Abstract
Base analogs are powerful antimetabolites and dangerous mutagens generated endogenously by oxidative stress, inflammation, and aberrant nucleotide biosynthesis. Human inosine triphosphate pyrophosphatase (ITPA) hydrolyzes triphosphates of noncanonical purine bases (i.e., ITP, dITP, XTP, dXTP, or their mimic: 6-hydroxyaminopurine (HAP) deoxynucleoside triphosphate) and thus regulates nucleotide pools and protects cells from DNA damage. We demonstrate that the model purine base analog HAP induces DNA breaks in human cells and leads to elevation of levels of ITPA. A human polymorphic allele of the ITPA, 94C->A encodes for the enzyme with a P32T amino-acid change and leads to accumulation of nonhydrolyzed ITP. The polymorphism has been associated with adverse reaction to purine base-analog drugs. The level of both spontaneous and HAP-induced DNA breaks is elevated in the cell line with the ITPA P32T variant. The results suggested that human ITPA plays a pivotal role in the protection of DNA from noncanonical purine base analogs.
1. Introduction
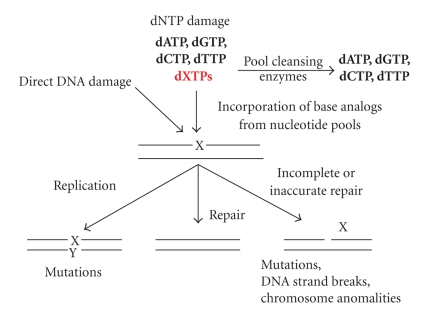
Modified bases in DNA pose a severe threat for genome integrity [1], (Figure 1). DNA could be directly damaged by environmental factors such as ionizing radiation, chemical mutagens or endogenous factors, such as oxidative stress and inflammation [2–4]. Many of these factors also damage nucleotides in DNA precursor pools [5–7]. Additionally, cellular metabolism per se also contributes to the contamination of pools by nucleobase-analogs. Base-analogs in the deoxyribonucleosidetriphosphate form are incorporated into DNA by DNA polymerases and are the source of genetic changes [8]. Potent repair systems remove not only the lesions from DNA, but also the harmful triphosphates from the DNA precursor pools (Figure 1, [1, 8]). Defects of these protection mechanisms lead to hypermutagenesis [9] or hyperrecombination [10–12] and result in genome instability, which predisposes individuals to diseases like cancer [13, 14]. Base-analogs are clinically important and are widely used as immunosuppressants as well, antiviral and anticancer agents. The determination of the reasons for individual sensitivity/resistance is of high priority.
Figure 1.
Base-analog DNA cycle. Environmental and intrinsic mutagens can damage DNA directly or can damage DNA precursor pools. When cleansing is inefficient, base-analogs are incorporated into DNA. Damaged bases lead to mutations in replication cycles or can be correctly repaired by base excision repair. Intermediates of this repair can lead to mutagenesis, DNA breaks, and chromosome changes.
The major mutagenic modified purine bases include 8-oxoguanine and 2-hydroxyadenine [15]. The 8-oxoguanine in deoxyribonucleoside triphosphate form can be incorporated into DNA by replicative as well as specialized DNA polymerases [16–18]. It can form base pairs with cytosine and adenine and, therefore, can lead to transversion mutations [19]. The MutT protein of E. coli hydrolyzes 8-oxodGTP to 8-oxodGMP, preventing incorporation into DNA [17]. Mutational inactivation of this gene leads to a 10.000-fold increase in the rates of transversions [20], but no elevation of DNA fragmentation is detected in mutT strains [21]. The 2-hydroxyadenine also has strong mutagenic potential [3]. Deamination of normal purines, as well as disregulation of the purine biosynthesis, leads to contamination of nucleotide pools with deoxyinosine triphosphate and deoxyxanthine triphosphate. Inflammation induces various types of base damage from oxidative stress and elevated lipid peroxidation [22]. The latter can generate mutagenic derivatives of adenine and guanine, HAP and its 2-amino derivative in vitro [23]. HAP might be generated by monooxygenation of adenine [24]. HAP also can be generated by adenylosuccinate synthase, an enzyme of the de novo purine biosynthesis pathway wherein hydroxylamine is provided instead of aspartate in the reaction with IMP [25]. Thus, HAP can be a natural contaminant of dNTP pools [23, 26], however the literature does not report direct measurements of HAP derivatives in nucleotide pools. HAP is strongly mutagenic in bacteria and yeast [26, 27]. The dHAPTP is incorporated into DNA by various DNA polymerases in vitro [28, 29]. Overwhelming indirect genetic evidence in microorganisms suggests that the mutagenic effect is mediated by dHAPTP incorporation into DNA [26]. There is only fragmentary information about the effects of endogenous and exogenous base-analogs, such as HAP, in multicellular eukaryotes.
There are a number of evolutionarily conserved enzymatic systems that sanitize the nucleotide pool by selectively breaking deoxynucleoside triphosphate forms of base-analog DNA precursors, either to monophosphate [17, 30] or to nucleoside [31, 32] or diphosphate [33]. Additional systems affect quality of dNTP pools [34]. One of them, NUDT16, destroys abnormal diphosphates [35]. Polymorphisms in the genes encoding protective enzymes are associated with an increased risk of cancer [36], a predisposition to base-analog-associated adverse drug reactions [37] or a modulation of response to therapy in hepatitis C patients [38].
ITPA is a prominent enzyme protecting from base-analogs [27, 39]. ITPA orthologs from humans, yeast (encoded by the HAM1 gene), and bacteria (encoded by the rdgB gene) control levels of ITP, dITP and dHAPTP by hydrolyzing ITP/dITP to PPi and IMP/dIMP [27, 30, 40]. One report documents that yeast ITPA can hydrolyze pyrimidine analogs as well [41]. ITPA is highly conserved among species [26, 40, 42]. In E. coli, the rdgB mutation is synthetically lethal with the recA mutation abolishing homologous recombination [27, 39]. The rdgB mutation sensitizes to the mutagenic and recombinogenic effects of HAP in molybdenum-cofactor defective strain background (another system protecting from HAP [43–45]) because of a massive accumulation of breaks in DNA [27, 39, 46]. DNA damage is caused by intermediates in the repair of base-analogs in DNA by Endo V encoded by the nfi gene. This has been proven by the viability of triple rdgB recA nfi mutants [27].
HAP is very mutagenic in yeast but does not induce recombination [48]. A defect in the yeast homolog of ITPA, Ham1, leads to elevated HAPmutagenesis, but it does not affect spontaneous mutagenesis [49]. The natural substrate, dITP, seems to be non-mutagenic when it is incorporated in DNA, because hypoxanthine base pairs properly with cytosine [50], and yeast apparently do not possess an enzyme able to recognize hypoxanthine, xanthine and HAP in DNA similar to Endo V. Therefore, DNA with these bases is not nicked and no recombination and DNA breakage are seen. The situation in yeast might be atypical. Inosine and xanthine in DNA are recognized in most organisms by a specialized repair system initiated by the orthologs of endonuclease V [51] and elicit DNA repair reactions that lead to DNA fragmentation and genomic instability when the level of analogues is high [3, 39, 46].
