Abstract
Computational studies of the mutational effects at the C3, G8, and G5 positions of the hammerhead ribozyme (HHR) are reported based on a series of twenty four 100-ns molecular dynamics simulations of the native and mutated HHR in the reactant state and in an activated precursor state (G8:2′OH deprotonated). Invoking the assumptions that G12 acts as the general base while the 2′OH of G8 acts as a general acid, the simulations are able to explain the origins of experimentally observed mutational effects, including several that are not easily inferred from the crystal structure. Simulations suggest that the Watson-Crick base-pairing between G8 and C3, the hydrogen bond network between C17 and G5, and the base stacking interactions between G8 and C1.1, collectively, are key to maintaining an active site structure conducive for catalytic activity. Mutation-induced disruption of any of these interactions will adversely affect activity. The simulation results predict that the C3U/G8D double mutant, where D is 2,6-diaminopurine, will have a rescue effect relative to the corresponding single mutations. Two general conclusions about the simulations emerge from this work. Firstly, mutation simulations may require 30 ns or more to suitably relax such that the mutational effects become apparent. Secondly, in some cases, it is necessary to look beyond the reactant state in order to interpret mutational effects in terms of catalytically active structure. The present simulation results lead to better understanding of the origin of experimental mutational effects, and provide insight into the key conserved features necessary to maintain the integrity of the active site architecture.
Introduction
The study of RNA enzymes, or ribozymes, is of considerable interest due to their importance in biology,1–4 their evolutionary implications,5–9 and potential impact for the design of drugs10 and new biomedical technology.11, 12 The hammerhead ribozyme (HHR)1, 13, 14 is a small self-cleaving ribozyme derived from RNA discovered in satellites of various plant virus genomes.15–17 Catalysis results from in-line nucleophilic attack of the 2′O of C17 to the adjacent scissile phosphate, followed by cleavage of the P-O5′ bond of C1.1 to produce a 2′,3′-cyclic phosphate and a 5′ hydroxy-terminus in two RNA product strands. The catalytic activity of HHR motifs have promise in a number of biomedical applications. These include use as a tool for gene regulation,18, 19 ribozyme-based target discovery,20 and therapeutic agents10, 21–23
Consequently, much effort has been devoted to understand the details of the HHR mechanism both experimentally3, 4, 13, 24 and theoretically.25–29 The HHR is formed from a 3-way junction with a conserved catalytic core30 that contains the active site (Figure 1), and requires tertiary interactions between stems I and II for optimal activity.31, 32 Mutagenesis experiments suggest that three conserved residues, G5, G8 and C3, among others, are critical for catalytic activity.24, 30, 33–37 Mutants of G5 and G8 exhibit different pH- dependent behavior,36 and it has been shown recently that HHR constructs can tolerate double-mutants that preserve base pairing between the positions 3 and 8, but that these mutations have some sequence dependence and are sensitive to the identity of residue 17.38, 39 The roles of these residues, however, are not evident from analysis of the minimal sequence HHR crystal structures,40–42 that, despite being active in the crystal, are in a conformation that requires considerable re-arrangement to arrive to a catalytically active structure. It was not until the recent full length HHR structure43, 44 was solved that a sound structural basis emerged that could serve as a departure point for theoretical investigations aimed at understanding the origin of mutational effects and helping to reconcile the HHR mechanistic debate.
Figure 1.
Schematic representations of the hammerhead ribozyme active sites and the two potentially important hydrogen bond networks between C3-G8 and between G5 and C17. All key structural indexes calculated are also labeled.
