Abstract
One of the proposed functions of sleep is to replenish energy stores in the brain that have been depleted during wakefulness. Benington and Heller formulated a version of the energy hypothesis of sleep in terms of the metabolites adenosine and glycogen. They postulated that during wakefulness, adenosine increases and astrocytic glycogen decreases reflecting the increased energetic demand of wakefulness. We review recent studies on adenosine and glycogen stimulated by this hypothesis. We also discuss other evidence that wakefulness is an energetic challenge to the brain including the unfolded protein response, the electron transport chain, NPAS2, AMP-activated protein kinase, the astrocyte-neuron lactate shuttle, production of reactive oxygen species and uncoupling proteins. We believe the available evidence supports the notion that wakefulness is an energetic challenge to the brain, and that sleep restores energy balance in the brain, although the mechanisms by which this is accomplished are considerably more complex than envisaged by Benington and Heller.
Introduction
Sleep has been investigated for over a century. Yet, the fundamental question of why we need to sleep remains unanswered. In particular, what physiological functions are fulfilled by sleep? One hypothesis is that sleep is necessary to replenish energy stores in the brain that are depleted during wakefulness. This theory posits that during waking, a relatively active metabolic period in the brain, energy stores become progressively diminished, thereby promoting sleep. During sleep, there is recovery of energy stores and thus restoration of energy balance.
Based on this concept, in 1995, Benington and Heller proposed that the energy-related substrates glycogen and adenosine are key sleep regulators (Benington and Heller, 1995). They suggested that alterations in astrocytic glycogen and extracellular adenosine in the brain both reflect metabolic alterations that occur during wakefulness and sleep and can influence the amount and quality of subsequent sleep. In particular, this model proposes that glycogen depletion during wakefulness leads to an increase in extracellular adenosine, which facilitates sleepiness and influences delta power during sleep, a measure of sleep homeostasis (Borbely and Achermann, 1999).
This hypothesis led to extensive studies and, over the ensuing years, numerous studies have examined, in relation to sleep and wakefulness, the role of glycogen and adenosine in addition to other aspects of regulation of brain energetics. We review the recent studies on adenosine and glycogen and place them in the context of earlier studies and Benington and Heller’s formulation of the energy hypothesis of sleep. We also discuss changes in other aspects of energy regulation in the brain with wakefulness and sleep. In particular, we discuss new information on the electron transport chain, AMP-activated protein kinase, NPAS2 and clock, uncoupling proteins, reactive oxygen species, and the unfolded protein response. We believe that available evidence does support the notion that wakefulness provides an energetic challenge to the brain, and one of the functions of sleep is to allow for recovery from this energy-challenged state, thereby allowing needed synthetic processes in the brain to occur during sleep. The Benington-Heller hypothesis has been fruitful in terms of stimulating research. The situation is, however, more complex than they proposed.
A. Adenosine
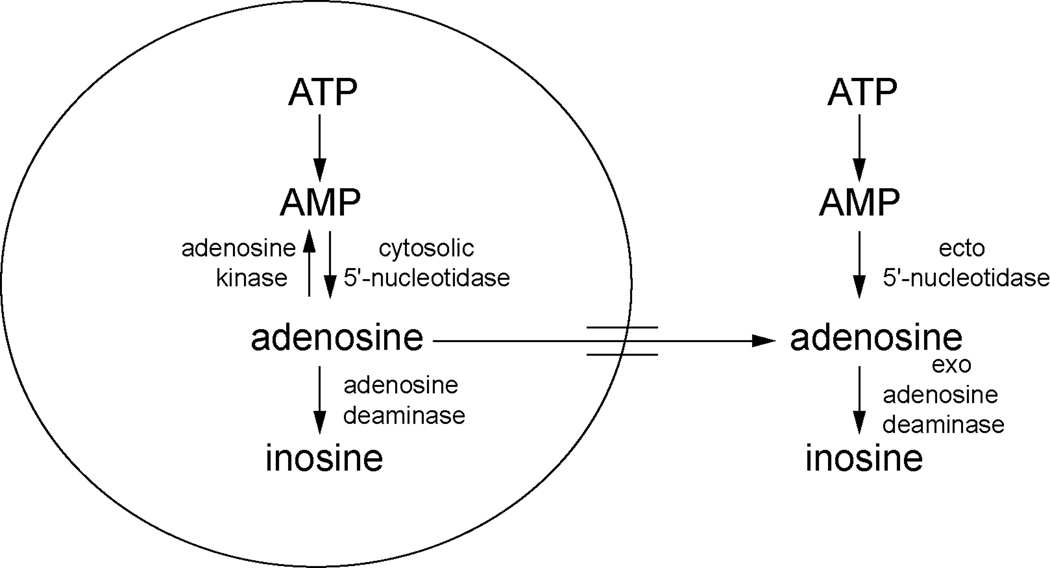
The production of adenosine has been linked to energy depletion. As the major cellular energy molecule ATP is depleted, the byproduct, AMP, is produced. AMP can be further metabolized to adenosine. Therefore, an increase in adenosine may reflect increased degradation of ATP, and a net decrease in the availability of cellular energy stores (Figure 1).
Figure 1.
Adenosine metabolic pathways. In the cell, ATP is metabolized to AMP and then to adenosine by cytosolic 5’-nucleotidase. Adenosine can be converted back to AMP by adenosine kinase or metabolized to inosine by adenosine deaminase. Extracellularly, ATP is metabolized to AMP and then to adenosine by ecto 5’-nucleotidase. Adenosine can be converted to inosine by exo adenosine deaminase.
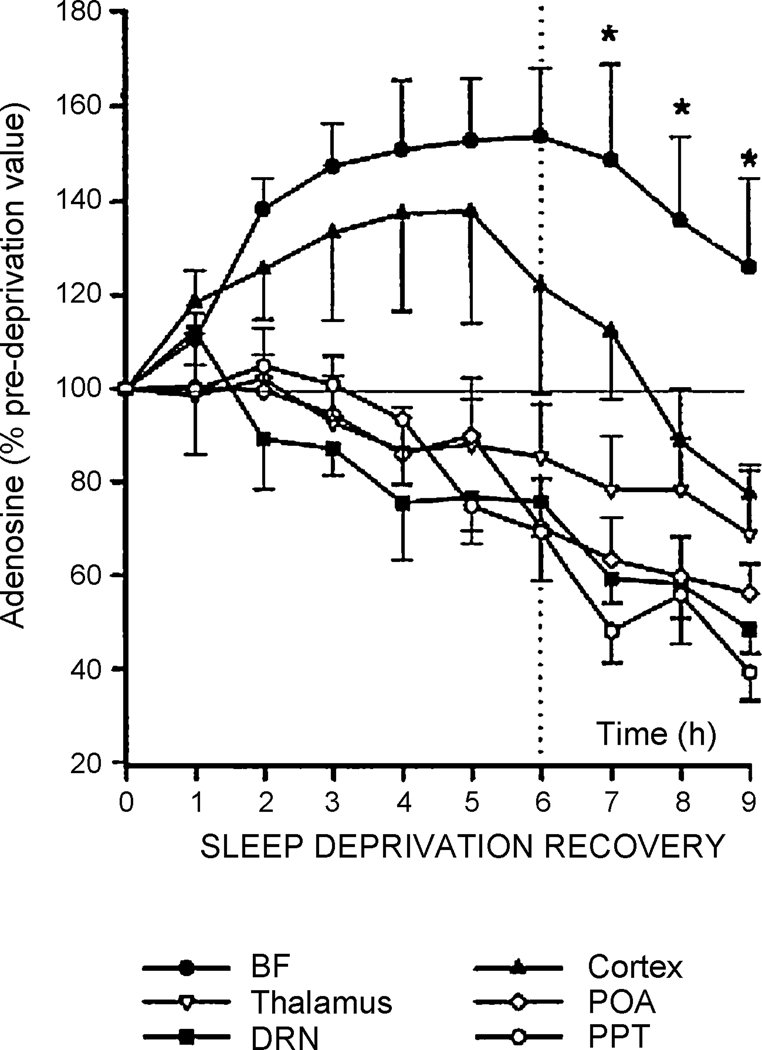
One of the proposed functions of sleep is to replenish energy stores in the brain that are depleted during wake. This theory would, therefore, predict that as energy stores diminish, adenosine would increase progressively during wake, thereby increasing sleepiness and then subsequently decrease during resultant sleep (Benington and Heller, 1995). Indeed, alterations in adenosine levels during normal sleep/wake have been demonstrated. Extracellular adenosine in the cat, as measured by microdialysis, is lower during NREM and REM sleep compared to wakefulness in the basal forebrain, cortex, thalamus and preoptic area of the hypothalamus (Porkka-Heiskanen et al., 2000). However, with 6 hours of sleep deprivation, extracellular adenosine increases only in the basal forebrain and cortex but in the other areas measured (thalamus, preoptic area of the hypothalamus, dorsal raphe nucleus, pedunculopontine tegmental area) it either remains the same or decreases (Figure 2). The reason why adenosine increases with sleep deprivation in certain brain regions but not others is unknown. It likely reflects that unlike cortex, the metabolism of these areas shows little difference between sleep and wake. Nonetheless, these findings indicate that adenosine may act, at least in the basal forebrain or the cortex, to alter aspects of sleep (Porkka-Heiskanen et al., 2000).
Figure 2.
Alterations in extracellular adenosine measured by microdialysis following sleep deprivation and in subsequent recovery sleep in cats. Cats were sleep deprived for 6 hours and then allowed to sleep for 3 hours. Extracellular adenosine was measured at the beginning of the experiment by microdialysis and then in 1 hour intervals for the duration of the experiment. Adenosine is presented as a percentage of the baseline value. Extracellular adenosine increases during sleep deprivation only in the basal forebrain and cortex. In the other areas studied, adenosine levels progressively decline. Adenosine stays elevated during recovery sleep only in the basal forebrain. BF, basal forebrain; POA, preoptic area of the hypothalamus; DRN, dorsal raphe nucleus; PPT, pedunculopontine tegmental area. (Porkka-Heiskanen et al. 2000; reprinted with permission).
Early studies showed that intracerebroventricular infusion of adenosine leads to a decrease in wake and an increase in sleep including slow wave sleep (Virus et al., 1983). Also, caffeine, a non-specific antagonist of adenosine receptors, increases wake, and decreases both NREM and REM sleep including slow wave sleep (Yanik et al., 1987). These and other early studies led Radulovacki to initially propose that adenosine is a sleep-promoting molecule (Radulovacki, 1985). Several early studies using different pharmacologic agents demonstrated that adenosine in the brain increases the homeostatic drive to sleep reflected in both the amount of sleep as well as in the quality of sleep as indicated by the increase in slow wave sleep during NREM sleep (reviewed in Radulovacki, 1985). Furthermore, systemic or intracerebroventricular administration of the A1 receptor agonist CPA increases EEG delta power during NREM sleep (Benington et al., 1995). The demonstration that activation of the A1 receptor directly leads to increases in delta power in sleep suggested a putative mechanism for how changes in cellular metabolism could alter sleep homeostasis since increases in EEG delta power are thought to be a measure of the homeostatic response to sleep deprivation (Borbely and Achermann, 1999). This evidence connecting adenosine to EEG delta power led Benington and Heller to propose that adenosine increases during wakefulness in cortex increase sleepiness and that when sleep occurs there is an increase in delta power (Benington and Heller, 1995). They did not envisage that adenosine played a role in the switch from wakefulness to sleep although others have proposed this (see further below).
A1. Adenosine Receptors
The effects of adenosine are mediated by G-protein coupled adenosine receptors of which there are 4 subtypes (reviewed in Dunwiddie and Masino, 2001). The A1, A2A and A3 receptors have a widespread distribution in the brain while the A2B has a more limited distribution with high expression primarily in the striatum, olfactory tubercle and nucleus accumbens (reviewed in Dunwiddie and Masino, 2001). The A1 and A3 receptors couple negatively to adenylyl cyclase and positively to phospholipase C. The latter may be an indirect effect. The A2A and A2B receptors couple positively to adenylyl cyclase (reviewed in Dunwiddie and Masino, 2001). Adenosine receptors can act on a number of ion channels including potassium and calcium channels (reviewed in Dunwiddie and Masino, 2001). The A1 receptor is primarily inhibitory while the A2A receptor is primarily excitatory (reviewed in Dunwiddie and Masino, 2001).
Both the A1 and A2A receptors have been implicated in the regulation of sleep. Intraperitoneal administration of the A1 receptor agonist CHA leads to an increase in slow wave and REM sleep (Radulovacki et al., 1984). Similarly, stimulation of the A1 receptor by either intraperitoneal or intracerebroventricular administration of the A1 receptor agonist CPA increases slow wave activity during NREM sleep (Benington et al., 1995). Consistent with these results implicating the A1 receptor, antagonizing the A1 receptor by intraperitoneal administration of CPT causes the opposite effect, namely, an increase in wake and a decrease in slow wave and REM sleep (Virus et al., 1990). The A2A receptor is also involved in the regulation of sleep since infusion into the subarachnoid space or the ventricular system of the A2A selective agonist CGS21680 increases NREM and REM sleep (Gerashchenko et al., 2000; Satoh et al., 1998; Satoh et al., 1999). These effects of the A1 receptor and A2A are likely mediated by actions of adenosine in different brain regions (see further below).
However, mice with genetic deletion of the A1 receptor have the same amounts of NREM and REM sleep as wild-type mice (Stenberg et al., 2003). Furthermore, both the wild-type and knockout mice show the same rebound in NREM and delta power following 6 hours of sleep deprivation indicating that sleep homeostasis is normal in the A1 receptor knockout mice (Stenberg et al., 2003). Thus, these results would argue that A1 receptor is not essential for sleep homeostasis. A2A receptor knockout mice have not been as well characterized but are reported to have impairments in NREM rebound following sleep deprivation (Urade et al., 2003). A2A receptor knockout mice do not respond to intraperitoneal administration of caffeine but A1 receptor knockout mice display a normal response to caffeine. This suggests that the A2A receptor is likely the more important mediator of the effects of caffeine on sleep (Huang et al., 2005). Although these results from knockout mice suggest that the A2A receptor is the key receptor for sleep/wake control and the effects of caffeine, constitutive lack of a gene can lead to compensatory changes, which may confound interpretation of the data. Genetic deletion of the A1 and A2A receptors in a spatially and temporally controlled manner would help elucidate the particular functions of A1 and A2A receptors in sleep homeostasis. Local administration of antisense oligonucleotides to selectively reduce expression of adenosine receptors (Thakkar et al., 2003b) has led to additional insights as we discuss more fully below, where we consider the action of adenosine in different neuronal groups.
A2. Adenosine Metabolism
Studies have also assessed whether adenosine metabolism is altered during sleep. Adenosine can be metabolized either intracellularly or extracellularly. A schematic of the adenosine metabolic pathways is presented in Figure 1. Within the cell, adenosine can be formed from AMP in a reaction catalyzed by cytosolic 5’-nucleotidase (Meghji, 1991). Also within a cell, adenosine can be converted back to AMP via adenylate kinase or converted to inosine via adenosine deaminase (Meghji, 1991). In the extracellular space, ATP, ADP and AMP can be converted to adenosine by ecto-5’-nucleotidases (Cunha et al., 1998; Dunwiddie et al., 1997).
In a study from our laboratory, we assessed activities of both the cytosolic and extracellular forms of 5’-nucleotidase using a new method to assess their activities (Mackiewicz et al., 2000), as well as activity of adenosine deaminase and adenosine kinase. We found that while activity of these enzymes did exhibit diurnal variation in a number of different brain regions, sleep deprivation did not alter the activity of any of these enzymes (Mackiewicz et al., 2003). Similarly, others have reported that activity of 5’-nucleotidase is altered following only 4 days of REM sleep deprivation but not following 2 days of REM sleep deprivation, suggesting that 5’-nucleotidase activity is not likely altered during spontaneous wake (Thakkar and Mallick, 1996). These studies suggest that modulation of the activities of adenosine enzymes is not a likely mechanism to explain the increase in adenosine that occurs during wakefulness. The activity of extracellular 5’-nucleotidase is, however, increased with age in cortex (Mackiewicz et al., 2006). This likely explains the increase in extracellular adenosine with age in basal forebrain that has been found using microdialysis in rat (Murillo-Rodriguez et al., 2004).