The repair of purine base-analogs in humans has not been studied. Mutations in the human ITPA gene lead to the accumulation of ITP in erythrocytes but do not show a clear disease phenotype, perhaps due to compensation by other cleansing enzymes [52, 53]. Human ITPA P32T variant, abolishing the ITPA activity in erythrocytes, has been associated in most publications with adverse reactions to purine analogues used in the treatment of blood cancers, transplant, and inflammatory bowel diseases [37, 54–59]. The ITPA P32T variant causes sensitivity to mercaptopurine used for the treatment of acute lymphoblastic leukemia [60].
Knockout of the Itpa gene in mice is lethal primarily because of heart failure [53]. Primary embryonic fibroblasts exhibit moderate chromosome instability phenotype [61], and the inviability of the Itpa knockout mice suggests that the enzyme performs essential functions in addition to the prevention of the misincorporation of purine base-analogs in DNA. If we assume that the role of ITPA is the same in humans, the ITPA P32T variant should result in an incomplete loss of activity, at least in the heart. Indeed, ITPA with the change possesses enzymatic activity but is thermally unstable [62, 63], suggesting a possibility that the levels of the protein and its activity could vary among tissues.
In this study, we demonstrate by comet assay that the model purine base-analog, HAP, induces DNA strand breaks in three different human cell lines. This suggests that there is active recognition of incorporated base-analogs and nicking of DNA. The inducibility of the system of HAP repair was indicated by the shape of HAP dose response on the frequency of DNA strand breaks. We suggest and provide the evidence that one component of the repair system is ITPA. Consistent with this, the level of spontaneous and base-analog-induced DNA damage was elevated in cell line P32T with compromised ITPA activity. The levels and distribution of ITPA P32T as determined by immunostaining have been changed in the P32T cell line. The results suggest that patients with the 94C->A polymorphism in the ITPA in addition to drug intolerance might possess increased predisposition to diseases resulting from DNA repair defects.
2. Materials and Methods
2.1. Human Cell Cultures
The normal, diploid, human lung fibroblast cell line, WI-38, (ATCC CCL-75) was kindly provided by Dr. Vera Gorbunova (University of Rochester, NY). The human fibroblast cell line (Coriell Institute Biorepository (GMO1617), called here P32T, is homozygous for a C>A transversion of nucleotide 94 (94C>A) in exon 2 of the ITPA gene. This leads to a proline to threonine substitution at codon 32 (P32T). These untransformed cell lines were cultivated as a monolayer in MEM (Invitrogen, USA) supplemented with 10% fetal calf serum (GIBCO) and 1 mM sodium pyruvate (Invitrogen, USA) at 37°C in a 5% CO2 atmosphere. For the fibroblasts, cells at early passages (<25 passages) were used in all experiments to avoid complications of replicative senescence because WI-38 cells have a mean lifespan of approximately 45 to 60 population doublings.
The epithelial colorectal cancer cell line HCT116 (ATCC, CCL247, kindly provided by Dr. Robert E. Lewis, UNMC), was cultivated in DMEM (Invitrogen, USA) containing 10% fetal calf serum at 37°C in a 5% CO2 atmosphere. The colorectal cancer HCT116 cells are defective in mismatch repair due to a nonsense mutation in the MLH1 gene [64]. For the experiment addressing the specificity of our antibodies against ITPA, we transfected colorectal carcinoma 293 cells by pEGFP plasmid (obtained from Dr. A. Rizzino, UNMC) with ITPA cloned into BamH1-EcoRI sites in frame with EGFP.
2.2. Characterization of P32T Fibroblast Cell Line
We verified the presence of the 94C->A change in the ITPA gene by sequencing of exon 2 amplified from genomic DNA (as exon 2–4 fragment) or from RNA. For genomic DNA isolation, 1 × 106 cells were harvested for fibroblasts bearing wild-type (WI-38) or mutant (P32T) ITPA, and DNA was isolated according to the Fermentas Inc. protocol: (http://www.fermentas.com/en/support/application-protocols). Briefly, cell pellets were lysed with SDS and Proteinase K. After incubation with NaCl, DNA was phenol:chloroform extracted and precipitated with ethanol. DNA pellets thus obtained were resuspended in nuclease-free water. For RNA extraction we have used the RNeasy kit (Qiagen, USA). The cDNA synthesis was performed using the qScript DNA synthesis kit (Quanta Biosciences #95047-025).
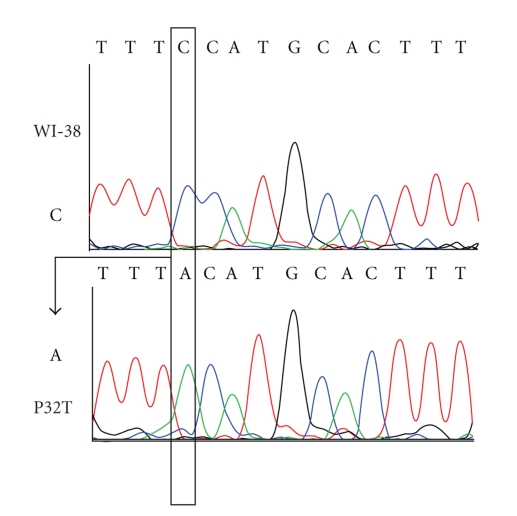
For amplification of the specific ITPA region encompassing the site of the 94C->A change, either genomic DNA or cDNA was diluted 100-fold and exons 2 through 4 were amplified using Exons 2, 3, 4 forward and reverse primers and conditions [65]. Sequencing of the genomic fragment was performed by the same primers and the sequence of the cDNA fragment was performed using ITPA-sN 5′TCATTGGTGGGGAAGAAGATC and ITPA-sC 5′AAGCTGCCAAACTGCCAAA. The sequencing confirmed that the P32T cell line possesses 94C->A transversions (Figure 2).
Figure 2.
Verification of the genotype of the P32T cell line. Electropherogram of the DNA sequence of the region with C94A mutation is shown. The experiment was performed as described in Section 2.
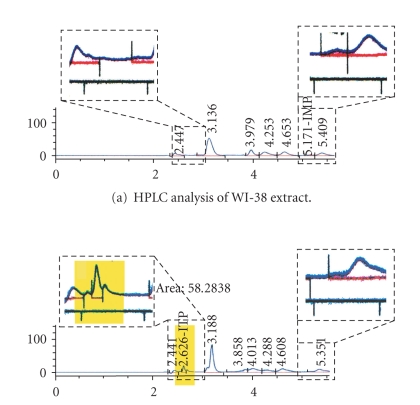
We also detected the hallmark accumulation of ITP in the P32T cell line by HPLC, confirming that ITPA activity is compromised in this cell line [65] (Figure 3).
Figure 3.