The purpose of the present work is to extend our preliminary studies27 to elucidate the origin of mutational effects in the hammerhead ribozyme. In a series of twenty four 100-ns molecular dynamics (MD) simulations, we explore the structure and dynamics of HHR mutants involving C3, G5 and G8 positions in the reactant and activated precursor (deprotonated 2′ OH) states. This work greatly extends the scope of our previous communication27 that utilized 600 ns of cumulative simulation (compared to 2.4 μs in the current work) to focus on a subset of mutations involving only the C3 and G8 positions. A key result that emerges is that, in performing computational mutagenesis simulations of ribozymes, one must consider time scales greater than 30 ns for relaxation of mutant structures, and in some instances look beyond the reactant state along the catalytic reaction coordinate in order to reconcile the origin of mutational effects. Consistent with the crystallographic structure, our results indicate that Watson-Crick base-pairing between G8 and C3 stabilize the stem-stem interactions and position the 2′OH of G8 so as to act as a general acid catalyst. More detailed examination reveals a hydrogen bond network between C17 and G5, and base stacking interactions between G8 and C1.1 that position the nucleophile and leaving group in an in-line orientation conducive for catalytic activity. In addition, the simulation results reported here offer several new insights into mutational effects that cannot be reasonably inferred from inspection of the crystallographic structure, and make a prediction of a compensatory mutation that has not yet been measured.
Methods
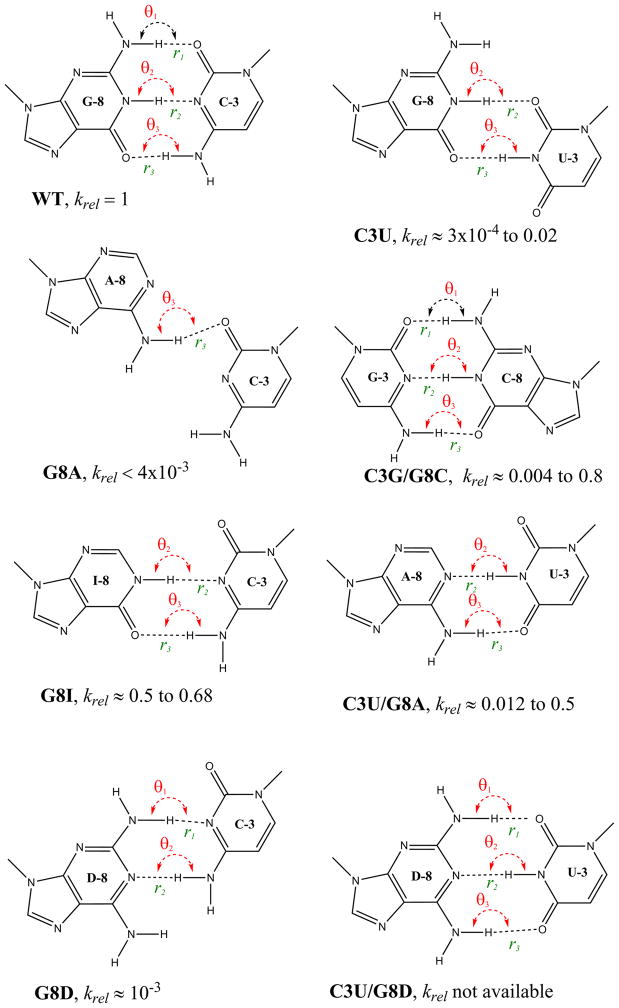
Total 12 mutants have been simulated: WT, C3U, G8A, G8I, G8D, C3G/G8C, C3U/G8A, C3U/G8D, G5I, G5A, G5D, and U7C (control). Each mutant was simulated in two different states (the reactant state and the C17:O2′-deprotonated “activated” precursor state). Hence 24 simulations were performed in total, with each having 10 ns solvent/ion equilibration, followed by 10 ns solute equilibration and 100 ns production simulations. Here the standard RNA nucleotide notations are used for A (adenine), G (guanine), C (cytosine), and U (uracil); while ”D” stands for 2,6-diaminopurine, and ”I” stands for inosine. The native “wild type” sequence simulation is designated “WT”. Mutant simulations are indicated by their particular mutation. For example, ”C3U” refers to the mutant with the native cytosine at position 3 replaced by a uridine. Double mutants are denoted by the combination of two mutations separated by a slash, e.g., C3G/G8C.