Adenosine has been extensively studied because increases in extracellular adenosine may reflect depletion of ATP in the cell concomitant with energy depletion. However, the connection between cellular metabolism and extracellular adenosine levels is not straightforward since there are numerous sources for extracellular adenosine. Adenosine can be produced in the cell and transported across the cell membrane by a number of transporters (reviewed in Cass et al., 1998). But adenosine can also be produced in the extracellular space by conversion of ATP, ADP and AMP (Cunha et al., 1998; Dunwiddie et al., 1997). ATP is co-localized with neurotransmitters such as acetylcholine, norepinephrine, serotonin and dopamine and can be released upon stimulation (reviewed in Dunwiddie and Masino, 2001). Impairment of synaptic transmission selectively in astrocytes by spatially and temporally restricted expression of a dominant negative SNARE domain leads to decreased ATP release, which decreases extracellular adenosine implicating astrocytes as a source of extracellular adenosine in the brain (Pascual et al., 2005). This is particularly interesting since mutant mice with the dominant negative SNARE have impaired sleep homeostasis, i.e., an attenuated increase in NREM sleep and delta power following sleep deprivation (Halassa et al., 2007). In particular, in these mice the increase in delta power following sleep deprivation is short and not sustained (Halassa et al., 2007). Furthermore, in basal forebrain, impairing adenosine transport with NBTI increases extracellular adenosine (Porkka-Heiskanen et al., 1997) suggesting that, at least in basal conditions in this region, the net flow of adenosine is from the extracellular space into the cell. Therefore, adenosine in the extracellular space may be produced within the cell or outside it, and the observed increase in extracellular adenosine induced by sleep deprivation does not necessarily reflect cellular energy depletion occurring with prolonged wakefulness. However, lesions of the cholinergic cells in the basal forebrain (see section A3a) completely negate the increase in adenosine with sleep deprivation in this region suggesting that during sleep deprivation adenosine is coming into the extracellular space from these neurons, i.e., it has an intracellular source. Thus, the source of adenosine in baseline conditions and during sleep deprivation may be different. It is also possible that the lesioned animals could have abnormalities in astrocytes, which may be responsible for the decrease in adenosine. Damage to the central nervous system is accompanied by changes to astrocytes, a process known as reactive gliosis (reviewed in Pekny et al., 2007). It is possible, therefore, that lesioning of the cholinergic neurons of the basal forebrain leads to alterations in astrocytes, which results in alteration in extracellular adenosine. Indeed, the enzyme adenosine kinase, which metabolizes adenosine to AMP, is increased in astrocytes during the process of reactive gliosis (Boison, 2006), which could contribute to the decreased extracellular adenosine observed in the lesioned animals. Nonetheless, the increase in adenosine following sleep deprivation is correlated with the restorative processes occurring during NREM sleep and may be caused by the synthesis of glycogen.
A3. Action of Adenosine in Different Brain Regions
Manipulation of the adenosine system in a number of different brain regions alters sleep/wake states. The basal forebrain, laterodorsal tegmental area, pontine reticular formation and ventrolateral preoptic area have all been implicated as important regions mediating the effects of adenosine on sleep. Following is an examination of the role of adenosine in each of these systems that are involved in sleep regulation and the adenosine receptor subtypes involved.
A3a. Basal Forebrain
The basal forebrain is involved in sleep/wake regulation. The term “basal forebrain” has become synonymous with the magnocellular cholinergic system in the medial septum, vertical and horizontal limbs of the diagonal band of Broca, magnocellular preoptic area, the substantia innominata and the nucleus basalis of Meynert (Szymusiak, 1995). Other cell types, however, are contained in the basal forebrain including GABA-ergic and various peptidergic neurons and these may be of functional significance (Szymusiak, 1995). In this review, we will refer to the basal forebrain as the anatomical region containing both the cholinergic neurons and noncholinergic neurons. The cholinergic neurons of the basal forebrain project diffusely and play a role in the cortical activation associated with wake and REM sleep (Jones, 2005; Szymusiak, 1995). Given the role of the cholinergic system of the basal forebrain in sleep/wake regulation, a number of studies have examined whether the basal forebrain is the “adenosine sensor” of the brain that is responsible for the adenosinergic modulation of sleep/wake.
If the adenosine sensor is in the basal forebrain, adenosine should increase with wakefulness in the basal forebrain. Indeed, extracellular adenosine increases during wake in a number of brain regions including the basal forebrain (Porkka-Heiskanen et al., 2000; Porkka-Heiskanen et al., 1997). Of these areas, extracellular adenosine increases only in the basal forebrain and cortex during sleep deprivation. During 3 hours of sleep recovery, adenosine declines in the cortex below pre-sleep deprivation levels. In basal forebrain it also decreases during recovery sleep but is still elevated above pre-sleep deprivation levels even after 3 hours of recovery sleep (Porkka-Heiskanen et al., 2000) (Figure 2). Therefore, McCarley and colleagues have suggested that the basal forebrain may be uniquely situated to detect changes in adenosine occurring with sleep and wake and be able to modulate sleep accordingly (Basheer et al., 2004; Porkka-Heiskanen et al., 2000). It should be noted, however, that a single location of action of adenosine is unlikely to explain the many effects that are observed with sleep deprivation and the recovery from sleep deprivation. In particular, it cannot explain the local increase in slow wave activity during sleep in brain areas specifically activated during wakefulness (Vyazovskiy and Tobler, 2008).
As further proof of the role of the basal forebrain in mediating the effects of adenosine on sleep/wake, modulation of the adenosinergic system in the basal forebrain can alter sleep. Administration of the adenosine transport inhibitor NBTI increases extracellular adenosine and concomitantly decreases wake and increases slow wave and REM sleep as well as delta power during NREM sleep (Methippara et al., 2005; Porkka-Heiskanen et al., 1997). Similarly, perfusion of adenosine into the basal forebrain by microdialysis decreases wake and increases NREM, REM and delta power (Basheer et al., 1999; Portas et al., 1997). These studies demonstrate that increasing adenosine levels in the basal forebrain can increase both the amount of sleep and the quality of sleep as reflected in the delta power, which is a sensitive indicator of the homeostatic component of sleep (Borbely and Achermann, 1999).
A1 and A2A receptors mediate the effects of adenosine on sleep/wake (see section A1). In the basal forebrain, A1 receptor mRNA increases following sleep deprivation but A2A receptor expression is undetectable (Basheer et al., 2001a). Reduction of A1 receptor in the basal forebrain by antisense oligonucleotides increases wake and decreases NREM and REM sleep (Thakkar et al., 2003b), although one needs to be aware of the potential for non-specific effects of antisense oligonucleotides (reviewed in Lebedeva and Stein, 2000). Since this intervention has the opposite effect to activating the A1 receptor, the results are compatible with the concept that adenosine promotes sleep in this region through the effects of its action on the A1 receptor. Furthermore, animals with their A1 receptors reduced in basal forebrain by antisense oligonucleotides show decreases in NREM and delta power following sleep deprivation, suggesting that it is the A1 receptor in the basal forebrain that mediates sleep homeostasis (Thakkar et al., 2003b). This result is incongruent with that from studies of mice with knockout of specific adenosine receptors that we described above.
A number of studies have examined the electrophysiological mechanisms by which adenosine can modulate the neuronal activity responsible for sleep/wake alteration. In the basal forebrain, microdialysis perfusion of adenosine, the adenosine transport inhibitor NBTI (which leads to an increase in extracellular adenosine) (see above), or the A1 receptor agonist CHA, decreases the neuronal activity of wake-active neurons (Alam et al., 1999; Thakkar et al., 2003a). Consistent with this result, the A1 receptor antagonists CPDX or CPT, also delivered by microdialysis perfusion into the basal forebrain, increase the discharge of wake-active neurons (Alam et al., 1999; Thakkar et al., 2003a). Administration of the A2A receptor agonist CGS had no effect on these wake-active neurons (Thakkar et al., 2003a). Therefore, adenosine decreases the activity of wake-active neurons in the basal forebrain by acting on the A1 receptor. One study reported a decrease in the firing rate of sleep-active neurons in this region with adenosine activation by NBTI (Alam et al., 1999), and one study with adenosine activation by the A1 receptor agonist CHA did not observe a change in the firing rate of these sleep-active neurons (Thakkar et al., 2003a).
The A1 receptor decreases neuronal activity by acting on inwardly rectifying potassium channels or the hyperpolarization activated cation current (Rainnie et al., 1994). In in vitro brain slices, adenosine inhibits the cholinergic neurons of the basal forebrain by activating an inwardly rectifying potassium channel (Arrigoni et al., 2006). This effect is blocked by the A1 receptor antagonist CPT (Arrigoni et al., 2006). Some of the non-cholinergic neurons of the basal forebrain are also inhibited by adenosine (Arrigoni et al., 2006). In contrast to the cholinergic neurons, however, the non-cholinergic neurons decrease activity in response to adenosine by inhibiting the hyperpolarization-activated cation current (Arrigoni et al., 2006). Therefore, although both cholinergic and non-cholinergic neurons are inhibited by adenosine, this inhibition is mediated by different ion currents; for the former it is an inwardly rectifying potassium current and the latter it is the hyperpolarization-activated cation current.
The A1 receptor mediates intracellular events by acting on cell signaling pathways, in particular by increasing the activity of phospholipase C (reviewed in Dunwiddie and Masino, 2001). Phospholipase C catalyzes the production of inositol triphosphate, which leads to activation of calcium channels and increases in intracellular calcium. Consistent with phospholipase C activation, adenosine causes an increase in intracellular calcium in cholinergic neurons of the basal forebrain by acting primarily through the A1 receptor and to a lesser extent, the A3 receptor (Basheer et al., 2002). This calcium is released from internal stores via inositol triphosphate receptors (Basheer et al., 2002).
Phospholipase C activation leads to activation of protein kinase C (Alberts et al., 2002). Protein kinase C alters transcription by acting on a number of downstream targets (reviewed in Ventura and Maioli, 2001) including NF-κB (reviewed in Weil and Israel, 2006). NF-κB translocates to the nucleus when one of its binding partners, I-κB, is phosphorylated and detaches from it (Alberts et al., 2002). A number of different kinases can phosphorylate I-κB including protein kinase C (reviewed in Siebenlist et al., 1994). In basal forebrain brain slices, adenosine increases NF-κB DNA binding activity in nuclear extracts compatible with it translocating to the nucleus and pretreatment with the A1 receptor selective antagonist CPT decreases NF-κB DNA binding activity (Basheer et al., 2001b). NF-κB translocation to the nucleus following sleep deprivation as well as following microinjection into the basal forebrain of either adenosine or the A1 receptor agonist CHA occurs almost exclusively in the cholinergic cells of the basal forebrain (Ramesh et al., 2007). Furthermore, blocking the nuclear translocation of NF-κB with an inhibitor peptide, SN50, reduces delta power following sleep deprivation (Ramesh et al., 2007). These observations are consistent with an A1 receptor-mediated activation of the phospholipase C signaling cascade in the basal forebrain and suggest that NF-κB may be an important mediator of both the molecular and behavioral effects of adenosine in this region.
The behavioral, electrophysiological and molecular studies of McCarley and colleagues lead to the hypothesis that the basal forebrain is a critical site for the effects of adenosine on sleep homeostasis (reviewed in Basheer et al., 2004). Benington and Heller posited that adenosine produced sleepiness and subsequently increased delta power during sleep by its action in cortex (Benington and Heller, 1995). McCarley and colleagues redefined the Benington-Heller hypothesis by positing that adenosine mediates it effect on sleep by acting focally in the basal forebrain. They specifically proposed that the wake-promoting cholinergic neurons of the basal forebrain are mediators of the action of adenosine on wakefulness and sleep (Porkka-Heiskanen et al., 1997). Although McCarley and coworkers provided much evidence for their hypothesis, they did not test it directly. In particular, is the increase in adenosine in the basal forebrain required for the homoeostatic response to sleep deprivation? Furthermore, are the cholinergic neurons of the basal forebrain required for the increases in sleep and EEG delta power following sleep deprivation?
A recent study (Blanco-Centurion et al., 2006) addressed these critical questions by destroying the cholinergic neurons of the basal forebrain by intracerebroventricular injection of the neurotoxin saporin conjugated to an antibody that is taken up by the p75 nerve growth factor receptor, which is selectively expressed in cholinergic neurons of the basal forebrain (Heckers et al., 1994). Lesioning of the cholinergic neurons of the basal forebrain eliminates the increase in extracellular adenosine induced by sleep deprivation in this region. However, surprisingly, loss of the cholinergic neurons of the basal forebrain does not alter baseline sleep/wake or recovery sleep following 6 or 12 hours of sleep deprivation. The increase in EEG delta power following sleep deprivation was also normal indicating an intact sleep homeostatic response (Blanco-Centurion et al., 2006). The lack of an increase in adenosine following sleep deprivation while the homeostatic response stays intact indicates that the increase in adenosine in the basal forebrain that occurs during sleep deprivation is not required for sleep homeostasis. Furthermore, this provocative study indicates that the cholinergic neurons of the basal forebrain are not necessary for sleep/wake control or for sleep homeostasis.
There is, however, some controversy surrounding this study and the role of the cholinergic neurons of the basal forebrain in mediating sleep homeostasis (for commentaries, see Heller, 2006; Kalinchuk et al., 2006; Noor Alam et al., 2006; Radulovacki, 2006). It is possible that since there is redundancy in the processes regulating sleep homeostasis, the loss of one component, i.e., the cholinergic cells of the basal forebrain, may not alter overall sleep homeostasis (Noor Alam et al., 2006). However, lesioning of the histamine neurons in the tuberomammillary nucleus and the noradrenergic locus ceruleus neurons in addition to the cholinergic neurons of the basal forebrain decreases wake and increases NREM sleep during only the first 4 hours of the dark period but over a full 24 hour period produces no significant change in wake or NREM (Blanco-Centurion et al., 2007). This result challenges the notion that the lack of an effect on total sleep in the animals with a lesion of the cholinergic basal forebrain is due to compensatory changes in other wake-promoting systems since lesioning of other wake-promoting neurons also fails to alter overall sleep. Since the triple lesioned animals had increased bout lengths for both wake and NREM, the role of these systems may be to alter the architecture of sleep rather than the total amount of NREM and wake (BlancoCenturion et al., 2007).
It is also possible that when the conjugated saporin is administered intracerebroventricularly, as was done in the study of Blanco-Centurion et al. (Blanco-Centurion et al., 2006), there are different effects than would be found if the saporin was delivered locally to the basal forebrain (Kalinchuk et al., 2006). This could be the result of the toxin acting in regions other than basal forebrain. Indeed, injection of saporin conjugated to an antibody that is taken up by the p75 nerve growth factor receptor as was used in the study of Blanco-Centurion et al. directly into the basal forebrain causes a decrease in recovery NREM sleep and delta power following sleep deprivation, indicating impaired sleep homeostasis (Kaur et al., 2008). Adenosine was not measured, however, in this study (Kaur et al., 2008) and the cause for the different results from local microinjection and ICV injection is currently not known. While much remains to be learned, the notion of the exclusivity of the cholinergic neurons of the basal forebrain as the adenosine sensor and mediator of the effects of adenosine on sleep homeostasis is seriously challenged, and can no longer be considered as a tenable hypothesis.
A3b. Laterodorsal Tegmental Nucleus
The laterodorsal tegmental nucleus is a sleep-promoting region with a particular role in REM sleep generation (Monti and Monti, 2000). The laterodorsal tegmental nucleus includes cholinergic and glutamatergic neurons (Lee et al., 2003; Monti and Monti, 2000). Microdialysis perfusion of adenosine into the laterodorsal tegmental nucleus in cats decreases wake and increases slow wave sleep and REM sleep (Portas et al., 1997).In vitro in brain slices, adenosine and the A1 receptor agonist CHA inhibit the firing of both cholinergic and non-cholinergic neurons in the laterodorsal tegmental nucleus by activating an inwardly rectifying potassium current and inhibiting the hyperpolarization-activated cation current (Rainnie et al., 1994). This study did not identify whether these two currents were active in both the cholinergic and noncholinergic cells or whether the effect of adenosine on cholinergic and non-cholinergic neurons was specifically mediated by one of the channels as was observed in the basal forebrain (see above). Adenosine also inhibits glutamatergic transmission in the laterodorsal tegmental nucleus by activating presynaptic A1 receptors (Arrigoni et al., 2001). Therefore, adenosine may modulate sleep/wake by acting on the neurons of the laterodorsal tegmental nucleus. If so, the precise role of adenosine in this region in sleep/wake control in vivo is currently unknown.
A3c. Pontine Reticular Formation
Another region to consider is the pontine reticular formation. The pontine reticular formation has a well-established role in the modulation of REM sleep (Monti and Monti, 2000). Microinjection of the A1 receptor agonist CHA into the pontine reticular formation of the rat decreases wake and increases REM (Marks and Birabil, 1998; Marks et al., 2003) and to a lesser extent NREM (Marks and Birabil, 1998). Microinjection of the A2A receptor agonist CGS into the pontine reticular formation also increases REM sleep (Coleman et al., 2006; Marks et al., 2003) and NREM sleep (Coleman et al., 2006), and decreases wake (Marks et al., 2003). The pontine reticular formation receives cholinergic projections from the laterodorsal and pedunculopontine tegmental nuclei, which are important for REM sleep generation (Lydic and Baghdoyan, 2005). Interestingly, CGS injected into the pontine reticular formation increases acetylcholine release in the pontine reticular formation (Coleman et al., 2006). These data suggest that adenosine may also act in the pontine reticular formation to increase acetylcholine release and thereby increase REM sleep.