Accumulation of ITP in the P32T cell line. Areas of chromatogram corresponding to ITP and IMP are enlarged in boxes above the actual printout. Whole-cell lysates corresponding to 100 μg of protein were deproteinized by acid extraction with 1 N HCl followed by alkali treatment with an equal volume of 1 N NaOH. The samples were centrifuged at 14,000 RPM for 10 min. The volume of aqueous fraction (supernatant) was adjusted to 500 uL with HPLC-grade water. The samples were run on an Alltech hypersil BDS C18 5u column. The HP 1100 HPLC connected to an autosampler and a DAD detector (which was set at 262 nm) was used. Flow rate was kept constant at 1.1 mL/min, and a gradient flow was used: 0 to 5 minutes, 100% (a). 5–5 : 10 ramp up to 100% (b), 5 : 10–8 : 10, 100% (b), 8 : 10–8 : 20 ramp down to 100% (a). In the preliminary experiments we have determined the retention of pure 0.5 mM solutions of ITP (~2.6) and IMP (~5.2).
2.3. The Alkaline Comet Assay
All the required chemicals were purchased from Trevigen, Inc. (USA). The comet assay was carried out under alkaline conditions, as described in the attached Trevigen instructions. Cells (WI-38, HCT116 and P32T) with or without H2O2 or HAP treatment were suspended in 1% low melting point agarose in 1xPBS, pH 7.4, at 37°C and immediately pipetted onto a CometSlide. The agarose was allowed to set at 4°C for 10–30 min and the slide was immersed in a lysis solution at 4°C for 50 min to remove cellular proteins. Slides were then placed at 0.3 M NaOH and 1 mM EDTA for 45 min at room temperature before electrophoresis at 300 mA for 60 min at 4°C. The slides were then washed two times for 10 min each with water and then dehydrated in 70% ethanol for 5 min before staining with 1xSYBR Green I staining solution. To prevent background DNA damage, handling samples and all the steps included in the preparation of the slides for the comet assay were conducted under yellow light or in the dark.
The slides were examined using an epifluorescence microscope “Nikon Eclipse 80i.” A total of 100 comets per slide were scored. Comets were randomly captured at a constant depth of the gel, avoiding the edges of the gel, occasional dead cells, and superimposed comets. The percent (%) of DNA in the comet tail was used in this study as the measure of DNA damage. Consistent with the Trevigen Inc. application guide, the average content of DNA in the comet tail of untreated normal cells is less than ten percent. The amount of DNA in the comet tail was estimated by computerized image analysis of selected comets using CometScore software. Six independent experiments were performed. Then, the average and standard error was determined. The statistical significance of differences was estimated by Student's t-criterion.
2.4. Chromosome Analysis
For metaphase chromosome spreads, WI-38, HCT116, and P32T cells (treated for 23 h with DMSO or HAP in DMSO) were arrested in metaphase by a 1 h treatment with 0.5 μg/ml colcemide (Gibco URL, USA), treated hypotonically with 0.075 M KCl, fixed three times in a 3 : 1 methanol-acetic acid mixture, spread on glass slides, and air dried. Twenty spreads for each culture were analyzed by CytoVision software (Genetix Corp., CA); only the total number and size of the chromosomes were determined.
2.5. Western Blot Analysis
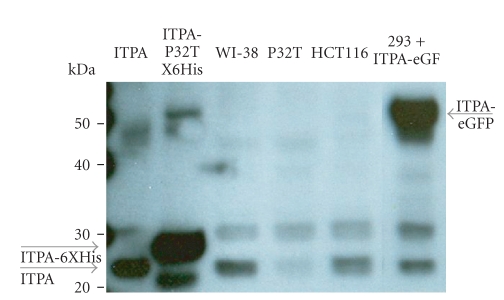
HCT116, WI-38, and P32T cells were cultivated until subconfluence (5 × 105 cells per plate). The cells were harvested and resuspended in the lysis buffer (1xPBS containing a protease inhibitor cocktail (Roche Biochemicals, IN, USA), pH = 7.4). Each pellet was mechanically sheared using a pestle. The lysate was cleared by centrifugation and protein content determined by Bradford reagent from BioRad. The lysate equivalent to 100 μg of protein was boiled in Laemli's buffer (Invitrogen, USA). The protein samples were resolved on a 10%–20% Tris-Glycine gel (Invitrogen, USA) by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked overnight in commercially available blocking buffer (Thermo Scientific, USA). Membranes were then incubated in 1 : 500 dilutions of primary antibody against ITPA (the in-house polyclonal antibodies against ITPA are described elsewhere [55]) and GAPDH (Cell Signaling #2118) for one hour at room temperature. The membranes were then washed with commercially available washing buffer (Thermoscientific, USA) five times for 10 min each. This was followed by incubation with secondary antibody (1 : 1500 dilution) (Cell signaling #7074) for 40 min at room temperature. This was followed by washing (5 times for 10 min each) and detection by the ECL system (ThermoScientific, USA) according to the manufacturer's instructions. The results are presented in Figure 4. A major visible band, corresponding to ITPA (compare with lane with pure ITPA), is prominent in WI-38 fibroblasts, and cancer HCT116 cell extracts (gray arrow) but is less pronounced in P32T cells, as we described before [55]. The intensity of the non-specific 30 kDa band is similar in all three cell lines. The position of the immunoreactive band was shifted up, closer to the 30 kDa marker, in lane with 6 His-tagged pure ITPA-P32T (predicted molecular mass 23.6 kDa [47]). We also analyzed the extract of 293 cell lines transfected with a plasmid expressing the gene for ITPA-EGFP fusion protein (51.4 kDa) and have found that a major band was detected at a position corresponding to 50 kDA. The data unequivocally prove that antibodies react primarily with ITPA.
Figure 4.
Specificity of polyclonal antibodies against ITPA in Western blot. Western blots were performed as described in Section 2. Antibodies against ITPA react largely with ITPA (a band closer to 20 kDa marker, see compared with lane where pure ITPA was loaded, and predicted molecular mass was 21.4 kDa) and, less intense, with an unrelated protein running at 30 kDa. The level of ITPA, but not of this non-specific protein, is decreased almost 10-fold in the soluble fraction of the P32T cell line. In the control reprobing experiment the amount of GAPDH was the same in all three cell lines (not shown). The position of the immunoreactive band was shifted up, closer to the 30 kDa marker in the lane with the 6 His-tagged pure ITPA-P32T (predicted molecular mass 23.6 kDa [47]). We also analyzed the extracts of 293 cell lines transfected with expressing ITPA-EGFP fusion protein (51.4 kDa) and have found that the major band was detected at a position corresponding to 50 kD.