Seven mutants were chosen to explore the C3 and G8 positions: C3U, G8A, G8I, G8D, C3G/G8C, C3U/G8A, and C3U/G8D (Figure 2). Among these mutants, G8A, and G8D result in reducing the number of basepair hydrogen bonds and changing their relative positions while G8I only alters the number of base-pair hydrogen bonds. The number of hydrogen bonds and their relative positions are all kept in the double mutants C3G/G8C and C3U/G8A; while the relative hydrogen bond positions are changed for C3U/G8D. For the G5 position, three mutants were chosen G5I, G5A and G5D (Figure 3). Only one hydrogen bond (to C17) is kept in G5A and G5I; while there are two hydrogen bonds (in different positions) for G5D. U7C was chosen as the control simulation since it has been shown to be a benign mutation34 and this mutation does not cause any change of hydrogen bond patterns at the G5/C3/G8 positions.
Figure 2.
Schematic representations of the mutants and the hydrogen bonding network of the C3 and G8 positions. krel is the experimental cleavage constant relative to the wild-type. The relevant references are listed in Table 6.
Figure 3.
Schematic representations of the mutants of the G5 position and the hydrogen bonding network between the G5 and C17 positions. krel is the experimental cleavage constant relative to the wild-type. The relevant references are listed in Table 6.
Simulation Setup
The initial structure used in the simulations were based on a 2.0 Å crystal structure with Mn2+ ions and solvent resolved (PDB: 2OEU),44 with Mn2+ ions replaced by native Mg2+ ions in the simulations. The positions of hydrogen atoms and atoms of the mutated residues were determined using the VMD program (version 1.8.6).45
The ribozyme was then immersed in a cubic cell of 60×60×100 Å3 filled with pre-equilibrated TIP3P46 waters. The ribozyme molecule was rotated so that the longest molecular axis was aligned with the longest water cell axis and it was centered about the site, and pruned such that any water molecule within 2.8 Å from the solute was removed. The ion atmosphere consisted of Na+ and Cl− ions that were added to neutralize the system and reach the physiologic (extracellular) concentration of 0.14 M. The ions positions were kept initially at least 5.0 Å away from any solute atoms. The resulting system (wild-type) contained 36,534 atoms: 11,463 water molecules, 5 Mg2+, 83 Na+, 29 Cl−, and 2,021 RNA atoms.
Simulations were performed with the NAMD simulation package (version 2.6)47 using the all-atom Cornell et al. force field (parm94)48 in CHARMM format, provided in the AMBER 9 package49, 50. The TIP3P water model was used.46 Periodic boundary conditions were used along with the isothermal-isobaric ensemble (NPT) at 1 atm and 298 K using extended system pressure algorithm51 with effective mass of 500.0 amu and Nosé-Hoover thermostat52, 53 with effective mass of 1000.0 kcal/mol-ps2, respectively. The smooth particle mesh Ewald (PME) method54, 55 was employed with a B-spline interpolation order of 6, the default k value (0.258 Å −1), and 60, 60, and 100 FFT grid points were used for the x, y, and z cubic lattice directions, respectively. Non-bonded interactions were treated using an atom-based cutoff of 12 Å with switching of the non-bonded potential beginning at 10 Å. Numerical integration was performed using the leap-frog Verlet algorithm with 1 fs time step.56 Covalent bond lengths involving hydrogen were constrained using the SHAKE algorithm.57
Equilibration Protocol
The following equilibration procedures (total 10 ns) was applied to the system prior the production simulations. The positions of the solute atoms, including the Mg2+ ions, were first kept by a harmonic potential of 50 kcal/mol/Å 2 in the equilibration stages.
Pre-annealing Stage
Water and ions were first energy-optimized for 2,000 steps then underwent a constant volume simulated annealing: The temperature was increased from 0 K to 298K at the rate of 1 K per ps. The system then was kept at 298 K for 500 ps.