A3d. Preoptic Area of the Hypothalamus
The preoptic area of the hypothalamus is an important sleep-promoting region. In particular, a population of neurons in the ventrolateral preoptic area shows increased fos levels (Sherin et al., 1996), and increased neuronal firing activity during sleep consistent with a sleep-promoting function (McGinty and Szymusiak, 2001; reviewed in Saper et al., 2005). However, despite this key difference that neurons in the ventrolateral preoptic area are sleep-active, microinjection of adenosine into the preoptic area increases total sleep, slow wave sleep and REM sleep as it does in the basal forebrain, pontine reticular formation and laterodorsal tegmental nucleus (Ticho and Radulovacki, 1991). This suggests that adenosine is excitatory to sleep-active neurons in the ventrolateral preoptic area in contrast to the inhibitory action on the wake-active cells in the basal forebrain. Local administration of the adenosine transport inhibitor NBTI into the lateral preoptic area, which in the basal forebrain has been shown to increase extracellular adenosine levels and sleep (Porkka-Heiskanen et al., 1997), decreases NREM and increases wake (Methippara et al., 2005). Thus, administration of adenosine (Ticho and Radulovacki, 1991) or NBTI (Methippara et al., 2005) produces opposite effects. But we do not know the source of adenosine in the preoptic area, i.e., extracellular or intracellular (see above). If in this region the net flux of adenosine through the transporter is from intracellular to extracellular space, NBTI could lead to a decline in extracellular adenosine. In comparison to studies described above for basal forebrain, the effect of local administration of NBTI on extracellular adenosine levels in the preoptic area has not been assessed. There may, moreover, be issues about the precise location of injection sites in this small region that could affect results. This seems likely since studies report opposite results for the administration of the A1 receptor agonist CPA into the preoptic area of the hypothalamus, which in one study decreases NREM and increases wake (Methippara et al., 2005) and in the other study increases sleep (Ticho and Radulovacki, 1991). The discrepancy in these results may reflect that although the ventrolateral preoptic area has a population of sleep-active neurons, injection of drugs cannot specifically target these neurons and may act on other populations of cells.
Different studies have also led to different conclusions about the role of the A2A receptor in the lateral preoptic area. Administration of the less selective A2 receptor agonist CV-1808 does not alter sleep or wake (Ticho and Radulovacki, 1991), but the relatively selective A2A receptor agonist CGS increases NREM and decreases wake (Methippara et al., 2005). The specificities of these drugs for the adenosine receptor subtypes are different (Hutchison et al., 1989), and therefore the results of these studies need to be interpreted with caution. Nonetheless, the latter study suggests that A2A receptor activation, which is excitatory in the lateral preoptic area, increases sleep. Neuronal excitation of sleep-active neurons mediated by the A2A receptor is a plausible explanation for the sleep-promoting role of adenosine in the ventrolateral preoptic area.
Adenosine reduces spontaneous GABA release onto neurons of the ventrolateral preoptic area suggesting a presynaptic mechanism for ventrolateral preoptic area activation, whereby adenosine disinhibits sleep-promoting neurons of the ventrolateral preoptic area (Chamberlin et al., 2003; Morairty et al., 2004). In a separate study, two groups of GABA-ergic sleep-promoting neurons in the ventrolateral preoptic area have been identified that are both inhibited by adenosine and the A1 receptor agonist CPA via a post-synaptic mechanism (Gallopin et al., 2005). The discrepancy between the former and latter studies may have to do with method of recording (patch clamp in whole cell recording versus loose cell-attached configuration). However, the latter study went further by also assessing the role of the A2A receptor in the two types of neurons that are inhibited by adenosine. The A2A receptor agonist CGS excites only one type (Gallopin et al., 2005) demonstrating that different neurons in the ventrolateral preoptic area have the capacity to respond differently to adenosine. There may be a subgroup of neurons in this region that are responsible for the sleep-promoting role of adenosine.
The A2A receptor may influence ventrolateral preoptic area neurons indirectly as well. Infusion of the A2A receptor agonist CGS into the subarachnoid space increases fos expression in the ventrolateral preoptic area suggesting that activation of A2A receptors in the leptomeninges or nucleus accumbens may also increase NREM sleep by increasing the activity of sleep-active ventrolateral preoptic area neurons (Scammell et al., 2001). Therefore, adenosine likely acts on the A2A receptor both directly in the preoptic area and indirectly to excite the ventrolateral preoptic area sleep-promoting neurons thereby increasing sleep.
A4. Synthesis of Studies of Adenosine
While the idea that adenosine might be a sleep-promoting molecule did not originate with Benington-Heller (Benington and Heller, 1995), but rather as a result of important earlier studies (Feldberg and Sherwood, 1954; Haulica et al., 1973; Marley and Nistico, 1972; Radulovacki, 1985; Virus et al., 1983), their manuscript did provide a cogent synthesis that stimulated research in this area. Benington et al. observed that activation of the A1 receptor leads to increases in EEG delta power during NREM sleep (Benington et al., 1995). This finding was a stimulus for Benington and Heller (1995) to propose that changes in adenosine in the cortex could promote sleepiness and subsequently, when sleep occurred, alter delta power, the main electrophysiologic correlate of sleep homeostasis (Borbely and Achermann, 1999). They did not envisage that adenosine played a role in the wakefulness to sleep switch. However, other investigators have examined whether changes in adenosine in other brain regions could alter not only sleep homeostasis as indicated by delta power, but also the total amounts of sleep and wake, i.e., could adenosine also play a role in the initiation of sleep or wake? There is now evidence that at least in some brain areas, e.g., the basal forebrain, extracellular adenosine increases in relationship to the duration of prior wakefulness (Basheer et al., 1999; Porkka-Heiskanen et al., 2000; Porkka-Heiskanen et al., 1997). However, this does not occur uniformly throughout the brain and it appears that in some wake-active brain regions such as the dorsal raphe nucleus, adenosine levels actually decline as wakefulness is prolonged (Porkka-Heiskanen et al., 2000). The basis for this result is unknown but likely reflects local differences in the metabolic changes between sleep and wakefulness. Moreover, it may be somewhat artifactual, since determining extracellular adenosine by microdialysis in these small neuronal populations is challenging.
There is extensive evidence that adenosine can alter sleep/wake states. To date, the basal forebrain, the laterodorsal tegmental nucleus, the pontine reticular formation and the preoptic area have been implicated as sites of adenosine action on sleep/wake control. The role of adenosine in sleep/wake control may result from inhibition of basal forebrain wake-active neurons (Thakkar et al., 2003a) or activation of preoptic area sleep-active neurons, the latter effect being mediated by A2A receptors (Gallopin et al., 2005). The role of the basal forebrain adenosinergic system in altering sleep has been the most studied. McCarley and co-workers have suggested that cholinergic neurons in the basal forebrain sense alterations of adenosine that occur during wake or sleep and alter the quantity and quality of sleep appropriately (reviewed in Basheer et al., 2004).
This modification of the Benington-Heller hypothesis, i.e., that for its role in control of sleep/wake, adenosine is specifically “sensed” by cholinergic neurons in the basal forebrain, while intriguing, now seems disproven (Blanco-Centurion et al., 2006). A challenge to this hypothesis from the beginning is that it has never been shown in what way cholinergic neurons of the basal forebrain had any special or unique properties with respect to the effects of adenosine on their function, i.e., what was special about the basal forebrain neurons that allowed them to play this role? There is no evidence, for example, that the regulation of adenosine enzymes controlling adenosine metabolism across the day or with sleep deprivation is different in the basal forebrain compared to other brain regions (Mackiewicz et al., 2003).
Other cell types in the basal forebrain or in other areas of the brain are likely also to be important for the mechanisms of action of adenosine on sleep/wake control. As discussed earlier, the laterodorsal tegmental nucleus, the pontine reticular formation and the preoptic area have all been shown to alter sleep upon modulation of their intrinsic adenosinergic system. These areas may work independently or, more likely, in concert with other regions of the brain to alter sleep. One intriguing possibility is that adenosine acts in the cortex to increase slow wave activity (Benington and Heller, 1995) while adenosine acts in other regions such as the basal forebrain to initiate sleep.
A recent exciting discovery is the role of astrocytes in response to sleep deprivation. These cells are involved in modulating effects of synapses- the tripartite synapse (reviewed in Fellin et al., 2006). Since a single astrocyte can contain greater than 100,000 synapses (Bushong et al., 2002), they provide the ability to coordinate responses over a large numbers of synapses. Astrocytes release neurochemicals that can affect synaptic transmission (reviewed in Fellin et al., 2006). One particular neurochemical that is released is ATP that is metabolized extracellularly to adenosine (Pascual et al., 2005). Thus, astrocytes can modulate adenosine “tone” of synapses and are likely to play a key role in the coupling of adenosine to the regulation of sleep. This discovery as to the role of astrocytes is likely to lead to more intensive investigation of the biology of astrocytes in relation to sleep/wake control and the response to sleep deprivation.
Pharmacologic studies implicate adenosine as a modulator of sleep/wake with the effects mediated by the A1 and A2A receptors. Antisense oligonucleotide reduction of the A1 receptor produces alterations in sleep homeostasis consistent with the pharmacologic data (Thakkar et al., 2003b). However, genetic deletion of the A1 receptor produces no alterations in sleep or in the response to sleep deprivation (Stenberg et al., 2003), while genetic deletion of the A2A receptor has been reported to produce sleep alterations in mice such as unresponsiveness to the wake-promoting effects of caffeine (Huang et al., 2005; Urade et al., 2003). Pharmacologic and genetic tools each have, however, their limitations. Pharmacologic agents are confounded by issues of specificity, delivery and dosage. However, given the number of drugs tested and the number of different experimental models used, it is extremely unlikely that non-specific effects can explain the sleep/wake alterations observed in these studies. These pharmacologic experiments implicate the adenosine system as being a modulator of both the quantity and quality of sleep.
Global deletion of a gene may produce developmental defects (for example see Lee et al., 2007) or lead to compensatory changes that may mask the physiological functions of the deleted gene when compared to temporally and/or spatially restricted deletion (for example see Balschun et al., 2003). Genetic tools are now available that allow for spatial and temporal control of gene expression (for reviews, see Mallo, 2006; Miyoshi and Fishell, 2006). These elegant genetic tools avoid many of the problems associated with pharmacologic agents and deletion of a target gene prior to development. Given the recent controversy of the role of the basal forebrain in mediating the effects of adenosine (Blanco-Centurion et al., 2006; Heller, 2006; Kalinchuk et al., 2006; Noor Alam et al., 2006; Radulovacki, 2006), future studies utilizing these genetic tools will allow for more precisely controlled experiments and should prove illuminating.
There is, however, no doubt that adenosine does play some role in sleep/wake control, and likely more prominently in the behavioral response to sleep deprivation. Adenosine is, however, not likely to be the major player but rather one of many mechanisms used to promote sleep when wakefulness is prolonged. Microarray studies conducted by us in both Drosophila (Zimmerman et al., 2006) and mice (Mackiewicz et al., 2007) show that the most common changes in gene expression with sleep and wakefulness are downregulation of expression of many genes during sleep deprivation, supporting the notion that multiple mechanisms are used to limit wakefulness. The adenosine studies to date are consistent with a role of adenosine in regulation of sleep but are insufficient on their own to explain how changes in brain energy are linked to control of sleep.
B. Glycogen
The other component of the Benington-Heller hypothesis is alteration in glycogen. In addition to being stored and produced in the liver, there is a relatively small amount of glycogen that is synthesized and stored in astrocytes of the brain. Benington and Heller proposed that glycogen stores in brain are depleted during wakefulness and restored during sleep (Benington and Heller, 1995). We first describe how glycogen is regulated and then review the evidence for changes in glycogen in brain with sleep/wake and sleep deprivation.
B1. Metabolic Regulation of Glycogen
There are many recent in-depth reviews of glycogen metabolism and the regulation of glycogen synthesis and degradation (Brown, 2004; Brown et al., 2003; Brushia and Walsh, 1999; Greenberg et al., 2006; Gruetter, 2003; Newgard et al., 2000; Oikonomakos, 2002). Briefly follows a summary of glycogen regulation in the brain. Glycogen is present in astrocytes in the brain. The synthesis of glycogen chains from glucose molecules is catalyzed by glycogen synthase and the release of glucose-1-phosphate from glycogen is catalyzed by glycogen phosphorylase. The activities of both of these enzymes are dependent on their state of phosphorylation, but in opposite directions. Glycogen phosphorylase is activated when phosphorylated and glycogen synthase is activated when dephosphorylated. Therefore, dephosphorylation of these 2 enzymes favors the synthesis of glycogen removing glucose from the cell energy pool and phosphorylation favors the release of glucose and availability of energy for the cell.
Both glycogen phosphorylase and glycogen synthase are dephosphorylated by the same enzyme, protein phosphatase 1. In the brain, complexes are formed that place all three of these enzymes in close proximity to glycogen by protein targeting to glycogen (PTG), a scaffolding molecule. PTG is a targeting subunit of protein phosphatase 1, which directs it to its substrates (Brady et al., 1997; Ou et al., 2005; Printen et al., 1997). Unlike other targeting subunits of protein phosphatase 1, the binding of PTG to protein phosphatase 1 is not regulated by phosphorylation (Brady et al., 1997). The concentration of PTG alters the magnitude of the increase in glycogen synthesis (Green et al., 2004).
Glycogen phosphorylase is phosphorylated by phosphorylase kinase (Brushia and Walsh, 1999), while glycogen synthase is a substrate of multiple enzymes that lead to its phosphorylation (Larner et al., 1979; Lawrence et al., 1997). The change in phosphorylation state is rapid and can lead to degradation of glycogen within minutes (Cruz and Dienel, 2002; Hutchins and Rogers, 1970; Karnovsky et al., 1983). This makes accurately measuring the glycogen content of any tissue problematic, and especially difficult in the brain as glycogen levels are lower than in other tissues, although estimates of glycogen in brain may be artifactually low as a result of tissue isolation techniques during which time glycogen is degraded (Cruz and Dienel, 2002). Regardless of the basal amount of glycogen in brain tissue, measuring glycogen levels in the brain requires methods that inactivate kinase and phosphatase pathways instantly.
B2. Changes in Brain Glycogen with Sleep/Wake and Sleep Deprivation
Two general strategies have been used to preserve the glycogen content in brain tissue, thereby permitting assessment of its amount. One strategy is extreme heat, generated by exposure to high-energy focused microwave irradiation in mammalian studies (Franken et al., 2003, 2006b; Gip et al., 2002; Gip et al., 2004; Kong et al., 2002; Swanson et al., 1992) or immersion in boiling water in a Drosophila study (Zimmerman et al., 2004). The other strategy is extreme cold achieved by immersion in liquid nitrogen (Cruz and Dienel, 2002; Hutchins and Rogers, 1970; Karnovsky et al., 1983). These strategies have been used to assess changes in brain glycogen occurring with short durations of wakefulness or sleep, diurnal variations and changes following sleep deprivation.
Glycogen stores can be depleted and repleted rapidly during wake or sleep, respectively. In one study, sleep/wake state was assessed in rats and then whole animals were dropped into liquid nitrogen after a specified duration of sleep or wake. A significant decrease in whole brain glycogen is seen after 2 to 5 minutes of spontaneous wake compared to sleep levels. In contrast, 5 to 11 minutes of spontaneous sleep is sufficient to restore glycogen levels (Karnovsky et al., 1983). These studies, which have not been repeated, imply that rapid changes in glycogen take place in the brain in wake or sleep, and likely result from changes in the phosphorylation state of glycogen synthase and glycogen phosphorylase. However, since freezing in liquid nitrogen is not sufficient to prevent post-sacrifice changes in glycogen (Kong et al., 2002), these results need to be interpreted with caution.
Other studies have indicated longer-term alterations in brain glycogen. There is a diurnal variation in total brain glycogen content, which correlates with the activity of animals, both in mice (Hutchins and Rogers, 1970) and flies (Zimmerman et al., 2004). In the fly, brain glycogen content increases during the consolidated rest period to the highest level at ZT 18 and then significantly declines between ZT 18 to ZT 22; this is the beginning of the early active period for the fly (Zimmerman et al., 2004). In flies, the glycogen content of the entire head (including brain) and the rest of the body stores do not demonstrate any significant diurnal variation; only the brain exhibits diurnal changes (Zimmerman et al., 2004). In male albino mouse brains, glycogen content also rises during a period of low locomotor activity, ZT 18 (6 hours after lights off) to ZT 22, and the greatest drop in glycogen levels is from ZT 10 to ZT 14 (2 hours after the beginning of their locomotor active period) (Hutchins and Rogers, 1970). That diurnal variation in brain glycogen correlates with overall activity, both in mouse and fly, is in agreement with studies that show stimulation of facial vibrissae in the rat leads to glycogen depletion in the lateral forebrain structures, which are known to be activated by such stimulation (Swanson, 1992; Swanson et al., 1992). Thus, it is hypothesized that increased neuronal activity results in reduced glycogen levels (Swanson, 1992; Swanson et al., 1992).