For the analysis of ITPA induction by HAP by Western blots we cultivated HCT116 cells with or without HAP until subconfluence (5 × 105 cells per plate). The cells were harvested and resuspended in the lysis buffer (50 mM Tris, pH = 8.0, 1 mM PMSF, 10% (v/v) glycerol, 0.5% Triton X-100) and disrupted and processed as before with the following modifications to improve the quality and resolution. The lysate equivalent to 30 μg of protein was boiled in Laemli's buffer containing 100 mM DTT and loaded on a 10%–20% Tris-Glycine gel by SDS-PAGE. After trial run and Coomassie staining the amount of extracts was further adjusted to produce equal amount of loaded protein in the control and treatment and run in the new gel. After transfer, nitrocellulose membranes were blocked overnight in 5% dry milk/1x PBST. Membranes were incubated in 1 : 2000 dilutions of primary antibody against ITPA described above for one hour at room temperature. The membranes were washed with 1x PBST three times for 15 min each, and, incubated with secondary HRP-linked antibody (1 : 100000 dilution) (Cell Signaling #7074) for 1 hour at room temperature. Membrane was washed three times for 15 min each and signals were detected by SuperSignal West Femto Maximum Sensitivity Substrate (ThermoScientific, USA) according to the manufacturer's instructions.
2.6. Immunocytochemistry
Cells (WI-38, HCT116 and P32T) with or without HAP treatment were fixed with methanol acetic acid mixture (3 : 1). All procedures were performed at room temperature. To prevent non-specific binding in the consequent antibody detection, samples were blocked in 1xPBS containing 5% BSA and 0.05% Tritone X-100. We have used the primary antibodies against ITPA described in the previous section and a goat-antirabbit antibody Alexa Fluor 488 nm conjugated (Thermoscientific, USA). Both antibodies were diluted 1 : 1000 in 1xPBS containing 5% BSA and 0.05% Triton X-100. Slides were washed three times in 1xPBST buffer between incubations with primary and secondary antibodies and after incubations. After washing, the cells were counterstained with DAPI, mounted in antifade medium and analyzed by fluorescent microscopy. No fluorescence was detected when primary antibodies were omitted from the protocol, and very low signal was detected when preimmune rabbit serum was used in place of primary antibody, suggesting that fluorescence signals were absolutely dependent on antiITPA antibodies. Three independent repeats of this experiment have been done.
2.7. Microscopy and Image Analysis
After the comet assay or immunocytochemistry procedures, cells were examined on a Nikon Eclipse 80i microscope. Images were recorded separately by a CCD device Photometrics CoolSnap cf and merged using Adobe Photoshop software.
3. Results
3.1. HAP Does not Cause Chromosome Hyperfragmentation in Three Human Cell Lines
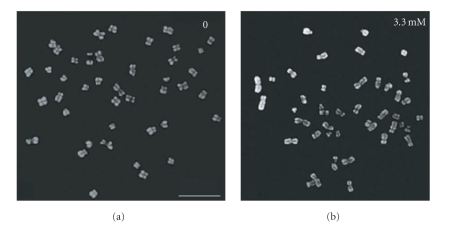
It has been reported previously that treatment of human epidermoid carcinoma cells with 1 mM HAP lead to massive chromosome fragmentation (Figure 2 in [66]). We examined HAP effects on chromosomes in WI-38, HCT116, and P32T lines. We studied chromosome spreads of untreated WI-38, HCT116, and P32T cells versus the same cells treated with 3.3 mM HAP. There were no more chromosomal abnormalities after treatment with 3.3 mM HAP than in the untreated HCT116 chromosome spreads (Figure 5). We did not observe any striking differences in the rates of chromosomal abnormalities in WI-38 and P32T chromosome spreads as well (data not shown). It is possible that the cell line used in earlier studies was hypersensitive to HAP.
Figure 5.
HAP does not induce massive chromosome fragmentation. The untreated HCT116 metaphase spreads (a) and HCT116 metaphase spreads obtained at the end of treatment by 3.3 mM HAP (b) were counterstained with DAPI (grey). Bar—10 μm.
3.2. The Increase in HAP Dose Proportionally Elevated the Rate of DNA Breaks in WI-38, HCT116, and P32T Cells
We investigated the effect of 24 h treatment by different HAP concentrations on the frequency of single-stranded breaks in WI-38, HCT116 and P32T cell cultures by single cell electrophoresis at pH > 13. Under our experimental conditions and with doses of mutagens used, no substantial cell killing occurred. The percent of DNA in the comet tail (thereafter named tail DNA) of the total DNA was used in the study as the estimate of the amount of single-stranded breaks. The mean tail DNA was statistically significantly different in the three lines: 8.42% in untreated normal WI-38 fibroblasts, 12.4% in colorectal cancer cell culture HCT116, and the highest 17.6%, in P32T fibroblast cell culture, a large change for this type of assay two-fold increase over normal fibroblasts (Figure 6(a)). The increased level of tail DNA in untreated cells likely indicates persistent unrepaired endogenous damage in HCT116 and P32T cells.
Figure 6.
HAP induces DNA breaks in human cells. The effect of HAP and 100 μM hydrogen peroxide on the frequency of single-stranded breaks recorded at the end of treatment in WI-38, HCT116 and P32T cell cultures by single-cell electrophoresis at pH > 13. (a) Analysis of the concentration-dependent effects of HAP and hydrogen peroxide on the frequency of DNA strand breaks in WI-38, HCT116 and P32T cells. The damaging DNA-agent dose responses are plotted on the x-axis. The mean percent DNA in the comet tail is plotted on the y-axis as a ratio of the amount of DNA in the comet tail/the amount of DNA in the whole comet. The insert in the upper part of the plot is a linear plot of this correlation for each culture. Results are shown as means ± standard error of results of six different experiments. The level of statistical significance was set at P < .05. (b) Micrographs of WI-38 (a, b, c), HCT116 (d, e, f), and P32T (g, h, i) cells treated with different doses of HAP: 0 (a, d, g), 0.66 mM (b, e, h) and 3.3 mM (c, f, i) for 24 hr. Bar—10 μm.
WI-38 normal fibroblasts were quite resistant to HAP: the highest concentration of 3.3 mM HAP increased tail DNA two-fold to 20.5%, much less than the positive control hydrogen peroxide (42.8% tail DNA) (Figure 6(a)). The induction curve had quite a gentle slope with a small hump/plateau of resistance when the dose increased from 0.66 to 1.32 (seen clearly in the insert in Figure 6(a)). The comet tails in WI-38 cells after treatment with 0.66 mM–3.3 mM of HAP were the shortest among the variables studied (Figure 6(b)). The response of the HCT116 cells to the hydrogen peroxide treatment was similar to that of the WI-38 cells (tail DNA was 42.9%). HAP produced more DNA damage than in WI-38 cells. An initial eight percent increase at 0.66 mM was followed by the plateau of around 25% tail DNA at doses 1.32 mM–2.64 mM (Figure 6(a), insert). This apparent resistance to the induction of breaks was finally concurred by 3.3 mM of HAP and tail DNA reached 30.3% (Figure 6(a)). P32T cells were most sensitive to HAP, while the sensitivity to hydrogen peroxide was similar to other cell lines (43.4%). The lowest dose of HAP induced as much tail DNA in P32T as the highest dose in normal fibroblasts (21%). The 1.98 mM HAP produced 30.7% tail DNA in P32T, exceeding the maximum of tail DNA induced by HAP in other cell cultures studied at the same dose. After 3.3 mM HAP treatment of these cells, the level of tail DNA reached 52.7% and the tail was much longer than in WI-38 and HCT116 under the same conditions (Figures 6(a) and 6(b)). In summary, HAP induced DNA breaks in human cells. HAP was moderately active in normal WI-38 fibroblasts, HCT116 cancer cells were more sensitive than WI-38 cells and P32T ITPA-deficient fibroblasts were the most susceptible.