Annealing Stage
First step: The temperature was increased from 298K to 600 K at the rate of 1 K/ps, then was kept at 600 K for 500 ps with constant volume. Second step: The temperature decreased from 600 K to 298K at the rate of 1 K/ps, then was kept at 298K for 1,500 ps with constant volume. Third step: The system was kept at 298K for 3,000 ps at constant pressure (1 atm). The whole annealing cycle was then repeated twice before the post-annealing stage.
Solute Relaxation Stage
After the annealing stage, the solute atoms were energy-optimized and then were allowed to move under harmonic restraints over 500 ps simulation at 298 K under constant pressure of 1 atm. The harmonic force constant (in kcal mol−1 Å−2) on each heavy atom was obtained from the empirical formula ki = 25+2×103/Bi where ki is the force constant for atom i and Bi is the corresponding crystallographic B-value. The restraints were exponentially released over 500 ps with a half-life decay parameter of 100 ps. At the end of the 500 ps simulation, the restraints were reduced to about 3 percent of the initial restraints.
Production Simulation
After the 10 ns of solvent equilibration, the whole system was energy-optimized and unconstrained dynamics simulation began from 0 K under constant pressure of 1 atm. The temperature was increased to 298 K at the rate of 1 K/ps and then kept fixed at 298 K. The same equilibration process was applied for each simulation.
At the first 10 ns simulation (solute equilibration), two harmonic restraints of 20 kcal mol−1 Å −2 were added to keep the Mg2+ ion binding to G10.1:N7 and A9:OP2 position. Another three harmonic restraint of 20 kcal mol−1 Å−2 were used: the distances between G8:HO′5 and C1.1:OO2′, and between G12:H1 and C17:O2′, were kept around 1.8 Å to ensure the initial hydrogen bonding; the distance between A9:OP2 and C1.1:OP2 was kept at 4.3 Å (crystal distance). After 10 ns, all restraints are removed.
The motions and relaxation of solvent and counter-ions are notoriously slow to converge in nucleic acid simulations,58 and careful equilibration is critical for reliable simulations. In summary, for each simulation, a total of 20 ns of equilibration (10 ns of solvent/ion relaxation and 10 ns of solvent and structure relaxation) has been carried out before 100 ns of data sampling.
To catch the overall picture of the mutational effect of each mutant, clustering analysis was performed based on RMSD of heavy atoms with a cut-off 1.5 Å for each mutant. For all mutants, the largest cluster is chosen to calculate the average structure, and every chosen cluster has a population of over 90% of the whole trajectory. The cluster-average structures, in PDB format, from both the reactant and the activated precursor state simulations of all mutants are available as Supporting Information. Mutants exhibit significantly visible mutational effects are shown in Figures 4, 5, and 6.
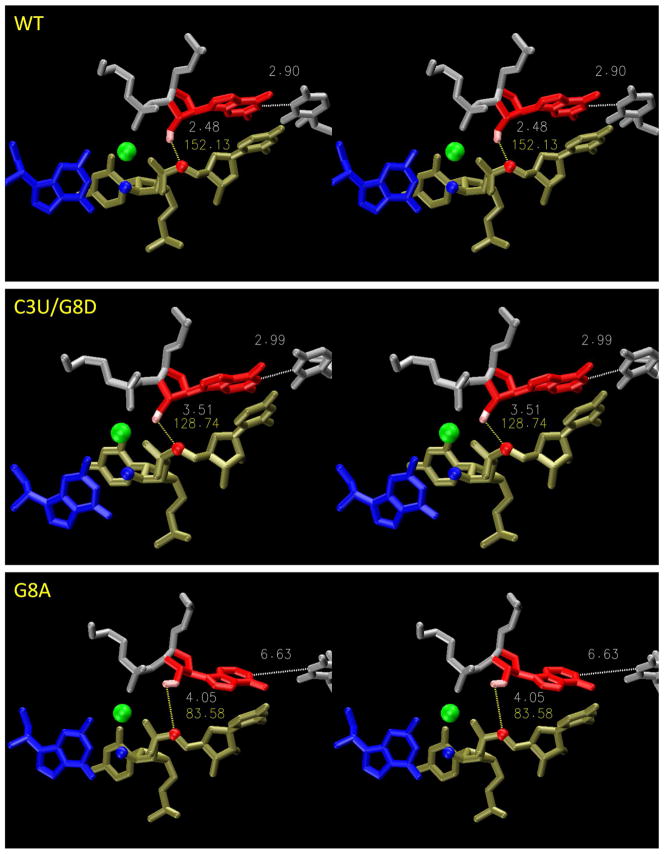
Figure 4.