However, the studies of the effects of sleep deprivation on brain glycogen have produced seemingly contradictory results both in rats and mice. One study examined the effects of prolonged sleep deprivation on 1-month-old Sprague-Dawley rats sacrificed using high-energy focused microwave irradiation. Whole brains minus cerebellum and brainstem from rats sleep deprived for 6, 12 and 24 hours were examined for glycogen content and compared to that in rats left undisturbed, allowed to sleep normally, and sacrificed at the same diurnal time (Kong et al., 2002). No differences in brain glycogen were seen between groups after 6 hours of sleep deprivation but significant decreases were observed after 12 and 24 hours of sleep deprivation (Kong et al., 2002). A separate study using 24-day-old Long-Evans rats sacrificed with focused microwave irradiation also found no changes in brain glycogen after 6 hours of sleep deprivation in the cerebral cortex, but did find a significant decrease in the cerebellum (Gip et al., 2002). (The cerebellum was not examined in the study of Kong et al. (2002).) However, after 12 hours of sleep deprivation in 59-day-old rats, glycogen had actually increased in the cortex but was unchanged in the cerebellum (Gip et al., 2002). Thus, in rats, there are regional differences in the brain with respect to changes in glycogen following sleep deprivation. Both decreases and increases have been reported and the changes observed depend on rat strain and age. Changes in glycogen in one brain region, i.e., cerebral cortex, are summarized in Table 1.
Table 1.
Effect of sleep deprivation on glycogen in cerebral cortex. Various studies assessing glycogen in the cortex of mouse and rat. Glycogen increases, decreases or stays the same depending on species, strain and length of sleep deprivation.
| Sleep Deprivation (h) | C57BL/6J Mouse | AKR/J Mouse | DBA/2J Mouse | Sprague-Dawley Rat | Long-Evans Rat (24 day old) | Long-Evans Rat (59 day old) |
|---|---|---|---|---|---|---|
| 1 | same1 | |||||
| 3 | ↑1 | |||||
| 6 | ↑↑1,2 | same2 | same2 | same3 | same4 | |
| 12 | ↓3 | ↑4 | ||||
| 24 | ↓3 |
Franken et al. 2006
Note: Kong et al. 2002 assessed tissue consisting mostly of cortex but other brain regions were present as well.
Several mouse strains have also been examined for the effects of sleep deprivation on glycogen content. In C57BL/6J mice sacrificed by focused microwave irradiation, there is no change in glycogen content in cerebral cortex after 1 hour of sleep deprivation (Franken et al., 2006b), a significant increase after 3 hours of sleep deprivation, and a further increase after 6 hours of sleep deprivation (Franken et al., 2003, 2006b). No significant changes were observed in the glycogen content of the cerebellum or brainstem after 6 hours of sleep deprivation (Franken et al., 2003). Two other strains, AKR/J and DBA/2J, demonstrated brain glycogen changes very similar to those observed in Long-Evans rats, i.e., no change in glycogen in the cortex and a decrease in the cerebellum and brainstem following 6 hours of sleep deprivation (Franken et al., 2003). Therefore, the changes in glycogen are dependent on the strain of mouse and the region of the brain (Franken et al., 2003).
In Drosophila, the effects of sleep deprivation on glycogen content are also not straightforward. Brain glycogen levels are decreased slightly but significantly after 3 hours of sleep deprivation, but, after 6 hours of sleep deprivation, glycogen levels are unaltered compared to sleeping controls sacrificed at the same diurnal time point (Zimmerman et al., 2004). Regional differences in glycogen content in the brain were not assessed in Drosophila so the extent to which regional differences contribute to these effects remains unknown.
The majority of studies to date examining the effects of sleep deprivation on glycogen in the brain have in common a study involving 6 hours of sleep deprivation (see Table 1). Following 6 hours of sleep deprivation, the changes in glycogen levels in vivo are different in rats and mice, different strains of rats and mice and different regions of the brain (see Table 1).
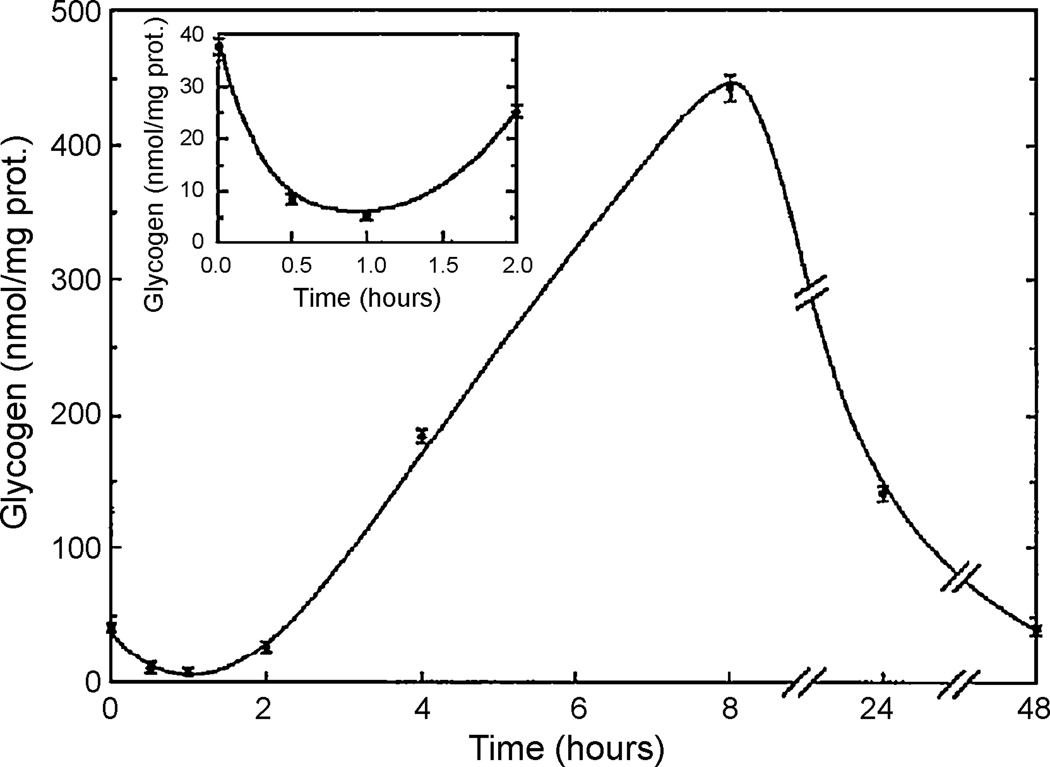
In vitro studies of glycogen utilization in primary cultures of astrocytes may hold the key to understanding these seemingly contradictory in vivo results. Exposure of primary cell cultures of mouse cerebral cortical astrocytes to wake-active neurotransmitters such as vasoactive intestinal peptide and noradrenaline depletes their glycogen content in minutes (Sorg and Magistretti, 1991), similar to the in vivo studies (Karnovsky et al., 1983) (see Figure 3). Within 30 minutes, the glycogen content drops to 25% of starting levels. However, thereafter there is synthesis of glycogen such that after 2 hours from application of the wake-active neurotransmitter, glycogen has returned to baseline levels (i.e., no change) (see Figure 3). At 8 hours following administration of the neurotransmitter, there is synthesis of glycogen and the glycogen content of the cells actually exceeds baseline levels by a substantial amount. This synthesis of glycogen is dependent upon cAMP signaling and protein synthesis (Allaman et al., 2000; Sorg and Magistretti, 1992). Finally, at 48 hours, glycogen levels return to baseline (Sorg and Magistretti, 1992) (Figure 3). Therefore, under constant exposure to wake-active neurotransmitters simulating the effects of wakefulness, there is a three-phase dynamic response: an initial glycogen degradation followed by synthesis to levels eventually above the starting levels followed by depletion once again. A similar temporal pattern of glycogen levels is found in vitro after brief exposures to wake-active neurotransmitters, although the changes are not as large (Sorg and Magistretti, 1992).
Figure 3.
Changes in glycogen in astrocytic cell culture following application of vasoactive intestinal peptide. Primary cultures of cerebral cortical astrocytes were treated with vasoactive intestinal peptide. Glycogen (in nmol/mg protein) was measured at 0.5, 1, 2, 4, 8, 24 and 48 hours after treatment. There is an initial decrease in glycogen (see inset) followed by an increase, which peaks at 8 hours. Levels of glycogen then decline again at 24 hours and 48 hours post-treatment. (Sorg and Magistretti, 1992; reprinted with permission, copyright 1992 by the Society for Neuroscience.)
The synthesis of glycogen some time after exposure to wake-active neurotransmitters (see above) is likely mediated by the scaffolding protein PTG. PTG is abundant in astrocytes and transcription of this gene is induced in primary cell cultures of astrocytes after exposure to wake-active neurotransmitters (Allaman et al., 2000). Interestingly,in vivo, PTG mRNA increases in the cerebral cortex of mice following 6 hours of sleep deprivation (Petit et al., 2002). Therefore, there is evidence in vivo that extended wakefulness, similar to the in vitro exposure of wake-active neurotransmitters, leads to an increase in PTG, which would promote glycogen synthesis.
An extension of the in vitro observations to the in vivo situation would predict that the change in glycogen in vivo, i.e., increase, decrease, or no change, depends on the temporal dynamics of two processes: glycogen depletion and synthesis (Gip et al., 2002). These two processes are predicted to occur at different rates in different species, strains of the same species or brain regions within an animal. Therefore, measurement of glycogen at any given time is a measurement of the current balance of the two opposite processes of glycogen depletion and synthesis. Differing rates of glycogen depletion and synthesis could explain the species, strain and regional variations in glycogen content observed after sleep deprivation.
B3. Conclusions about Brain Glycogen and Sleep/Wake Control
The hypothesis proposed by Benington-Heller (Benington and Heller, 1995) had the advantage that it was testable. It led to a series of studies primarily by Heller and collaborators (Franken et al., 2003, 2006b; Gip et al., 2002; Gip et al., 2004), and by our laboratory and collaborators (Kong et al., 2002; Zimmerman et al., 2004), to address whether glycogen was depleted with prolonged wakefulness as the hypothesis suggested.
While at first sight it may seem that disparate data arose from these studies, the results of the various studies can be rationalized when one considers the complex time course that has been demonstrated in vitro in response to administration of wake-active neurotransmitters such as noradrenaline (Sorg and Magistretti, 1991) that we have just described. In addition, the different levels of glycogen depletion and synthesis in vivo between strains of the same rodent at a single duration of sleep deprivation may be due to differential glucocorticoid response to the stress of sleep deprivation. Indeed, 34-day-old adrenalectomized Long-Evans rats have increased glycogen levels in the cerebral cortex after 6 hours of sleep deprivation compared to no significant change in intact animals (Gip et al., 2004). This effect of adrenalectomy may be explained by in vitro data that shows that the addition of a synthetic glucocorticoid to primary astrocyte cultures suppresses the synthesis of glycogen normally induced by noradrenaline (Allaman et al., 2004).
Studies of the time course of glycogen change in vivo are not as complete as the in vitro data, but the results are compatible with the major aspects: early depletion of glycogen, later return to baseline, still later repletion of glycogen with increased activity of glycogen synthase, and much later depletion again (Figure 3). That the in vivo data are not as complete is not surprising since, as discussed above, measurement of glycogen in vivo is challenging due to the rapid decline in its level post-sacrifice, making detailed time course measurements difficult.
The early depletion of glycogen on awakening was demonstrated in a seminal study (Karnovsky et al., 1983). This is compatible with sudden activation through phosphorylation of the degradative enzyme glycogen phosphorylase, mediated by increased cyclic AMP and activation of PKA. This suggests that early depletion of glycogen is likely part of the arousal mechanism. On sudden awakening, there is a rapid increase in neuronal firing, which will require immediately available ATP. Glycogen depletion is likely part of this as increased neuronal activity leads to increased glycogen degradation (Swanson, 1992; Swanson et al., 1992).
After this early depletion, it appears that glycogen returns to control levels as it does in vitro. Studies in rodents (Franken et al., 2003; Gip et al., 2002; Gip et al., 2004; Kong et al., 2002) and Drosophila (Zimmerman et al., 2004) show no alterations in glycogen from baseline in the cortex of most strains of rodents or whole brain of the fly after 6 hours of sleep deprivation (Table 1). Other brain areas were examined in some but not all of these studies. However, in two studies a significant increase in glycogen was observed in cortex after 3 and 6 hours of deprivation in C57BL/6J mice (Franken et al., 2003) and 12 hours in the cortex of 59-day-old Long-Evans rats (Gip et al., 2002).
The concept that glycogen is repleted even as wakefulness continues is supported by the C57BL/6J data (Franken et al., 2006b). This glycogen repletion is likely mediated by the increase in PTG (Allaman et al., 2000; Petit et al., 2002) that enhances the function of glycogen synthase (Green et al., 2004). It is surprising that given the increased energy demand of wakefulness that there would be increased synthesis of glycogen when there is continued need for increased ATP. Therefore, could this repletion of glycogen play a role in reducing ATP levels and increasing adenosine? One possible mechanism would be that synthesizing glycogen would utilize ATP and lead to increased adenosine that would promote sleepiness as predicted by Benington and Heller. If so, this would argue that timed repletion of glycogen is one of the mechanisms serving to promote sleepiness, rather than depletion of glycogen as envisaged by Benington-Heller (Benington and Heller, 1995). But this is likely to be only one of many mechanisms since multiple mechanisms seem to be used to limit wakefulness when it is prolonged (see Mackiewicz et al., 2007; Zimmerman et al., 2006; and above).
Finally, it seems that with even longer-term sleep deprivation glycogen is again depleted, i.e., after 24 hours of sleep deprivation in rat. This duration of sleep deprivation has, however, been evaluated in only one study and this needs to be replicated (Kong et al., 2002). Moreover, these data need to be interpreted with caution since longer term sleep deprivation may lead to a greater stress response, mediated by the hypothalamic-pituitary-adrenal axis, since keeping rodents awake for this long period of time is challenging due to the increasing pressure for sleep.
Thus, some data about changes in brain glycogen are compatible with the Benington-Heller hypothesis but control of glycogen in relationship to sleep/wake and sleep deprivation is more complex than they envisaged. Our analyses lead to two additional concepts about changes in brain glycogen. First, sudden depletion of glycogen is part of the arousal mechanism. Second, repletion of glycogen, which occurs as wakefulness is prolonged, is, we propose, part of the mechanism used to contribute to sleepiness.
C. Other Aspects of Energy Regulation in the Brain
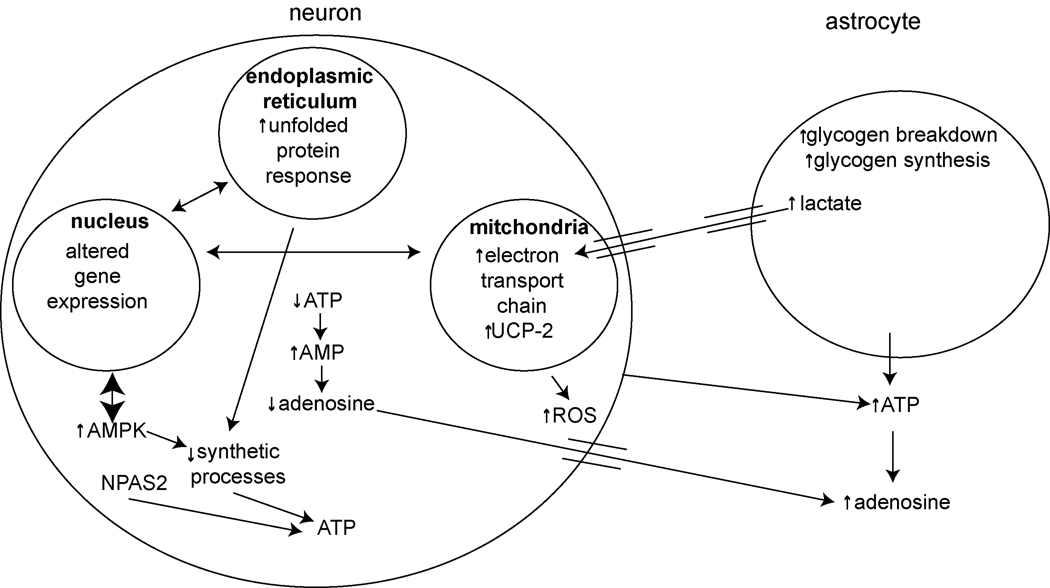
While the Benington-Heller hypothesis specifically focused on changes in adenosine and glycogen, regulation of cellular energetics in the brain is more complex and likely involves many other mediators. In this section, we discuss, albeit briefly, other energy-related mechanisms that may play a role in sleep/wake control.