3.3. ITPA Cellular Levels and Distribution Change Differently after HAP Treatment of WI-38, HCT116 and P32T Lines
The presence of the plateau in the dose-response curves for HAP indicated that wild-type cells might possess an inducible protection system, which is activated at 0.66–1.32 mM HAP. One possible candidate is ITPA and we studied ITPA distribution after HAP treatment in WI-38, HCT116 and P32T cells by whole-cell immunostaining with specific antibodies against ITPA (specificity of antibodies has been verified by Western Blot, because of the shift of detected GFP-tagged ITPA to higher position, Figure 4).
ITPA was not readily detected in untreated WI-38 fibroblasts, and cells were stained very weakly (Figure 7(a)). The detected ITPA amount increased after the treatment with 0.66 mM and 1.32 mM HAP. The untreated HCT116 cancer cells were stained with antibody against ITPA somewhat more efficiently than WI-38 cells (Figure 7(a)). The ITPA amount increased after 0.66 mM and 1.32 mM HAP treatment, the latter dose producing strikingly bright cytoplasm (Figure 7(a)). Maybe this phenomenon simply relates to the smaller size of cytoplasm relative to nucleus. Some ITPA was detected in untreated ITPA-deficient P32T fibroblasts, similar to HCT116 cells (Figure 7(a), lower row). After treatment with 0.66 mM HAP, the ITPA amount increased, so induction occurred at lower dose.
Figure 7.
Intracellular localization and induction of ITPA. (a) The effect of HAP on ITPA levels and distribution in the WI-38, HCT116 and P32T cells. Immunofluorescence of ITPA in the WI-38, upper row, HCT116, middle row and P32T, lower row. Immunostaining and counterstaining were performed with antirabbit monoclonal antibody (green) and DAPI (blue), respectively. Bar—10 μm Representative images are shown. (b) Details of immunofluorescence pattern of ITPA in the HAPtreated WI-38 (upper row, 1.32 mM HAP) and P32T (lower row, 0.66 mM HAP) cells. Left column—DAPI only, middle column—detection of ITPA antibodies, and right column—merged picture.
Fluorescence in all cases was mostly in cytoplasm and was generally uneven. The brighter fluorescent regions in WI-38 cells were of three types. One type resembled a net structure adjacent to nuclei. Another type was small granules localized in nuclei (Figure 7(b), upper row). The third was one big bright granule, typically at the border of nuclei and cytoplasm or localized in the nuclei close to its edge. In HCT116, the heterogeneity was less pronounced and the net structure was not observed (compare to Figure 7(a)). It is possible that it is undetectable due to a small cytoplasmic part of the cell and very bright fluorescence. In P32T fibroblasts the distribution was different. Net-like structure has not been observed. Some brighter regions adjacent to nuclei were present but they were unstructured (Figure 7(b), lower row).
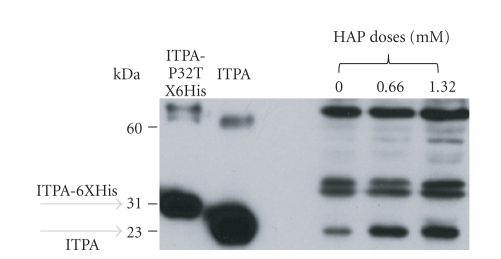
We also have used Western blot analysis in HCT116 cells to confirm and quantify induction of ITPA by HAP (Figure 8). Clearly, HAP treatment leads to up to 2.6-fold elevation of the amount of ITPA.
Figure 8.
HAP treatment leads to the increase of ITPA levels in HCT116 cell extracts. Western blots were performed as described in Section 2. Two controls are in the left part: first lane—His-tagged pure ITPA (30 kDA), second lane:—pure ITPA (23 kDa). Third, fourth, and fifth lane shows the levels of ITPA (23 kDA) in extract of untreated cells, and cells treated by 0.66 mM HAP and 1.32 mM HAP. To estimate the induction of ITPA, we measured the ratio of the intensity of the band corresponding to monomeric ITPA to nonspecific bands. Estimated by this method, the induction relative to the control was 2.2 times at 0.66 mM and 2.6 times at 1.32 mM.
4. Discussion
4.1. Response of Human Cells to Base-analog HAP Treatment
It is known that HAP is mutagenic, clastogenic and carcinogenic in hamster cells [67, 68]. Using Comet assay we have shown that HAP produced DNA breaks in the human cell lines tested. The most logical explanation of this and other currently available data is that the base-analog was incorporated into DNA and either human Endo V homolog (encoded by the LOC284131, see description of closely related mouse homolog in [51]) or other an unknown glycosylase/endonuclease recognized incorporated HAP and incised DNA strands (Figure 1).
Analysis of the dose-response curves revealed a hump/plateau region indicative of the induction of a cellular protective response/repair system. The HAPtreated WI-38 fibroblasts with wild-type ITPA had the lowest rates of DNA breaks, and untreated WI-38 cells had the lowest levels of spontaneous breaks among cell lines studied. The hump in the dose-response curve indicated that there is an induction of the repair activity at 0.66–1.32 mM HAP.
We found moderately elevated levels of spontaneous breaks in HCT116. It remains to be determined whether this effect is related to the defect of a mismatch-repair system. The levels of DNA breaks by HAP in HCT116 cells were higher than in WI-38 fibroblasts and the analysis of the dose-response curve indicated that the repair system most likely was activated completely only by 1.32 mM HAP.
The P32T ITPA-deficient fibroblasts were the most sensitive to the induction of DNA breaks by HAP treatment. We propose that deoxyribonucleoside triphosphates of HAP, usually removed by ITPA in wild-type cell lines, remain in the detectable amount in the DNA precursor pools of ITPA P32T fibroblasts. They are incorporated in DNA causing the excess of DNA breaks. This observation correlates well with the accumulation of ITP only in this cell line (Figure 3). It is known that ITPA P32T produced ectopically protects bacteria and yeast from HAP to the same extent as wild-type ITPA, but the level of ITPA P32T in human cell extracts is much lower than in normal fibroblasts [62]. Apparently, the human repair system does not work properly in P32T cells alleviating DNA damage by downstream repair enzymes. The spontaneous level of DNA breaks in this cell line was the highest, raising the possibility that there is persistent endogenous DNA damage in these cells. It is tempting to speculate that the damage is caused by endogeous dITP/dXTP.
The next set of experiments explored one possible component of the HAPinduced repair system.
4.2. ITPA Levels and Distribution in HAP Treated Human Cells
We have analyzed in detail the levels and intracellular distribution of ITPA. We confirmed that the enzyme is located mainly in cytoplasm [69]. We have found that ITPA levels increased significantly upon treatment with HAP in all the investigated cells, although it was significantly lower in the ITPA P32T fibroblasts than in WI-38 and HCT116 cells. P32T ITPA-deficient fibroblasts responded by the production of ITPA to much lower doses of HAP, because the destruction of HAPTP is less efficient in these cells and effective concentrations of substrates for the ITPA are higher.
