatology. 2006;43(4):826–836. doi: 10.1002/hep.21108. [DOI] [PubMed] [Google Scholar]
- 261.Rushmore TH, Kong A-NT. Pharmacogenomics, regulation and signaling pathways of phase I and II drug metabolizing enzymes. Current Drug Metabolism. 2002;3(5):481–490. doi: 10.2174/1389200023337171. [DOI] [PubMed] [Google Scholar]
- 262.Bock KW. Vertebrate UDP-glucuronosyltransferases: functional and evolutionary aspects. Biochemical Pharmacology. 2003;66(5):691–696. doi: 10.1016/s0006-2952(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 263.König J, Nies AT, Cui Y, Leier I, Keppler D. Conjugate export pumps of the multidrug resistance protein (MRP) family: localization, substrate specificity, and MRP2-mediated drug resistance. Biochimica et Biophysica Acta. 1999;1461(2):377–394. doi: 10.1016/s0005-2736(99)00169-8. [DOI] [PubMed] [Google Scholar]
- 264.Voskoboinik I, Drew R, Ahokas JT. Differential effect of peroxisome proliferators on rat glutathione S-transferase isoenzymes. Toxicology Letters. 1996;87(2-3):147–155. doi: 10.1016/0378-4274(96)03371-1. [DOI] [PubMed] [Google Scholar]
- 265.James SI, Ahokas JT. Effect of peroxisome proliferators on glutathione-dependent sulphobromophthalein excretion. Xenobiotica. 1992;22(12):1425–1432. doi: 10.3109/00498259209056692. [DOI] [PubMed] [Google Scholar]