C1. Unfolded Protein Response
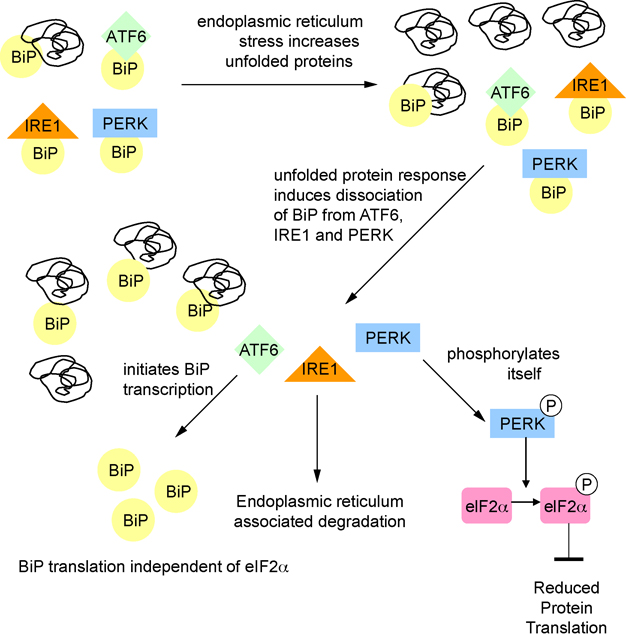
Folding of proteins in the endoplasmic reticulum is an energy-dependent process (Braakman et al., 1992; Dorner et al., 1990). When cells are energetically-challenged, there may be stress in the endoplasmic reticulum with protein misfolding ensuing (Kaufman, 2002; Lee, 2001; Schroder and Kaufman, 2005b). One of the defense mechanisms cells have to respond to misfolded proteins is to activate the unfolded protein response (reviewed in Harding et al., 2002; Ma and Hendershot, 2002; Schroder and Kaufman, 2005a; 2005b, 2006; Zhang and Kaufman, 2004). The unfolded protein response, acting through a master regulator, BiP (also known as Glucose Regulated Protein 78), and three transducers, PERK, IRE1, and ATF6, serves to reduce energy-costly protein translation, increase degradation of misfolded proteins, and to bolster the protein-folding capacity by increasing the expression of specific chaperones. BiP is an endoplasmic reticulum specific chaperone, which in addition to chaperoning misfolded proteins, plays an essential role in activating IRE1, PERK, and ATF6 in response to endoplasmic reticulum stress (Shen et al., 2002). Under conditions of endoplasmic reticulum stress, PERK phosphorylates the translation initiation factor eIF2α. When eIF2α is phosphorylated, the formation of the ternary translation initiation complex eIF2-GTP-tRNAMeti is prevented, leading to inhibition of protein translation.
Upregulation of BiP mRNA with acute short-term sleep deprivation has been described in rat cortex (Cirelli et al., 2004a; Terao et al., 2003), mouse cortex and hypothalamus (Mackiewicz et al., 2007), fly head (Shaw et al., 2000) and bird brain (Jones et al., 2008). BiP protein levels have also been shown to increase with short term sleep deprivation in mouse cortex and in fly brain (Naidoo et al., 2007; Naidoo et al., 2005). Following 6 hours of sleep deprivation, the increased BiP protein levels in fly brain decline slowly over the next 24 hours if the flies are left to sleep undisturbed (Naidoo et al., 2007). Furthermore, with 6 hours of sleep deprivation in mouse, there is dissociation of PERK from BiP, PERK activation through autophosphorylation and phosphorylation of eIF2α in the cerebral cortex (Naidoo et al., 2005) (Figure 4). These observations demonstrate activation of the unfolded protein response with sleep deprivation.
Figure 4.
Unfolded protein response during sleep deprivation. Conditions of endoplasmic reticulum stress such as sleep deprivation lead to increases in unfolded proteins. When proteins are misfolded, the chaperone BiP binds to them. BiP dissociates from ATF6, PERK, and IRE1. PERK autophosphorylates and then phosphorylates eIF2α leading to decreases in protein translation. ATF6 initiates transcription of BiP, which is translated independent of eIF2α.
While activation of the unfolded protein response is one mechanism to defend against the endoplasmic reticulum stress that occurs with extended wakefulness, the ultimate defense is to sleep so that the endoplasmic reticulum is no longer under stress. Sleep deprivation leads to increased sleep amounts in rodents (Franken et al., 1999) and in flies (Hendricks et al., 2000), i.e., there is recovery sleep. The unfolded protein response itself plays a role in determining the amount of recovery sleep. Flies overexpressing BiP show no alteration in baseline sleep/wake amounts but do show increased recovery sleep following 6 hours of sleep deprivation compared to wild-type control flies (Naidoo et al., 2007). Flies expressing a dominant negative of BiP, with reduced BiP activity, have reduced amounts of recovery sleep following sleep deprivation (Naidoo et al., 2007). These results may imply that increased BiP itself can promote sleep, although more likely is the effect of altered BiP on the unfolded protein response. Overexpression of BiP leads to a delay in the unfolded protein response since it delays the kinase PERK becoming free (Dorner et al., 1992). Thus, if the unfolded protein response is delayed, recovery sleep increases. The converse occurs with loss of function of BiP, i.e., earlier activation of the unfolded protein response and less subsequent recovery sleep.
These results indicate that the unfolded protein response is induced with sleep deprivation. The mechanism by which the unfolded protein response is activated during sleep deprivation is not known but an attractive explanation is that the unfolded protein response is activated by depletion of cellular energy occurring with extended wakefulness. We believe that the unfolded protein response is a mechanism by which the brain responds to the energetic challenge of extended wakefulness.
C2. Electron Transport Chain
Oxidative phosphorylation occurring in mitochondria is the main process by which eukaryotic cells produce ATP. In mammals, the oxidative phosphorylation system comprises five tightly regulated multi-subunit enzyme complexes, complexes I to V, transcripts of which originate from both nuclear and mitochondrial genomes and are expressed in a tissue-specific fashion (Lenka et al., 1998). Addition of electrons to complex I or II allows for the establishment of a proton gradient, which ultimately leads to ATP synthesis.
There is evidence of alterations in mitochondrial ATP production with sleep/wake. Increased transcription of components of the oxidative phosphorylation machinery from the mitochondrial genome (subunit I of cytochrome C oxidase, subunit 2 of NADH dehydrogenase, and 12S rRNA) has been shown in cortex after 3 hours of spontaneous wakefulness or sleep deprivation (Cirelli and Tononi, 1998). An early up-regulation of the mRNA of the mitochondrial encoded subunit 1 of cytochrome C oxidase with 3 hours of sleep deprivation has also been seen in fly heads (Shaw et al., 2000). In addition to transcriptional upregulation, increased activity of cytochrome C oxidase enzyme, a key enzyme in respiratory control, has been described after 3 hours of sleep deprivation in multiple brain regions in rat, including cortex (Nikonova et al., 2005b). Further studies extend that finding by demonstrating that cytochrome C oxidase enzyme activity in mouse cerebral cortex remains elevated even after 12 hours of sleep deprivation (Nikonova et al., 2005a). Both transcript and protein levels of mitochondrial-encoded cytochrome C oxidase subunit I and nuclear-encoded cytochrome C oxidase subunit IV are increased after 12 hours of sleep deprivation (Nikonova et al., 2005a). Somewhat surprisingly, an extensive microarray study in mouse found that many nuclear encoded components for the enzyme subunits of the various complexes in the electron transport chain are upregulated during sleep, not wakefulness (Mackiewicz et al., 2007). It is possible that mitochondrial and nuclear proteins are synthesized in temporally distinct profiles, which may simply reflect the fact that mitochondrial proteins can be made in areas of the cell that are very metabolically active such as the nerve terminal (Wong-Riley et al., 1997), whereas nuclear proteins need to be transported longer distances to their sites of action. Nonetheless, the increase in cytochrome C oxidase enzyme activity and the upregulation of subunits I and IV suggests that mitochondria increase energy production to meet the increased energy demand occurring during wakefulness.
C3. Transcriptional Factors of the Molecular Clock
The CLOCK protein is a key component of the circadian clock in the suprachiasmatic nucleus, the master circadian pacemaker. It is a transcription factor that is part of the negative feedback loop producing the circadian oscillation in gene expression (reviewed in Lowrey and Takahashi, 2004). Mice with a mutation of clock have increased activity during the normally quiescent light period and decreased activity during the active dark period (Turek et al., 2005). Furthermore, clock mutant mice have increased food intake and metabolic rate during the light period and decreased food intake and metabolic rate during the dark period (Turek et al., 2005). Overall, the clock mutant mice are hyperphagic and obese (Turek et al., 2005). These data suggest a link between circadian and energy-regulating processes. However, it should be noted, that the mechanism by which clock alters energy regulation is unknown, it may be a central or a peripheral process, e.g., in kidney or liver, since clock is expressed in many tissues (Chilov et al., 2001).
Another transcription factor involved in the clock mechanism is NPAS2. NPAS2 has been proposed as a specific energy sensor for control of rest/activity (sleep/wake) (Rutter et al., 2002). NPAS2 is a transcription factor that is involved in regulation of expression of circadian clock genes. Transcription of these genes is controlled by a transcription complex of CLOCK/BMAL or NPAS2/BMAL, i.e., NPAS2 acts in a similar fashion to CLOCK (Reick et al., 2001). The main transcription factor complex in the master clock, the suprachiasmatic nucleus, is CLOCK/BMAL, while NPAS2/BMAL functions outside the suprachiasmatic nucleus, including in other brain regions (Rutter et al., 2002).
The concept that NPAS2 may act as an energy sensor came from in vitro observations that the dimerization and the DNA binding of NPAS2:BMAL heterodimers are markedly affected by the energy status as assessed by the redox state of NAD cofactors (Rutter et al., 2001). NPAS2 alters sleep/wake amounts and entrainment of sleep/wake cycles to stimuli other than light as revealed by studies of the NPAS2 knockout mice (Dudley et al., 2003; Franken et al., 2006a). The NPAS2 knockout mouse has less sleep, specifically NREM sleep, and more wakefulness than wild-type controls during the night, which is the active period of the mouse (Franken et al., 2006a). During the night, wild-type mice have "siesta sleep" that is not found in NPAS2 knockout mice (Dudley et al., 2003; Franken et al., 2006a).
NPAS2 knockout mice also have markedly impaired entrainment to altered feeding patterns, i.e., restricted feeding (Dudley et al., 2003). In restricted feeding, mice are fed only during the day when they are normally inactive. Over a short period of days their rest/activity patterns shift such that mice become active during the day in anticipation of food. This phenomenon is described in both mice (Marchant and Mistlberger, 1997) and rats (Mistlberger et al., 1990). It occurs in suprachiasmatic nucleus lesioned animals and hence does not involve the suprachiasmatic nucleus (Marchant and Mistlberger, 1997). After entrainment to this new pattern of feeding, the diurnal rhythm of clock gene expression is altered in, for example, liver and heart, but not in the suprachiasmatic nucleus (Damiola et al., 2000; Stokkan et al., 2001; Watanabe et al., 2006). NPAS2 is very relevant to this process. Mice lacking NPAS2 have very delayed entrainment to restricted feeding; such knockout mice lose weight due to lack of food and some die before being entrained (Dudley et al., 2003). In NPAS2 knockout mice, entrainment to light (mediated by the suprachiasmatic nucleus) is normal, or indeed, enhanced (Dudley et al., 2003). Thus, there is a clock mechanism that involves NPAS2 outside the suprachiasmatic nucleus, thought to be in forebrain (Dudley et al., 2003) that can alter both the timing of rest/activity (sleep/wake) cycles as well as the amount of sleep. This mechanism may be affected by energy status and hence provides a direct mechanism to link the energy state of brain to sleep/wake control.
C4. AMP-activated protein kinase
Another mechanism by which a cell can respond to an energetic challenge is by the AMP-activated protein kinase (AMPK), which is a cellular adaptor to alterations in energy state. AMPK is activated by phosphorylation when energy consumed exceeds energy produced and the ratio of AMP to ATP increases. Activated AMPK preserves ATP by decreasing energy consuming processes and increasing energy producing processes (reviewed in Hardie et al., 2006).
Assessing the phosphorylation status of AMPK in vivo is challenging (Scharf et al., 2008). Preliminary results from our laboratory have shown that AMPK phosphorylation is greater following 6 hours of sleep deprivation than sleep in whole brain, cortex and basal forebrain consistent with the brain being energetically challenged in wakefulness (Scharf et al., 2006), and we are currently assessing changes in AMPK phosphorylation with sleep deprivation utilizing a methodology we developed to preserve in vivo AMPK phosphorylation (Scharf et al., 2008). Thus, AMPK may serve as a molecular sensor to shift the brain from energy-consuming synthetic processes that occur during sleep to catabolic energy-producing processes that occur during extended wakefulness.
C5. Astrocyte-to-Neuron Lactate Shuttle
Another postulated metabolic role for astrocytes is the astrocyte-to-neuron lactate shuttle although this idea has been controversial since it was first proposed (Pellerin and Magistretti, 1994). Recent reviews have either supported (Bergersen, 2007; Pellerin et al., 2007) or contested (Fillenz, 2005; Simpson et al., 2007) the hypothesis. The central tenet of the hypothesis is that during neuronal activation, astrocytes take up glucose from blood in adjacent capillaries and metabolize the glucose into lactate. The astrocytes release lactate, which is transported into neurons, where it serves as an energy source for neurons to meet increased energy demand occurring with neuronal activation (Pellerin and Magistretti, 1994).
Components that could mediate the lactate shuttle have been found to be upregulated during sleep deprivation. Lactate dehydrogenase 2 B protein, which is associated with astrocytes (Pellerin et al., 1998) and catalyzes the conversion of pyruvate to lactate (Bishop et al., 1972), is significantly increased in mouse cerebral cortex after 6 hours of sleep deprivation (Pawlyk et al., 2007). Furthermore, glucose transporter 1, associated with epithelial cells of the blood-brain barrier and astrocytes (reviewed in Duelli and Kuschinsky, 2001), shows increased gene expression in rat cerebral cortex following sleep deprivation, and is also increased in spontaneous wakefulness compared to sleep (Cirelli et al., 2004b; Cirelli and Tononi, 2000). Presumably, increased glucose transporter 1 would facilitate glucose transport into astrocytes. The observations of increased lactate dehydrogenase 2 B protein and glucose transporter 1 mRNA are consistent with increased activity of the astrocyte-neuron lactate shuttle during wakefulness, and suggest that this may be another mechanism by which the brain adapts to the increased metabolic demand of wakefulness.
C6. Reactive Oxygen Species
One of the consequences of increased metabolic activity and, hence, an increase in oxygen consumption, is an increase in production of reactive oxygen species. Ikeda et al. proposed that reactive oxygen species are produced during wakefulness and that increased antioxidant enzyme activity during sleep reduces their levels in preparation for subsequent wakefulness (Ikeda et al., 2005). Reactive oxygen species are difficult to measure in vivo since they are so labile. Hence, evidence that they change in the brain with sleep/wake is indirect.
The evidence for alteration in reactive oxygen species production with sleep/wake is limited. Activity of a key antioxidant enzyme, superoxide dismutase, decreases in hippocampus and brainstem following sleep deprivation (Ramanathan et al., 2002). Moreover, mRNA for a number of antioxidant enzymes, including superoxide dismutase, glutathione S-transferase, glutathione peroxidase, glutathione reductase, catalase, methionine sulfoxide reductase, thioredoxin reductase and the transcription factor Nfe2, which is involved in induction of genes encoding antioxidant proteins and certain detoxifying enzymes, are all upregulated during sleep in cerebral cortex as revealed by a microarray study in mouse (Mackiewicz et al., 2007).
There is also evidence that reactive oxygen species may also directly alter sleep. Intracerebroventricular infusion of oxidized glutathione increases slow wave and REM sleep in rats (Ikeda et al., 2005) although the mechanism by which it does so is unknown. Intracerebroventricular administration of an inhibitor of glutathione peroxidase, tert-butyl hydroperoxide (TBHP), which would attenuate metabolism of reactive oxygen species thereby increasing their level, also increases the amount of NREM and REM sleep (Ikeda et al., 2005). Although the data are limited, these studies are consistent with the notion that there are alterations in reactive oxygen species with sleep/wake.
C6. Uncoupling Proteins
The first described uncoupling protein, UCP1, was shown to increase the proton conductance of the inner mitochondrial membrane and, therefore, uncouple oxidative phosphorylation. Consistent with this uncoupling mechanism, UCP1 in brown adipose tissue was shown to catalyze adaptive thermogenesis by increasing heat production (Cannon and Nedergaard, 2004). Additional UCPs found in other tissues without a clear thermogenic function include UCP2 (Fleury et al., 1997), UCP3 (Boss et al., 1997), UCP4 (Mao et al., 1999) and UCP5 (BMCP1) (Sanchis et al., 1998). Interestingly, UCP2, UCP4 and UCP5 are expressed in brain (see Andrews et al., 2005). The role of these new UCPs, in particular in the brain, is a subject of debate, i.e., do they act to uncouple oxidative phosphorylation (see Kim-Han and Dugan, 2005)?