The antibody staining revealed a net-like structure adjacent to nuclei and other, granular structures in the WI-38 normal fibroblasts, when ITPA is induced by HAP. This is consistent with previous analyses of the localization of ITPA to purine biosynthetic complexes. It has been shown previously that human enzymes involved in de novo purine biosynthesis (for example, hTrifGART protein, and formylglycinamidine ribonucleotide synthase, PRPP amidotransferase, hPAICS, adenylosuccinate lyase and hATIC), colocalize and cluster in human cell cytoplasm [70]. This is consistent with the hypothesis that these clusters represent a “purinosome”—an organoid essential for purine biosynthesis. Usually these clusters localized throughout the whole cytoplasm volume but there were one or two big clusters on the border of nuclei and cytoplasm. One possible interpretation of our results is that ITPA, the enzyme of purine “salvage” pathway, constitutes a part of this purinosome. The candidate structure is a big fluorescent granule. Further colocalization experiments are needed to test our hypothesis.
Hypothetically, the net-like structure described here may represent a factory checking quality of dNTP pools. It is possible that this abnormal ITPA distribution is one of the reasons for a lack of ITPA function in P32T individuals, despite almost normal activity of the enzyme [62].
Taken together, our results suggest that human cells possess a repair system for purine base-analogs similar, in part, to bacteria. The genetically active derivative of HAP is deoxyribonucleoside triphosphate. Cells are protected from dHAPTP by hydrolysis by ITPA. Some dHAPTP is presumably incorporated into DNA, and subsequent repair of HAP leads to DNA strand breaks.
Acknowledgments
The authors are grateful to Dr. S. Kozmin, Dr. I. M. Spivak, and Dr. N. E. Enukashvili for critical reading of the manuscript and helpful comments. They thank Coriell Repository for the P32T cell line. The work was supported by an NE DHHS LB506 Grant, by a UNMC Eppley Cancer Center 010107 Pilot Grant, and in part, by NCI Grant R01 CA129925 to Youri I. Pavlov. Miriam R. Menezes was supported by a UNMC graduate student fellowship. I. S.-R. Waisertreiger and M. R. Menezes contributed equally to this work.
References
- 1.Friedberg EC. DNA Repair and Mutagenesis. Washington, DC, USA: ASM Press; 2006. [Google Scholar]
- 2.Ames BN, Gold LS. Endogenous mutagens and the causes of aging and cancer. Mutation Research. 1991;250(1-2):3–16. doi: 10.1016/0027-5107(91)90157-j. [DOI] [PubMed] [Google Scholar]
- 3.Kamiya H. Mutagenic potentials of damaged nucleic acids produced by reactive oxygen/nitrogen species: approaches using synthetic oligonucleotides and nucleotides. Nucleic Acids Research. 2003;31(2):517–531. doi: 10.1093/nar/gkg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dedon PC, Tannenbaum SR. Reactive nitrogen species in the chemical biology of inflammation. Archives of Biochemistry and Biophysics. 2004;423(1):12–22. doi: 10.1016/j.abb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 5.Hori M, Ishiguro C, Harashima H, Kamiya H. In vivo mutagenicities of damaged nucleotides produced by nitric oxide and ionizing radiation. Biological and Pharmaceutical Bulletin. 2005;28(3):520–522. doi: 10.1248/bpb.28.520. [DOI] [PubMed] [Google Scholar]
- 6.Haghdoost S, Sjölander L, Czene S, Harms-Ringdahl M. The nucleotide pool is a significant target for oxidative stress. Free Radical Biology and Medicine. 2006;41(4):620–626. doi: 10.1016/j.freeradbiomed.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Ohshima H, Sawa T, Akaike T. 8-Nitroguanine, a product of nitrative DNA damage caused by reactive nitrogen species: formation, occurrence, and implications in inflammation and carcinogenesis. Antioxidants and Redox Signaling. 2006;8(5-6):1033–1045. doi: 10.1089/ars.2006.8.1033. [DOI] [PubMed] [Google Scholar]
- 8.Mathews CK. DNA precursor metabolism and genomic stability. FASEB Journal. 2006;20(9):1300–1314. doi: 10.1096/fj.06-5730rev. [DOI] [PubMed] [Google Scholar]
- 9.Fowler RG, Schaaper RM. The role of the mutT gene of Escherichia coli cell in maintaining replication fidelity. FEMS Microbiology Reviews. 1997;21(1):43–54. doi: 10.1111/j.1574-6976.1997.tb00344.x. [DOI] [PubMed] [Google Scholar]
- 10.Kouzminova EA, Kuzminov A. Chromosomal fragmentation in dUTPase-deficient mutants of Escherichia coli and its recombinational repair. Molecular Microbiology. 2004;51(5):1279–1295. doi: 10.1111/j.1365-2958.2003.03924.x. [DOI] [PubMed] [Google Scholar]
- 11.Kouzminova EA, Kuzminov A. Fragmentation of replicating chromosomes triggered by uracil in DNA. Journal of Molecular Biology. 2006;355(1):20–33. doi: 10.1016/j.jmb.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 12.Kouzminova EA, Kuzminov A. Patterns of chromosomal fragmentation due to uracil-DNA incorporation reveal a novel mechanism of replication-dependent double-stranded breaks. Molecular Microbiology. 2008;68(1):202–215. doi: 10.1111/j.1365-2958.2008.06149.x. [DOI] [PubMed] [Google Scholar]
- 13.Jackson AL, Loeb LA. The mutation rate and cancer. Genetics. 1998;148(4):1483–1490. doi: 10.1093/genetics/148.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rai P, Onder TT, Young JJ, et al. Continuous elimination of oxidized nucleotides is necessary to prevent rapid onset of cellular senescence. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(1):169–174. doi: 10.1073/pnas.0809834106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nunoshiba T, Watanabe T, Nakabeppu Y, Yamamoto K. Mutagenic target for hydroxyl radicals generated in Escherichia coli mutant deficient in Mn- and Fe-superoxide dismutases and Fur, a repressor for iron-uptake systems. DNA Repair. 2002;1(5):411–418. doi: 10.1016/s1568-7864(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 16.Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G → T and A → C substitutions. The Journal of Biological Chemistry. 1992;267(1):166–172. [PubMed] [Google Scholar]
- 17.Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355(6357):273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 18.Pavlov YI, Minnick DT, Izuta S, Kunkel TA. DNA replication fidelity with 8-oxodeoxyguanosine triphosphate. Biochemistry. 1994;33(15):4695–4701. doi: 10.1021/bi00181a029. [DOI] [PubMed] [Google Scholar]
- 19.Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) Journal of Bacteriology. 1992;174(20):6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox EC. Mutator gene studies in Escherichia coli: the mutT gene. Genetics. 1973;73(1973):67–80. [PubMed] [Google Scholar]
- 21.Rotman E, Kuzminov A. The mutT defect does not elevate chromosomal fragmentation in Escherichia coli because of the surprisingly low levels of MutM/MutY-recognized DNA modifications. Journal of Bacteriology. 2007;189(19):6976–6988. doi: 10.1128/JB.00776-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pang B, Zhou X, Yu H, et al. Lipid peroxidation dominates the chemistry of DNA adduct formation in a mouse model of inflammation. Carcinogenesis. 2007;28(8):1807–1813. doi: 10.1093/carcin/bgm037. [DOI] [PubMed] [Google Scholar]
- 23.Simandan T, Sun J, Dix TA. Oxidation of DNA bases, deoxyribonucleosides and homopolymers by peroxyl radicals. Biochemical Journal. 1998;335(2):233–240. doi: 10.1042/bj3350233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clement B, Kunze T. Hepatic microsomal N-hydroxylation of adenine to 6-N-hydroxylaminopurine. Biochemical Pharmacology. 1990;39(5):925–933. doi: 10.1016/0006-2952(90)90209-4. [DOI] [PubMed] [Google Scholar]
- 25.Lieberman I. Enzymatic synthesis of adenosine-5′-phosphate from inosine-5′-phosphate. The Journal of Biological Chemistry. 1956;223(1):327–339. [PubMed] [Google Scholar]
- 26.Kozmin SG, Schaaper RM, Shcherbakova PV, et al. Multiple antimutagenesis mechanisms affect mutagenic activity and specificity of the base analog 6-N-hydroxylaminopurine in bacteria and yeast. Mutation Research. 1998;402(1-2):41–50. doi: 10.1016/s0027-5107(97)00280-7. [DOI] [PubMed] [Google Scholar]
- 27.Burgis NE, Brucker JJ, Cunningham RP. Repair system for noncanonical purines in Escherichia coli . Journal of Bacteriology. 2003;185(10):3101–3110. doi: 10.1128/JB.185.10.3101-3110.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdul-Masih MT, Bessman MJ. Biochemical studies on the mutagen, 6-N-hydroxylaminopurine. Synthesis of the deoxynucleoside triphosphate and its incorporation into DNA in vitro. The Journal of Biological Chemistry. 1986;261(5):2020–2026. [PubMed] [Google Scholar]
- 29.Pavlov YI, Suslov VV, Shcherbakova PV, et al. Base analog N6-hydroxylaminopurine mutagenesis in Escherichia coli: genetic control and molecular specificity. Mutation Research. 1996;357(1-2):1–15. doi: 10.1016/0027-5107(96)00060-7. [DOI] [PubMed] [Google Scholar]
- 30.Burgis NE, Cunningham RP. Substrate specificity of RdgB protein, a deoxyribonucleoside triphosphate pyrophosphohydrolase. The Journal of Biological Chemistry. 2007;282(6):3531–3538. doi: 10.1074/jbc.M608708200. [DOI] [PubMed] [Google Scholar]
- 31.Quirk S, Bhathnagar SK, Bessman MJ. Primary structure of the deoxyguanosine triphosphate triphosphohydrolase-encoding gene (dgt) of Escherichia coli . Gene. 1990;89(1):13–18. doi: 10.1016/0378-1119(90)90200-b. [DOI] [PubMed] [Google Scholar]
- 32.Gawel D, Hamilton MD, Schaaper RM. A novel mutator of Escherichia coli carrying a defect in the dgt gene, encoding a dGTP triphosphohydrolase. Journal of Bacteriology. 2008;190(21):6931–6939. doi: 10.1128/JB.00935-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng J, Singh VK, Jia Z. Identification of an ITPase/XTPase in Escherichia coli by structural and biochemical analysis. Structure. 2005;13(10):1511–1520. doi: 10.1016/j.str.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Budke B, Kuzminov A. Production of clastogenic DNA precursors by the nucleotide metabolism in Escherichia coli . Molecular Microbiology. 2010;75(1):230–245. doi: 10.1111/j.1365-2958.2009.06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iyama T, Abolhassani N, Tsuchimoto D, Nonaka M, Nakabeppu Y. NUDT16 is a (deoxy)inosine diphosphatase, and its deficiency induces accumulation of single-strand breaks in nuclear DNA and growth arrest. Nucleic Acids Research. 2010;2010:p. 12. doi: 10.1093/nar/gkq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohno T, Kunitoh H, Toyama K, et al. Association of the OGG1-Ser326Cys polymorphism with lung adenocarcinoma risk. Cancer Science. 2006;97(8):724–728. doi: 10.1111/j.1349-7006.2006.00240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marinaki AM, Duley JA, Arenas M, et al. Mutation in the ITPA gene predicts intolerance to azathioprine. Nucleosides, Nucleotides and Nucleic Acids. 2004;23(8-9):1393–1397. doi: 10.1081/NCN-200027639. [DOI] [PubMed] [Google Scholar]
- 38.Fellay J, Thompson AJ, Ge D, et al. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature. 2010;464(7287):405–408. doi: 10.1038/nature08825. [DOI] [PubMed] [Google Scholar]
- 39.Bradshaw JS, Kuzminov A. RdgB acts to avoid chromosome fragmentation in Escherichia coli . Molecular Microbiology. 2003;48(6):1711–1725. doi: 10.1046/j.1365-2958.2003.03540.x. [DOI] [PubMed] [Google Scholar]
- 40.Lin S, McLennan AG, Ying K, et al. Cloning, expression, and characterization of a human inosine triphosphate pyrophosphatase encoded by the itpa gene. The Journal of Biological Chemistry. 2001;276(22):18695–18701. doi: 10.1074/jbc.M011084200. [DOI] [PubMed] [Google Scholar]
- 41.Takayama S, Fujii M, Kurosawa A, Adachi N, Ayusawa D. Overexpression of HAM1 gene detoxifies 5-bromodeoxyuridine in the yeast Saccharomyces cerevisiae. Current Genetics. 2007;52(5-6):203–211. doi: 10.1007/s00294-007-0152-z. [DOI] [PubMed] [Google Scholar]
- 42.Hwang KY, Chung JH, Kim S-H, Sun Han Y, Cho Y. Structure-based identification of a novel NTPase from Methanococcus jannaschii . Nature Structural Biology. 1999;6(7):691–696. doi: 10.1038/10745. [DOI] [PubMed] [Google Scholar]
- 43.Kozmin SG, Pavlov YI, Dunn RL, Schaaper RM. Hypersensitivity of Escherichia coli D(uvrB-bio) mutants to 6- hydroxylaminopurine and other base analogs is due to a defect in molybdenum cofactor biosynthesis. Journal of Bacteriology. 2000;182(12):3361–3367. doi: 10.1128/jb.182.12.3361-3367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozmin SG, Schaaper RM. Molybdenum cofactor-dependent resistance to N-hydroxylated base analogs in Escherichia coli is independent of MobA function. Mutation Research. 2007;619(1-2):9–15. doi: 10.1016/j.mrfmmm.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozmin SG, Leroy P, Pavlov YI, Schaaper RM. YcbX and yiiM, two novel determinants for resistance of Escherichia coli to N-hydroxylated base analogues. Molecular Microbiology. 2008;68(1):51–65. doi: 10.1111/j.1365-2958.2008.06128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lukas L, Kuzminov A. Chromosomal fragmentation is the major consequence of the rdgB defect in Escherichia coli . Genetics. 2006;172(2):1359–1362. doi: 10.1534/genetics.105.051144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Porta J, Kolar C, Kozmin SG, Pavlov YI, Borgstahl GEO. Structure of the orthorhombic form of human inosine triphosphate pyrophosphatase. Acta Crystallographica Section F. 2006;62(11):1076–1081. doi: 10.1107/S1744309106041790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pavlov II. Mutants of Saccharomyces cerevisiae supersensitive to the mutagenic effect of 6-N-hydroxylaminopurine. Genetika. 1986;22(9):2235–2243. [PubMed] [Google Scholar]