There are no differences in UCP2 or UCP5 mRNA expression in the cerebral cortex between spontaneous wakefulness and sleep in rats (Cirelli and Tononi, 2004) (mRNA for UCP4 could not be detected). However, short-term sleep deprivation of 8 hours in rats leads to a 40–50% increase in mRNA for UCP2 and UCP5 in cerebral cortex (Cirelli and Tononi, 2004). Thus, increased expression of UCPs in brain may be, like the unfolded protein response, part of the mechanism to protect against the effects of extending wakefulness. Uncoupling of oxidative phosphorylation will likely result in decreased production of reactive oxygen species.
Conclusion
The Benington-Heller hypothesis stimulated a lot of useful research. While it has turned out to be an oversimplification, this does not detract from its value in providing a focus to investigate the role of energy utilization in determining sleep/wake states. As proposed, there is increasing evidence that adenosine does act to promote sleep. There are, however, questions about whether changes in adenosine with wakefulness reflect altered cellular energy states. The elaboration of the Benington-Heller hypothesis that adenosine changes are specifically “sensed” by cholinergic neurons of the basal forebrain to effect sleep/wake control now seems disproven. Adenosine is likely to act in different brain regions to alter both the amount of sleep as well as delta power. How adenosine regulates the quantity and quality of sleep and how adenosine exerts its effects in specific brain regions are questions that are being actively investigated. The emerging role of astrocytes in sleep homeostasis is likely to be important since astrocytes contribute to adenosinergic tone.
The predictions of Benington and Heller about glycogen changes with sleep/wake and sleep deprivation have been supported, at least in part. However, the dynamic changes are more complex than they proposed. A reassessment of data obtained in experiments stimulated by their hypothesis leads to new hypotheses: a) depletion of glycogen is part of the arousal response providing ATP for the sudden increase in energy required to support enhanced neuronal firing on sudden awakening; and b) it is repletion, not depletion, of glycogen that is likely the signal to promote sleepiness.
Control of energy utilization in the brain is, however, more complex than is simply reflected by changes in adenosine and glycogen. There are growing data that many aspects of the regulation of cellular energetics in the brain are altered with sleep/wake and sleep deprivation: the unfolded protein response; components of the electron transport chain; the astrocyte-to-neuron lactate shuttle; antioxidant enzymes; uncoupling proteins; clock transcription factors and the degree of activation of the cellular energy regulator AMPK. Available data suggest that there may be regulation of many components to provide the necessary amounts of ATP that are required during wakefulness and to provide responses to the energetic challenge of wakefulness. In turn, these pathways likely mediate the quantity and quality of sleep.
Altogether, the evidence reviewed here supports the fundamental tenet that extended wakefulness poses an energetic challenge to the brain. It would therefore appear likely that sleep is a stage of synthesis necessary to respond to the “insult” of wakefulness as we have proposed (Mackiewicz et al., 2007). We believe that energy-related pathways in addition to glycogen and adenosine are involved in transitioning the brain from the metabolically-deplete catabolic state of wakefulness to the metabolically-replete anabolic state of sleep. This elaboration of the energy hypothesis of sleep is illustrated in Figure 5. Hopefully, this new view of the energy hypothesis of sleep-wake control will stimulate as much helpful research as did the provocative review of Benington and Heller (Benington and Heller, 1995).
Figure 5.
Proposed model of alterations in energy homeostasis during extended wakefulness. In astrocytes, glycogen breakdown and synthesis occur concomitantly. Lactate is transported out of astrocytes and taken up by neurons where it enters the mitochondria and the krebs cycle to produce ATP. Also within the mitochondria, there is an increase in the activity of the electron transport chain to increase ATP production. Reactive oxygen species are produced as a byproduct of this increased activity of the electron transport chain. UCP-2 increases to attenuate production of reactive oxygen species. In the endoplasmic reticulum, the unfolded protein response occurs. This ultimately leads to transcriptional changes as well as decreases in energy-consuming synthetic processes. AMPK is activated, which also attenuates energy-consuming synthetic processes. NPAS2 most likely also acts to preserve energy. Multiple pathways lead to changes in gene expression. Adenosine increases in the extracellular space either by transport out of the cell and/or by conversion of ATP released from glia or neurons into adenosine.
Acknowledgements
We would like to thank Daniel Barrett and Jennifer Montoya for help in preparation of this manuscript. This research was supported by the National Institute of Aging Grant AG-17628 and National Heart, Lung, and Blood Institute Grants HL-60287 and HL-07953.
Abbreviations Used
- A1
adenosine 1
- A2A
adenosine 2A
- A2B
adenosine 2B
- A3
adenosine 3
- ADP
adenosine diphosphate
- AMP
adenosine monophosphate
- AMPK
AMP-activated protein kinase
- ATP
adenosine triphosphate
- CGS
2-[p-(2-carboxyethyl)phenethylamino]-5'-N-ethylcarboxamido adenosine
- CHA
N6-cyclo-hexyladenosine
- CPA
cyclopentyladenosine
- CPDX
8-cyclopentyl-1, 3-dimethylxanthine
- CPT
cyclopentyl-1-3-dimethylxanthine
- CV-1808
2-phenylaminoadenosine
- EEG
electroencephalogram
- GABA
gamma-amino butyric acid
- I-κB
inhibitory κ B
- NBTI
nitrobenzylthioinosine
- NF-κB
nuclear factor κ B
- NPAS2
neuronal Per-Arnt-Sim-type signal-sensor protein (PAS) domain protein 2
- NREM
non-rapid eye movement sleep
- PTG
protein targeting to glycogen
- REM
rapid eye movement sleep
- TBHP
tert-butyl hydroperoxide
- UCP
uncoupling protein
- ZT
zeitgeiber
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alam MN, Szymusiak R, Gong H, King J, McGinty D. Adenosinergic modulation of rat basal forebrain neurons during sleep and waking: neuronal recording with microdialysis. J Physiol. 1999;521(Pt 3):679–690. doi: 10.1111/j.1469-7793.1999.00679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 2002 [Google Scholar]
- Allaman I, Pellerin L, Magistretti PJ. Protein targeting to glycogen mRNA expression is stimulated by noradrenaline in mouse cortical astrocytes. Glia. 2000;30:382–391. [PubMed] [Google Scholar]
- Allaman I, Pellerin L, Magistretti PJ. Glucocorticoids modulate neurotransmitter-induced glycogen metabolism in cultured cortical astrocytes. J Neurochem. 2004;88:900–908. doi: 10.1046/j.1471-4159.2003.02235.x. [DOI] [PubMed] [Google Scholar]
- Andrews ZB, Diano S, Horvath TL. Mitochondrial uncoupling proteins in the CNS: in support of function and survival. Nat Rev Neurosci. 2005;6:829–840. doi: 10.1038/nrn1767. [DOI] [PubMed] [Google Scholar]
- Arrigoni E, Chamberlin NL, Saper CB, McCarley RW. Adenosine inhibits basal forebrain cholinergic and noncholinergic neurons in vitro. Neuroscience. 2006;140:403–413. doi: 10.1016/j.neuroscience.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Arrigoni E, Rainnie DG, McCarley RW, Greene RW. Adenosine-mediated presynaptic modulation of glutamatergic transmission in the laterodorsal tegmentum. J Neurosci. 2001;21:1076–1085. doi: 10.1523/JNEUROSCI.21-03-01076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balschun D, Wolfer DP, Gass P, Mantamadiotis T, Welzl H, Schutz G, Frey JU, Lipp HP. Does cAMP response element-binding protein have a pivotal role in hippocampal synaptic plasticity and hippocampus-dependent memory? J Neurosci. 2003;23:6304–6314. doi: 10.1523/JNEUROSCI.23-15-06304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basheer R, Arrigoni E, Thatte HS, Greene RW, Ambudkar IS, McCarley RW. Adenosine induces inositol 1,4,5-trisphosphate receptor-mediated mobilization of intracellular calcium stores in basal forebrain cholinergic neurons. J Neurosci. 2002;22:7680–7686. doi: 10.1523/JNEUROSCI.22-17-07680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basheer R, Halldner L, Alanko L, McCarley RW, Fredholm BB, Porkka-Heiskanen T. Opposite changes in adenosine A1 and A2A receptor mRNA in the rat following sleep deprivation. Neuroreport. 2001a;12:1577–1580. doi: 10.1097/00001756-200106130-00013. [DOI] [PubMed] [Google Scholar]
- Basheer R, Porkka-Heiskanen T, Stenberg D, McCarley RW. Adenosine and behavioral state control: adenosine increases c-Fos protein and AP1 binding in basal forebrain of rats. Brain Res Mol Brain Res. 1999;73:1–10. doi: 10.1016/s0169-328x(99)00219-3. [DOI] [PubMed] [Google Scholar]
- Basheer R, Rainnie DG, Porkka-Heiskanen T, Ramesh V, McCarley RW. Adenosine, prolonged wakefulness, and A1-activated NF-kappaB DNA binding in the basal forebrain of the rat. Neuroscience. 2001b;104:731–739. doi: 10.1016/s0306-4522(01)00111-7. [DOI] [PubMed] [Google Scholar]
- Basheer R, Strecker RE, Thakkar MM, McCarley RW. Adenosine and sleep-wake regulation. Prog Neurobiol. 2004;73:379–396. doi: 10.1016/j.pneurobio.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Benington JH, Heller HC. Restoration of brain energy metabolism as the function of sleep. Prog Neurobiol. 1995;45:347–360. doi: 10.1016/0301-0082(94)00057-o. [DOI] [PubMed] [Google Scholar]
- Benington JH, Kodali SK, Heller HC. Stimulation of A1 adenosine receptors mimics the electroencephalographic effects of sleep deprivation. Brain Res. 1995;692:79–85. doi: 10.1016/0006-8993(95)00590-m. [DOI] [PubMed] [Google Scholar]
- Bergersen LH. Is lactate food for neurons? Comparison of monocarboxylate transporter subtypes in brain and muscle. Neuroscience. 2007;145:11–19. doi: 10.1016/j.neuroscience.2006.11.062. [DOI] [PubMed] [Google Scholar]
- Bishop MJ, Everse J, Kaplan NO. Identification of lactate dehydrogenase isoenzymes by rapid kinetics. Proc Natl Acad Sci U S A. 1972;69:1761–1765. doi: 10.1073/pnas.69.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Centurion C, Gerashchenko D, Shiromani PJ. Effects of saporin-induced lesions of three arousal populations on daily levels of sleep and wake. J Neurosci. 2007;27:14041–14048. doi: 10.1523/JNEUROSCI.3217-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Centurion C, Xu M, Murillo-Rodriguez E, Gerashchenko D, Shiromani AM, Salin-Pascual RJ, Hof PR, Shiromani PJ. Adenosine and sleep homeostasis in the Basal forebrain. J Neurosci. 2006;26:8092–8100. doi: 10.1523/JNEUROSCI.2181-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boison D. Adenosine kinase, epilepsy and stroke: mechanisms and therapies. Trends Pharmacol Sci. 2006;27:652–658. doi: 10.1016/j.tips.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Borbely AA, Achermann P. Sleep homeostasis and models of sleep regulation. J Biol Rhythms. 1999;14:557–568. doi: 10.1177/074873099129000894. [DOI] [PubMed] [Google Scholar]
- Boss O, Samec S, Paoloni-Giacobino A, Rossier C, Dulloo A, Seydoux J, Muzzin P, Giacobino JP. Uncoupling protein-3: a new member of the mitochondrial carrier family with tissue-specific expression. FEBS Lett. 1997;408:39–42. doi: 10.1016/s0014-5793(97)00384-0. [DOI] [PubMed] [Google Scholar]
- Braakman I, Helenius J, Helenius A. Role of ATP and disulphide bonds during protein folding in the endoplasmic reticulum. Nature. 1992;356:260–262. doi: 10.1038/356260a0. [DOI] [PubMed] [Google Scholar]
- Brady MJ, Printen JA, Mastick CC, Saltiel AR. Role of protein targeting to glycogen (PTG) in the regulation of protein phosphatase-1 activity. J Biol Chem. 1997;272:20198–20204. doi: 10.1074/jbc.272.32.20198. [DOI] [PubMed] [Google Scholar]
- Brown AM. Brain glycogen re-awakened. J Neurochem. 2004;89:537–552. doi: 10.1111/j.1471-4159.2004.02421.x. [DOI] [PubMed] [Google Scholar]
- Brown AM, Tekkok SB, Ransom BR. Glycogen regulation and functional role in mouse white matter. J Physiol. 2003;549:501–512. doi: 10.1113/jphysiol.2003.042416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brushia RJ, Walsh DA. Phosphorylase kinase: the complexity of its regulation is reflected in the complexity of its structure. Front Biosci. 1999;4:D618–D641. doi: 10.2741/brushia. [DOI] [PubMed] [Google Scholar]
- Bushong EA, Martone ME, Jones YZ, Ellisman MH. Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J Neurosci. 2002;22:183–192. doi: 10.1523/JNEUROSCI.22-01-00183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Cass CE, Young JD, Baldwin SA. Recent advances in the molecular biology of nucleoside transporters of mammalian cells. Biochem Cell Biol. 1998;76:761–770. doi: 10.1139/bcb-76-5-761. [DOI] [PubMed] [Google Scholar]
- Chamberlin NL, Arrigoni E, Chou TC, Scammell TE, Greene RW, Saper CB. Effects of adenosine on gabaergic synaptic inputs to identified ventrolateral preoptic neurons. Neuroscience. 2003;119:913–918. doi: 10.1016/s0306-4522(03)00246-x. [DOI] [PubMed] [Google Scholar]
- Chilov D, Hofer T, Bauer C, Wenger RH, Gassmann M. Hypoxia affects expression of circadian genes PER1 and CLOCK in mouse brain. Faseb J. 2001;15:2613–2622. doi: 10.1096/fj.01-0092com. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Gutierrez CM, Tononi G. Extensive and Divergent Effects of Sleep and Wakefulness on Brain Gene Expression. Neuron. 2004a;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004b;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Tononi G. Differences in gene expression between sleep and waking as revealed by mRNA differential display. Brain Res Mol Brain Res. 1998;56:293–305. doi: 10.1016/s0169-328x(98)00057-6. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Tononi G. Gene expression in the brain across the sleep-waking cycle. Brain Res. 2000;885:303–321. doi: 10.1016/s0006-8993(00)03008-0. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Tononi G. Uncoupling proteins and sleep deprivation. Arch Ital Biol. 2004;142:541–549. [PubMed] [Google Scholar]
- Coleman CG, Baghdoyan HA, Lydic R. Dialysis delivery of an adenosine A2A agonist into the pontine reticular formation of C57BL/6J mouse increases pontine acetylcholine release and sleep. J Neurochem. 2006;96:1750–1759. doi: 10.1111/j.1471-4159.2006.03700.x. [DOI] [PubMed] [Google Scholar]
- Cruz NF, Dienel GA. High glycogen levels in brains of rats with minimal environmental stimuli: implications for metabolic contributions of working astrocytes. J Cereb Blood Flow Metab. 2002;22:1476–1489. doi: 10.1097/01.WCB.0000034362.37277.C0. [DOI] [PubMed] [Google Scholar]