- 49.Noskov VN, Staak K, Shcherbakova PV, et al. HAM1, the gene controlling 6-N-hydroxylaminopurine sensitivity and mutagenesis in the yeast Saccharomyces cerevisiae. Yeast. 1996;12(1):17–29. doi: 10.1002/(SICI)1097-0061(199601)12:1%3C17::AID-YEA875%3E3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 50.Budke B, Kuzminov A. Hypoxanthine incorporation is nonmutagenic in Escherichia coli . Journal of Bacteriology. 2006;188(18):6553–6560. doi: 10.1128/JB.00447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moe A, Ringvoll J, Nordstrand LM, et al. Incision at hypoxanthine residues in DNA by a mammalian homologue of the Escherichia coli antimutator enzyme endonuclease V. Nucleic Acids Research. 2003;31(14):3893–3900. doi: 10.1093/nar/gkg472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galperin MY, Moroz OV, Wilson KS, Murzin AG. House cleaning, a part of good housekeeping. Molecular Microbiology. 2006;59(1):5–19. doi: 10.1111/j.1365-2958.2005.04950.x. [DOI] [PubMed] [Google Scholar]
- 53.Behmanesh M, Sakumi K, Abolhassani N, et al. ITPase-deficient mice show growth retardation and die before weaning. Cell Death and Differentiation. 2009;16(10):1315–1322. doi: 10.1038/cdd.2009.53. [DOI] [PubMed] [Google Scholar]
- 54.Duley JA, Marinaki AM, Arenas M, Florin THJ. Do ITPA and TPMT genotypes predict the development of side effects to AZA? Gut. 2006;55(7):1048–1049. [PMC free article] [PubMed] [Google Scholar]
- 55.van Dieren JM, van Vuuren AJ, Kusters JG, Nieuwenhuis EES, Kuipers EJ, Van Der Woude CJ. ITPA genotyping is not predictive for the development of side effects in AZA treated inflammatory bowel disease patients. Gut. 2005;54(11):p. 1664. [PMC free article] [PubMed] [Google Scholar]
- 56.Marinaki AM, Ansari A, Duley JA, et al. Adverse drug reactions to azathioptine therapy are associated with polymorphism in the gene encoding inosine triphosphate pyrophosphatase (ITPase) Pharmacogenetics. 2004;14(3):181–187. doi: 10.1097/00008571-200403000-00006. [DOI] [PubMed] [Google Scholar]
- 57.Maeda T, Sumi S, Ueta A, et al. Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency in the Japanese population. Molecular Genetics and Metabolism. 2005;85(4):271–279. doi: 10.1016/j.ymgme.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 58.Shipkova M, Lorenz K, Oellerich M, Wieland E, Von Ansen N. Measurement of erythrocyte inosine triphosphate pyrophosphohydrolase (ITPA) activity by HPLC and correlation of ITPA genotype-phenotype in a Caucasian population. Clinical Chemistry. 2006;52(2):240–247. doi: 10.1373/clinchem.2005.059501. [DOI] [PubMed] [Google Scholar]
- 59.von Ahsen N, Armstrong VW, Behrens C, et al. Association of inosine triphosphatase 94C>A and thiopurine S-methyltransferase deficiency with adverse events and study drop-outs under azathioprine therapy in a prospective Crohn disease study. Clinical Chemistry. 2005;51(12):2282–2288. doi: 10.1373/clinchem.2005.057158. [DOI] [PubMed] [Google Scholar]
- 60.Stocco G, Cheok MH, Crews KR, et al. Genetic polymorphism of inosine triphosphate pyrophosphatase is a determinant of mercaptopurine metabolism and toxicity during treatment for acute lymphoblastic leukemia. Clinical Pharmacology and Therapeutics. 2009;85(2):164–172. doi: 10.1038/clpt.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abolhassani N, Iyama T, Tsuchimoto D, et al. NUDT16 and ITPA play a dual protective role in maintaining chromosome stability and cell growth by eliminating dIDP/IDP and dITP/ITP from nucleotide pools in mammals. Nucleic Acids Research. 2010;38(9):2891–2903. doi: 10.1093/nar/gkp1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stepchenkova EI, Tarakhovskaya ER, Spitler K, et al. Functional study of the P32T ITPA variant associated with drug sensitivity in humans. Journal of Molecular Biology. 2009;392(3):602–613. doi: 10.1016/j.jmb.2009.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herting G, Barber K, Zappala MR, Cunningham RP, Burgis NE. Quantitative in vitro and in vivo characterization of the human P32T mutant ITPase. Biochimica et Biophysica Acta. 2009;1802:269–274. doi: 10.1016/j.bbadis.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jacob S, Aguado M, Fallik D, Praz F. The role of the DNA mismatch repair system in the cytotoxicity of the topoisomerase inhibitors camptothecin and etoposide to human colorectal cancer cells. Cancer Research. 2001;61(17):6555–6562. [PubMed] [Google Scholar]
- 65.Sumi S, Marinaki AM, Arenas M, et al. Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency. Human Genetics. 2002;111(4-5):360–367. doi: 10.1007/s00439-002-0798-z. [DOI] [PubMed] [Google Scholar]
- 66.Biesele JJ. Some morphological effects of alkylating agents. Experimental Cell Research. 1963;9(supplement):525–534. doi: 10.1016/0014-4827(63)90293-3. [DOI] [PubMed] [Google Scholar]
- 67.Barrett JC. Induction of gene mutation in and cell transformation of mammalian cells by modified purines: 2-aminopurine and 6-N-hydroxylaminopurine. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(9):5685–5689. doi: 10.1073/pnas.78.9.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsutsui T, Maizumi H, Barrett JC. Induction by modified purines (2-aminopurine and 6-N-hydroxylaminopurine) of chromosome aberrations and aneuploidy in Syrian hamster embryo cells. Mutation Research. 1985;148(1-2):107–112. doi: 10.1016/0027-5107(85)90213-1. [DOI] [PubMed] [Google Scholar]
- 69.Behmanesh M, Sakumi K, Tsuchimoto D, et al. Characterization of the structure and expression of mouse Itpa gene and its related sequences in the mouse genome. DNA Research. 2005;12(1):39–51. doi: 10.1093/dnares/12.1.39. [DOI] [PubMed] [Google Scholar]
- 70.An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320(5872):103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]