- Cunha RA, Sebastiao AM, Ribeiro JA. Inhibition by ATP of hippocampal synaptic transmission requires localized extracellular catabolism by ecto-nucleotidases into adenosine and channeling to adenosine A1 receptors. J Neurosci. 1998;18:1987–1995. doi: 10.1523/JNEUROSCI.18-06-01987.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner AJ, Wasley LC, Kaufman RJ. Protein dissociation from GRP78 and secretion are blocked by depletion of cellular ATP levels. Proc Natl Acad Sci U S A. 1990;87:7429–7432. doi: 10.1073/pnas.87.19.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner AJ, Wasley LC, Kaufman RJ. Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. Embo J. 1992;11:1563–1571. doi: 10.1002/j.1460-2075.1992.tb05201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley CA, Erbel-Sieler C, Estill SJ, Reick M, Franken P, Pitts S, McKnight SL. Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science. 2003;301:379–383. doi: 10.1126/science.1082795. [DOI] [PubMed] [Google Scholar]
- Duelli R, Kuschinsky W. Brain glucose transporters: relationship to local energy demand. News Physiol Sci. 2001;16:71–76. doi: 10.1152/physiologyonline.2001.16.2.71. [DOI] [PubMed] [Google Scholar]
- Dunwiddie TV, Diao L, Proctor WR. Adenine nucleotides undergo rapid, quantitative conversion to adenosine in the extracellular space in rat hippocampus. J Neurosci. 1997;17:7673–7682. doi: 10.1523/JNEUROSCI.17-20-07673.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwiddie TV, Masino SA. The role and regulation of adenosine in the central nervous system. Annu Rev Neurosci. 2001;24:31–55. doi: 10.1146/annurev.neuro.24.1.31. [DOI] [PubMed] [Google Scholar]
- Feldberg W, Sherwood SL. Injections of Drugs Into the Lateral Ventricle of the Cat. J Physiol. 1954;123:148–167. doi: 10.1113/jphysiol.1954.sp005040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellin T, Pascual O, Haydon PG. Astrocytes coordinate synaptic networks: balanced excitation and inhibition. Physiology (Bethesda) 2006;21:208–215. doi: 10.1152/physiol.00161.2005. [DOI] [PubMed] [Google Scholar]
- Fillenz M. The role of lactate in brain metabolism. Neurochem Int. 2005;47:413–417. doi: 10.1016/j.neuint.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Fleury C, Neverova M, Collins S, Raimbault S, Champigny O, Levi-Meyrueis C, Bouillaud F, Seldin MF, Surwit RS, Ricquier D, Warden CH. Uncoupling protein-2: a novel gene linked to obesity and hyperinsulinemia. Nat Genet. 1997;15:269–272. doi: 10.1038/ng0397-269. [DOI] [PubMed] [Google Scholar]
- Franken P, Dudley CA, Estill SJ, Barakat M, Thomason R, O'hara BF, McKnight SL. NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: genotype and sex interactions. Proc Natl Acad Sci U S A. 2006a;103:7118–7123. doi: 10.1073/pnas.0602006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken P, Gip P, Hagiwara G, Ruby NF, Heller HC. Changes in brain glycogen after sleep deprivation vary with genotype. Am J Physiol Regul Integr Comp Physiol. 2003;285:R413–R419. doi: 10.1152/ajpregu.00668.2002. [DOI] [PubMed] [Google Scholar]
- Franken P, Gip P, Hagiwara G, Ruby NF, Heller HC. Glycogen content in the cerebral cortex increases with sleep loss in C57BL/6J mice. Neurosci Lett. 2006b. [DOI] [PubMed]
- Franken P, Malafosse A, Tafti M. Genetic determinants of sleep regulation in inbred mice. Sleep. 1999;22:155–169. [PubMed] [Google Scholar]
- Gallopin T, Luppi PH, Cauli B, Urade Y, Rossier J, Hayaishi O, Lambolez B, Fort P. The endogenous somnogen adenosine excites a subset of sleep-promoting neurons via A2A receptors in the ventrolateral preoptic nucleus. Neuroscience. 2005;134:1377–1390. doi: 10.1016/j.neuroscience.2005.05.045. [DOI] [PubMed] [Google Scholar]
- Gerashchenko D, Okano Y, Urade Y, Inoue S, Hayaishi O. Strong rebound of wakefulness follows prostaglandin D2- or adenosine A2a receptor agonist-induced sleep. J Sleep Res. 2000;9:81–87. doi: 10.1046/j.1365-2869.2000.00175.x. [DOI] [PubMed] [Google Scholar]
- Gip P, Hagiwara G, Ruby NF, Heller HC. Sleep deprivation decreases glycogen in the cerebellum but not in the cortex of young rats. Am J Physiol Regul Integr Comp Physiol. 2002;283:R54–R59. doi: 10.1152/ajpregu.00735.2001. [DOI] [PubMed] [Google Scholar]
- Gip P, Hagiwara G, Sapolsky RM, Cao VH, Heller HC, Ruby NF. Glucocorticoids influence brain glycogen levels during sleep deprivation. Am J Physiol Regul Integr Comp Physiol. 2004;286:R1057–R1062. doi: 10.1152/ajpregu.00528.2003. [DOI] [PubMed] [Google Scholar]
- Green AR, Aiston S, Greenberg CC, Freeman S, Poucher SM, Brady MJ, Agius L. The glycogenic action of protein targeting to glycogen in hepatocytes involves multiple mechanisms including phosphorylase inactivation and glycogen synthase translocation. J Biol Chem. 2004;279:46474–46482. doi: 10.1074/jbc.M405660200. [DOI] [PubMed] [Google Scholar]
- Greenberg CC, Jurczak MJ, Danos AM, Brady MJ. Glycogen branches out: new perspectives on the role of glycogen metabolism in the integration of metabolic pathways. Am J Physiol Endocrinol Metab. 2006;291:E1–E8. doi: 10.1152/ajpendo.00652.2005. [DOI] [PubMed] [Google Scholar]
- Gruetter R. Glycogen: the forgotten cerebral energy store. J Neurosci Res. 2003;74:179–183. doi: 10.1002/jnr.10785. [DOI] [PubMed] [Google Scholar]
- Halassa MM, Fellin T, Munoz JR, Haydon PG, Frank MG. Astrocytes regulate sleep homeostasis. Soc Neurosci Abs. 2007:631.16. [Google Scholar]
- Hardie DG, Hawley SA, Scott JW. AMP-activated protein kinase--development of the energy sensor concept. J Physiol. 2006;574:7–15. doi: 10.1113/jphysiol.2006.108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Calfon M, Urano F, Novoa I, Ron D. Transcriptional and translational control in the Mammalian unfolded protein response. Annu Rev Cell Dev Biol. 2002;18:575–599. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- Haulica I, Ababei L, Branisteanu D, Topoliceanu F. Letter: Preliminary data on the possible hypnogenic role of adenosine. J Neurochem. 1973;21:1019–1020. doi: 10.1111/j.1471-4159.1973.tb07549.x. [DOI] [PubMed] [Google Scholar]
- Heckers S, Ohtake T, Wiley RG, Lappi DA, Geula C, Mesulam MM. Complete and selective cholinergic denervation of rat neocortex and hippocampus but not amygdala by an immunotoxin against the p75 NGF receptor. J Neurosci. 1994;14:1271–1289. doi: 10.1523/JNEUROSCI.14-03-01271.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller HC. A global rather than local role for adenosine in sleep homeostasis. Sleep. 2006;29:1382–1383. doi: 10.1093/sleep/29.11.1382. discussion 1387–9. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Huang ZL, Qu WM, Eguchi N, Chen JF, Schwarzschild MA, Fredholm BB, Urade Y, Hayaishi O. Adenosine A2A, but not A1, receptors mediate the arousal effect of caffeine. Nat Neurosci. 2005;8:858–859. doi: 10.1038/nn1491. [DOI] [PubMed] [Google Scholar]
- Hutchins DA, Rogers KJ. Physiological and drug-induced changes in the glycogen content of mouse brain. Br J Pharmacol. 1970;39:9–25. doi: 10.1111/j.1476-5381.1970.tb09551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison AJ, Webb RL, Oei HH, Ghai GR, Zimmerman MB, Williams M. CGS 21680C, an A2 selective adenosine receptor agonist with preferential hypotensive activity. J Pharmacol Exp Ther. 1989;251:47–55. [PubMed] [Google Scholar]
- Ikeda M, Ikeda-Sagara M, Okada T, Clement P, Urade Y, Nagai T, Sugiyama T, Yoshioka T, Honda K, Inoue S. Brain oxidation is an initial process in sleep induction. Neuroscience. 2005;130:1029–1040. doi: 10.1016/j.neuroscience.2004.09.057. [DOI] [PubMed] [Google Scholar]
- Jones BE. Basic Mechanisms of Sleep-Wake States. In: Kryger MH, Roth T, Dement WC, editors. Principles and Practice of Sleep Medicine. 2005. pp. 136–153. [Google Scholar]
- Jones S, Pfister-Genskow M, Benca RM, Cirelli C. Molecular correlates of sleep and wakefulness in the brain of the white-crowned sparrow. J Neurochem. 2008;105:46–62. doi: 10.1111/j.1471-4159.2007.05089.x. [DOI] [PubMed] [Google Scholar]
- Kalinchuk AV, Porkka-Heiskanen T, McCarley RW. Basal forebrain and saporin cholinergic lesions: the devil dwells in delivery details. Sleep. 2006;29:1385–1387. doi: 10.1093/sleep/29.11.1385. discussion 1387–9. [DOI] [PubMed] [Google Scholar]
- Karnovsky ML, Reich P, Anchors JM, Burrows BL. Changes in brain glycogen during slow-wave sleep in the rat. J Neurochem. 1983;41:1498–1501. doi: 10.1111/j.1471-4159.1983.tb00853.x. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Orchestrating the unfolded protein response in health and disease. J Clin Invest. 2002;110:1389–1398. doi: 10.1172/JCI16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Junek A, Black MA, Semba K. Effects of ibotenate and 192IgG-saporin lesions of the nucleus basalis magnocellularis/substantia innominata on spontaneous sleep and wake states and on recovery sleep after sleep deprivation in rats. J Neurosci. 2008;28:491–504. doi: 10.1523/JNEUROSCI.1585-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Han JS, Dugan LL. Mitochondrial uncoupling proteins in the central nervous system. Antioxid Redox Signal. 2005;7:1173–1181. doi: 10.1089/ars.2005.7.1173. [DOI] [PubMed] [Google Scholar]
- Kong J, Shepel PN, Holden CP, Mackiewicz M, Pack AI, Geiger JD. Brain glycogen decreases with increased periods of wakefulness: implications for homeostatic drive to sleep. J Neurosci. 2002;22:5581–5587. doi: 10.1523/JNEUROSCI.22-13-05581.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larner J, Roach PJ, Huang LC, Brooker G, Murad F, Hazen R. Hormonal control of glycogen metabolism. Adv Exp Med Biol. 1979;111:103–123. doi: 10.1007/978-1-4757-0734-2_6. [DOI] [PubMed] [Google Scholar]
- Lawrence JC, Jr, Skurat AV, Roach PJ, Azpiazu I, Manchester J. Glycogen synthase: activation by insulin and effect of transgenic overexpression in skeletal muscle. Biochem Soc Trans. 1997;25:14–19. doi: 10.1042/bst0250014. [DOI] [PubMed] [Google Scholar]
- Lebedeva IV, Stein CA. Antisense oligonucleotides in cancer: recent advances. BioDrugs. 2000;13:195–216. doi: 10.2165/00063030-200013030-00005. [DOI] [PubMed] [Google Scholar]
- Lee AS. The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem Sci. 2001;26:504–510. doi: 10.1016/s0968-0004(01)01908-9. [DOI] [PubMed] [Google Scholar]
- Lee HS, Kim MA, Valentino RJ, Waterhouse BD. Glutamatergic afferent projections to the dorsal raphe nucleus of the rat. Brain Res. 2003;963:57–71. doi: 10.1016/s0006-8993(02)03841-6. [DOI] [PubMed] [Google Scholar]
- Lee JH, Koh H, Kim M, Kim Y, Lee SY, Karess RE, Lee SH, Shong M, Kim JM, Kim J, Chung J. Energy-dependent regulation of cell structure by AMP-activated protein kinase. Nature. 2007;447:1017–1020. doi: 10.1038/nature05828. [DOI] [PubMed] [Google Scholar]
- Lenka N, Vijayasarathy C, Mullick J, Avadhani NG. Structural organization and transcription regulation of nuclear genes encoding the mammalian cytochrome c oxidase complex. Prog Nucleic Acid Res Mol Biol. 1998;61:309–344. doi: 10.1016/s0079-6603(08)60830-2. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydic R, Baghdoyan HA. Sleep, anesthesiology, and the neurobiology of arousal state control. Anesthesiology. 2005;103:1268–1295. doi: 10.1097/00000542-200512000-00024. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. The mammalian endoplasmic reticulum as a sensor for cellular stress. Cell Stress & Chaperones. 2002;7:222–229. doi: 10.1379/1466-1268(2002)007<0222:tmeraa>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz M, Geiger JD, Pack AI. Simultaneous assessment of ecto- and cytosolic-5'-nucleotidase activities in brain micropunches. J Neurosci Methods. 2000;104:9–18. doi: 10.1016/s0165-0270(00)00314-9. [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Nikonova EV, Zimmerman JE, Galante RJ, Zhang L, Cater JR, Geiger JD, Pack AI. Enzymes of adenosine metabolism in the brain: diurnal rhythm and the effect of sleep deprivation. J Neurochem. 2003;85:348–357. doi: 10.1046/j.1471-4159.2003.01687.x. [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Nikonova EV, Zimmermann JE, Romer MA, Cater J, Galante RJ, Pack AI. Age-related changes in adenosine metabolic enzymes in sleep/wake regulatory areas of the brain. Neurobiol Aging. 2006;27:351–360. doi: 10.1016/j.neurobiolaging.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Shockley KR, Romer MA, Galante RJ, Zimmerman JE, Naidoo N, Baldwin DA, Jensen ST, Churchill GA, Pack AI. Macromolecule biosynthesis - a key function of sleep. Physiol Genomics. 2007 doi: 10.1152/physiolgenomics.00275.2006. [DOI] [PubMed] [Google Scholar]
- Mallo M. Controlled gene activation and inactivation in the mouse. Front Biosci. 2006;11:313–327. doi: 10.2741/1799. [DOI] [PubMed] [Google Scholar]
- Mao W, Yu XX, Zhong A, Li W, Brush J, Sherwood SW, Adams SH, Pan G. UCP4, a novel brain-specific mitochondrial protein that reduces membrane potential in mammalian cells. FEBS Lett. 1999;443:326–330. doi: 10.1016/s0014-5793(98)01713-x. [DOI] [PubMed] [Google Scholar]
- Marchant EG, Mistlberger RE. Anticipation and entrainment to feeding time in intact and SCN-ablated C57BL/6j mice. Brain Res. 1997;765:273–282. doi: 10.1016/s0006-8993(97)00571-4. [DOI] [PubMed] [Google Scholar]
- Marks GA, Birabil CG. Enhancement of rapid eye movement sleep in the rat by cholinergic and adenosinergic agonists infused into the pontine reticular formation. Neuroscience. 1998;86:29–37. doi: 10.1016/s0306-4522(98)00005-0. [DOI] [PubMed] [Google Scholar]
- Marks GA, Shaffery JP, Speciale SG, Birabil CG. Enhancement of rapid eye movement sleep in the rat by actions at A1 and A2a adenosine receptor subtypes with a differential sensitivity to atropine. Neuroscience. 2003;116:913–920. doi: 10.1016/s0306-4522(02)00561-4. [DOI] [PubMed] [Google Scholar]
- Marley E, Nistico G. Effects of catecholamines and adenosine derivatives given into the brain of fowls. Br J Pharmacol. 1972;46:619–636. doi: 10.1111/j.1476-5381.1972.tb06888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty D, Szymusiak R. Brain structures and mechanisms involved in the generation of NREM sleep: focus on the preoptic hypothalamus. Sleep Med Rev. 2001;5:323–342. doi: 10.1053/smrv.2001.0170. [DOI] [PubMed] [Google Scholar]
- Meghji P. Adenosine Production and Metabolism. In: Stone T, editor. Adenosine in the Nervous System Vol. 1. San Diego, CA: Academic Press Inc.; 1991. p. 278. [Google Scholar]
- Methippara MM, Kumar S, Alam MN, Szymusiak R, McGinty D. Effects on sleep of microdialysis of adenosine A1 and A2a receptor analogs into the lateral preoptic area of rats. Am J Physiol Regul Integr Comp Physiol. 2005;289:R1715–R1723. doi: 10.1152/ajpregu.00247.2005. [DOI] [PubMed] [Google Scholar]
- Mistlberger RE, Houpt TA, Moore-Ede MC. Food-anticipatory rhythms under 24-hour schedules of limited access to single macronutrients. J Biol Rhythms. 1990;5:35–46. doi: 10.1177/074873049000500104. [DOI] [PubMed] [Google Scholar]
- Miyoshi G, Fishell G. Directing neuron-specific transgene expression in the mouse CNS. Curr Opin Neurobiol. 2006;16:577–584. doi: 10.1016/j.conb.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Monti JM, Monti D. Role of dorsal raphe nucleus serotonin 5-HT1A receptor in the regulation of REM sleep. Life Sci. 2000;66:1999–2012. doi: 10.1016/s0024-3205(99)00649-9. [DOI] [PubMed] [Google Scholar]
- Morairty S, Rainnie D, McCarley R, Greene R. Disinhibition of ventrolateral preoptic area sleep-active neurons by adenosine: a new mechanism for sleep promotion. Neuroscience. 2004;123:451–457. doi: 10.1016/j.neuroscience.2003.08.066. [DOI] [PubMed] [Google Scholar]
- Murillo-Rodriguez E, Blanco-Centurion C, Gerashchenko D, Salin-Pascual RJ, Shiromani PJ. The diurnal rhythm of adenosine levels in the basal forebrain of young and old rats. Neuroscience. 2004;123:361–370. doi: 10.1016/j.neuroscience.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Casiano V, Cater J, Zimmerman J, Pack AI. A role for the molecular chaperone protein BiP/GRP78 in Drosophila sleep homeostasis. Sleep. 2007;30:557–565. doi: 10.1093/sleep/30.5.557. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Giang W, Galante RJ, Pack AI. Sleep deprivation induces the unfolded protein response in mouse cerebral cortex. J Neurochem. 2005;92:1150–1157. doi: 10.1111/j.1471-4159.2004.02952.x. [DOI] [PubMed] [Google Scholar]
- Newgard CB, Brady MJ, O'Doherty RM, Saltiel AR. Organizing glucose disposal: emerging roles of the glycogen targeting subunits of protein phosphatase-1. Diabetes. 2000;49:1967–1977. doi: 10.2337/diabetes.49.12.1967. [DOI] [PubMed] [Google Scholar]
- Nikonova EV, Frank BC, Mackiewicz M, Quackenbush J, Naidoo N, Pack AI. Changes in components of the electron transport chain in mouse cortex with increases in wakefulness. Soc Neurosci Abs. 2005a [Google Scholar]
- Nikonova EV, Vijayasarathy C, Zhang L, Cater JR, Galante RJ, Ward SE, Avadhani NG, Pack AI. Differences in activity of cytochrome C oxidase in brain between sleep and wakefulness. Sleep. 2005b;28:21–27. doi: 10.1093/sleep/28.1.21. [DOI] [PubMed] [Google Scholar]
- Noor Alam MD, Szymusiak R, McGinty D. Adenosinergic regulation of sleep: multiple sites of action in the brain. Sleep. 2006;29:1384–1385. doi: 10.1093/sleep/29.11.1384. discussion 1387–9. [DOI] [PubMed] [Google Scholar]
- Oikonomakos NG. Glycogen phosphorylase as a molecular target for type 2 diabetes therapy. Curr Protein Pept Sci. 2002;3:561–586. doi: 10.2174/1389203023380422. [DOI] [PubMed] [Google Scholar]
- Ou H, Yan L, Osmanovic S, Greenberg CC, Brady MJ. Spatial reorganization of glycogen synthase upon activation in 3T3-L1 adipocytes. Endocrinology. 2005;146:494–502. doi: 10.1210/en.2004-1022. [DOI] [PubMed] [Google Scholar]
- Pascual O, Casper KB, Kubera C, Zhang J, Revilla-Sanchez R, Sul JY, Takano H, Moss SJ, McCarthy K, Haydon PG. Astrocytic purinergic signaling coordinates synaptic networks. Science. 2005;310:113–116. doi: 10.1126/science.1116916. [DOI] [PubMed] [Google Scholar]
- Pawlyk AC, Ferber M, Shah A, Pack AI, Naidoo N. Proteomic analysis of the effects and interactions of sleep deprivation and aging in mouse cerebral cortex. J Neurochem. 2007;103:2301–2313. doi: 10.1111/j.1471-4159.2007.04949.x. [DOI] [PubMed] [Google Scholar]
- Pekny M, Wilhelmsson U, Bogestal YR, Pekna M. The role of astrocytes and complement system in neural plasticity. Int Rev Neurobiol. 2007;82:95–111. doi: 10.1016/S0074-7742(07)82005-8. [DOI] [PubMed] [Google Scholar]
- Pellerin L, Bouzier-Sore AK, Aubert A, Serres S, Merle M, Costalat R, Magistretti PJ. Activity-dependent regulation of energy metabolism by astrocytes: an update. Glia. 2007;55:1251–1262. doi: 10.1002/glia.20528. [DOI] [PubMed] [Google Scholar]
- Pellerin L, Magistretti PJ. Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proc Natl Acad Sci U S A. 1994;91:10625–10629. doi: 10.1073/pnas.91.22.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin L, Pellegri G, Bittar PG, Charnay Y, Bouras C, Martin JL, Stella N, Magistretti PJ. Evidence supporting the existence of an activity-dependent astrocyte-neuron lactate shuttle. Dev Neurosci. 1998;20:291–299. doi: 10.1159/000017324. [DOI] [PubMed] [Google Scholar]
- Petit JM, Tobler I, Allaman I, Borbely AA, Magistretti PJ. Sleep deprivation modulates brain mRNAs encoding genes of glycogen metabolism. Eur J Neurosci. 2002;16:1163–1167. doi: 10.1046/j.1460-9568.2002.02145.x. [DOI] [PubMed] [Google Scholar]
- Porkka-Heiskanen T, Strecker RE, McCarley RW. Brain site-specificity of extracellular adenosine concentration changes during sleep deprivation and spontaneous sleep: an in vivo microdialysis study. Neuroscience. 2000;99:507–517. doi: 10.1016/s0306-4522(00)00220-7. [DOI] [PubMed] [Google Scholar]
- Porkka-Heiskanen T, Strecker RE, Thakkar M, Bjorkum AA, Greene RW, McCarley RW. Adenosine: a mediator of the sleep-inducing effects of prolonged wakefulness. Science. 1997;276:1265–1268. doi: 10.1126/science.276.5316.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portas CM, Thakkar M, Rainnie DG, Greene RW, McCarley RW. Role of adenosine in behavioral state modulation: a microdialysis study in the freely moving cat. Neuroscience. 1997;79:225–235. doi: 10.1016/s0306-4522(96)00640-9. [DOI] [PubMed] [Google Scholar]
- Printen JA, Brady MJ, Saltiel AR. PTG, a protein phosphatase 1-binding protein with a role in glycogen metabolism. Science. 1997;275:1475–1478. doi: 10.1126/science.275.5305.1475. [DOI] [PubMed] [Google Scholar]
- Radulovacki M. Role of adenosine in sleep in rats. Rev Clin Basic Pharm. 1985;5:327–339. [PubMed] [Google Scholar]
- Radulovacki M. Adenosine and sleep homeostasis in the basal forebrain: commentary on Blanco-Centurion et al. Sleep. 2006;29:1381–1382. doi: 10.1093/sleep/29.11.1381b. discussion 1387–9. [DOI] [PubMed] [Google Scholar]
- Radulovacki M, Virus RM, Djuricic-Nedelson M, Green RD. Adenosine analogs and sleep in rats. J Pharmacol Exp Ther. 1984;228:268–274. [PubMed] [Google Scholar]
- Rainnie DG, Grunze HC, McCarley RW, Greene RW. Adenosine inhibition of mesopontine cholinergic neurons: implications for EEG arousal. Science. 1994;263:689–692. doi: 10.1126/science.8303279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan L, Gulyani S, Nienhuis R, Siegel JM. Sleep deprivation decreases superoxide dismutase activity in rat hippocampus and brainstem. Neuroreport. 2002;13:1387–1390. doi: 10.1097/00001756-200208070-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh V, Thatte HS, McCarley RW, Basheer R. Adenosine and sleep deprivation promote NF-kappaB nuclear translocation in cholinergic basal forebrain. J Neurochem. 2007;100:1351–1363. doi: 10.1111/j.1471-4159.2006.04314.x. [DOI] [PubMed] [Google Scholar]
- Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science. 2001;293:506–569. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu Rev Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- Sanchis D, Fleury C, Chomiki N, Goubern M, Huang Q, Neverova M, Gregoire F, Easlick J, Raimbault S, Levi-Meyrueis C, Miroux B, Collins S, Seldin M, Richard D, Warden C, Bouillaud F, Ricquier D. BMCP1, a novel mitochondrial carrier with high expression in the central nervous system of humans and rodents, and respiration uncoupling activity in recombinant yeast. J Biol Chem. 1998;273:34611–34615. doi: 10.1074/jbc.273.51.34611. [DOI] [PubMed] [Google Scholar]
- Saper CB, Cano G, Scammell TE. Homeostatic, circadian, and emotional regulation of sleep. J Comp Neurol. 2005;493:92–98. doi: 10.1002/cne.20770. [DOI] [PubMed] [Google Scholar]
- Satoh S, Matsumura H, Hayaishi O. Involvement of adenosine A2A receptor in sleep promotion. Eur J Pharmacol. 1998;351:155–162. doi: 10.1016/s0014-2999(98)00302-1. [DOI] [PubMed] [Google Scholar]
- Satoh S, Matsumura H, Koike N, Tokunaga Y, Maeda T, Hayaishi O. Region-dependent difference in the sleep-promoting potency of an adenosine A2A receptor agonist. Eur J Neurosci. 1999;11:1587–1597. doi: 10.1046/j.1460-9568.1999.00569.x. [DOI] [PubMed] [Google Scholar]
- Scammell TE, Gerashchenko DY, Mochizuki T, McCarthy MT, Estabrooke IV, Sears CA, Saper CB, Urade Y, Hayaishi O. An adenosine A2a agonist increases sleep and induces Fos in ventrolateral preoptic neurons. Neuroscience. 2001;107:653–163. doi: 10.1016/s0306-4522(01)00383-9. [DOI] [PubMed] [Google Scholar]
- Scharf MT, Mackiewicz M, Naidoo N, O'callaghan JP, Pack AI. AMP-activated protein kinase phosphorylation in brain is dependent on method of killing and tissue preparation. J Neurochem. 2008 doi: 10.1111/j.1471-4159.2007.05182.x. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf MT, Romer MA, Mackiewicz M, Pack AI. The role of AMP-activated protein kinase in sleep-wake state. Soc Neurosci Abs. 2006:458.11. [Google Scholar]
- Schroder M, Kaufman RJ. ER stress and the unfolded protein response. Mutat Res. 2005a;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005b;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. Divergent roles of IRE1alpha and PERK in the unfolded protein response. Curr Mol Med. 2006;6:5–36. doi: 10.2174/156652406775574569. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- Shen Y, Meunier L, Hendershot LM. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J Biol Chem. 2002;277:15947–15956. doi: 10.1074/jbc.M112214200. [DOI] [PubMed] [Google Scholar]
- Sherin JE, Shiromani PJ, McCarley RW, Saper CB. Activation of ventrolateral preoptic neurons during sleep. Science. 1996;271:216–219. doi: 10.1126/science.271.5246.216. [DOI] [PubMed] [Google Scholar]
- Siebenlist U, Franzoso G, Brown K. Structure, regulation and function of NF-kappa B. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- Simpson IA, Carruthers A, Vannucci SJ. Supply and demand in cerebral energy metabolism: the role of nutrient transporters. J Cereb Blood Flow Metab. 2007;27:1766–1791. doi: 10.1038/sj.jcbfm.9600521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg O, Magistretti PJ. Characterization of the glycogenolysis elicited by vasoactive intestinal peptide, noradrenaline and adenosine in primary cultures of mouse cerebral cortical astrocytes. Brain Res. 1991;563:227–233. doi: 10.1016/0006-8993(91)91538-c. [DOI] [PubMed] [Google Scholar]
- Sorg O, Magistretti PJ. Vasoactive intestinal peptide and noradrenaline exert long-term control on glycogen levels in astrocytes: blockade by protein synthesis inhibition. J Neurosci. 1992;12:4923–4931. doi: 10.1523/JNEUROSCI.12-12-04923.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg D, Litonius E, Halldner L, Johansson B, Fredholm BB, Porkka-Heiskanen T. Sleep and its homeostatic regulation in mice lacking the adenosine A1 receptor. J Sleep Res. 2003;12:283–290. doi: 10.1046/j.0962-1105.2003.00367.x. [DOI] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Swanson RA. Physiologic coupling of glial glycogen metabolism to neuronal activity in brain. Can J Physiol Pharmacol. 1992;(70 Suppl):S138–S144. doi: 10.1139/y92-255. [DOI] [PubMed] [Google Scholar]
- Swanson RA, Morton MM, Sagar SM, Sharp FR. Sensory stimulation induces local cerebral glycogenolysis: demonstration by autoradiography. Neuroscience. 1992;51:451–461. doi: 10.1016/0306-4522(92)90329-z. [DOI] [PubMed] [Google Scholar]
- Szymusiak R. Magnocellular nuclei of the basal forebrain: substrates of sleep and arousal regulation. Sleep. 1995;18:478–500. doi: 10.1093/sleep/18.6.478. [DOI] [PubMed] [Google Scholar]
- Terao A, Steininger TL, Hyder K, Apte-Deshpande A, Ding J, Rishipathak D, Davis RW, Heller HC, Kilduff TS. Differential increase in the expression of heat shock protein family members during sleep deprivation and during sleep. Neuroscience. 2003;116:187–200. doi: 10.1016/s0306-4522(02)00695-4. [DOI] [PubMed] [Google Scholar]
- Thakkar M, Mallick BN. Effect of rapid eye movement sleep deprivation on 5'-nucleotidase activity in the rat brain. Neurosci Lett. 1996;206:177–180. doi: 10.1016/s0304-3940(96)12453-8. [DOI] [PubMed] [Google Scholar]
- Thakkar MM, Delgiacco RA, Strecker RE, McCarley RW. Adenosinergic inhibition of basal forebrain wakefulness-active neurons: a simultaneous unit recording and microdialysis study in freely behaving cats. Neuroscience. 2003a;122:1107–1113. doi: 10.1016/j.neuroscience.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Thakkar MM, Winston S, McCarley RW. A1 receptor and adenosinergic homeostatic regulation of sleep-wakefulness: effects of antisense to the A1 receptor in the cholinergic basal forebrain. J Neurosci. 2003b;23:4278–4287. doi: 10.1523/JNEUROSCI.23-10-04278.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticho SR, Radulovacki M. Role of adenosine in sleep and temperature regulation in the preoptic area of rats. Pharmacol Biochem Behav. 1991;40:33–40. doi: 10.1016/0091-3057(91)90317-u. [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urade Y, Eguchi N, Qu WM, Sakata M, Huang ZL, Chen JF, Schwarzschild MA, Fink JS, Hayaishi O. Sleep regulation in adenosine A2A receptor-deficient mice. Neurology. 2003;61:S94–S96. doi: 10.1212/01.wnl.0000095222.41066.5e. [DOI] [PubMed] [Google Scholar]
- Ventura C, Maioli M. Protein kinase C control of gene expression. Crit Rev Eukaryot Gene Expr. 2001;11:243–267. [PubMed] [Google Scholar]
- Virus RM, Djuricic-Nedelson M, Radulovacki M, Green RD. The effects of adenosine and 2'-deoxycoformycin on sleep and wakefulness in rats. Neuropharmacology. 1983;22:1401–1404. doi: 10.1016/0028-3908(83)90231-9. [DOI] [PubMed] [Google Scholar]
- Virus RM, Ticho S, Pilditch M, Radulovacki M. A comparison of the effects of caffeine, 8-cyclopentyltheophylline, and alloxazine on sleep in rats. Possible roles of central nervous system adenosine receptors. Neuropsychopharmacology. 1990;3:243–249. [PubMed] [Google Scholar]
- Vyazovskiy VV, Tobler I. Handedness leads to interhemispheric EEG asymmetry during sleep in the rat. J Neurophysiol. 2008;99:969–975. doi: 10.1152/jn.01154.2007. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Kojima M, Tomida S, Nakamura TJ, Yamamura T, Nakao N, Yasuo S, Yoshimura T, Ebihara S. Peripheral clock gene expression in CS mice with bimodal locomotor rhythms. Neurosci Res. 2006;54:295–301. doi: 10.1016/j.neures.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Weil R, Israel A. Deciphering the pathway from the TCR to NF-kappaB. Cell Death Differ. 2006;13:826–833. doi: 10.1038/sj.cdd.4401856. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MT, Mullen MA, Huang Z, Guyer C. Brain cytochrome oxidase subunit complementary DNAs: isolation, subcloning, sequencing, light and electron microscopic in situ hybridization of transcripts, and regulation by neuronal activity. Neuroscience. 1997;76:1035–1055. doi: 10.1016/s0306-4522(96)00410-1. [DOI] [PubMed] [Google Scholar]
- Yanik G, Glaum S, Radulovacki M. The dose-response effects of caffeine on sleep in rats. Brain Res. 1987;403:177–180. doi: 10.1016/0006-8993(87)90141-7. [DOI] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ. Signaling the unfolded protein response from the endoplasmic reticulum. J Biol Chem. 2004;279:25935–25938. doi: 10.1074/jbc.R400008200. [DOI] [PubMed] [Google Scholar]
- Zimmerman JE, Mackiewicz M, Galante RJ, Zhang L, Cater J, Zoh C, Rizzo W, Pack AI. Glycogen in the brain of Drosophila melanogaster: diurnal rhythm and the effect of rest deprivation. J Neurochem. 2004;88:32–40. doi: 10.1046/j.1471-4159.2003.02126.x. [DOI] [PubMed] [Google Scholar]
- Zimmerman JE, Rizzo W, Shockley KR, Raizen DM, Naidoo N, Mackiewicz M, Churchill GA, Pack AI. Multiple mechanisms limit the duration of wakefulness in Drosophila brain. Physiol Genomics. 2006;27:337–350. doi: 10.1152/physiolgenomics.00030.2006. [DOI] [PubMed] [Google Scholar]