Stereo views of cluster-averaged structures of C3 or G8 mutants causing significant deviation at the general base step. Results are from the reactant state simulations. Upper: WT; middle C3U; lower: G8D. Residues are colored as follows: blue: G12 (general base); red: G8 (general acid); gray: A9 (middle) and C3 (right); tan: active site C17 and C1.1. All hydrogens are not show except the general base (light blue). The blue ball is C17:O2′ (the nucleophilic attacking group); the red ball is C1.1:O5′ (the leaving group); the green ball is the Mg2+ ion. The gray number next to the Mg2+ ion is the A9:O2P –C1.1:O2P distance (d0, see Figure 1). The gray and the orange numbers at the left corner are the general base hydrogen bond length (rHB) and angle (θHB), respectively. The gray number at the upper right corner is the N1-N3 distance between C3 and G8. Clustering analysis was performed based on RMSD of heavy atoms with a cut-off 1.5 Å. For all mutants, the largest cluster is chosen to calculate the average structure, and every chosen cluster has a population of over 90% of the whole trajectory.
Figure 5.
Stereo views of cluster-averaged structures of C3 or G8 mutants causing significant deviation at the general acid step. Results are from the activated precursor state simulations. Upper: WT; middle C3U/G8D; lower: G8A. Residues are colored as follows: blue: G12 (general base); red: G8 (general acid); gray: A9 (middle) and C3 (right); tan: active site C17 and C1.1. All hydrogens are not show except the general acid (pink). The blue ball is C17:O2′ (the nucleophilic attacking group); the red ball is C1.1:O5′ (the leaving group); the green ball is the Mg2+ ion. The gray and the orange numbers at the middle are the general acid hydrogen bond length (rHA) and angle (θHA, see Figure 1), respectively. The gray number at the upper right corner is the N1-N3 (rNN ) distance between C3 and G8. Clustering analysis was performed based on RMSD of heavy atoms with a cut-off 1.5 Å. For all mutants, the largest cluster is chosen to calculate the average structure, and every chosen cluster has a population of over 90% of the whole trajectory.
Figure 6.
Stereo views of cluster-averaged structures of G5 mutants causing significant deviation at the general acid step. Results are from the activated precursor state simulations. Upper: WT; upper middle G5I; lower middle: G5A; lower: G5D. Residues are colored as follows: blue: G12 (general base); red: G8 (general acid); gray: A9 (middle) and G5 (lower); tan: active site C17 and C1.1. All hydrogens are not show except the general acid (pink). The blue ball is C17:O2′ (the nucleophilic attacking group); the red ball is C1.1:O5′ (the leaving group); the green ball is the Mg2+ ion. The gray and the orange numbers at the upper middle corner are the general acid hydrogen bond length (rHA) and angle (θHA, see Figure 1), respectively. The gray numbers at the lower middle corner are the hydrogen distances between C1.1 and G5. The orange number at the left is the in-attack angle (θinl). Note that these views are from different angles compared to Figures 4 and 5. Clustering analysis was performed based on RMSD of heavy atoms with a cut-off 1.5 Å. For all mutants, the largest cluster is chosen to calculate the average structure, and every chosen cluster has a population of over 90% of the whole trajectory.
Results
In the present work, nucleobase mutations are explored at the C3, G8, G5 and U7 positions. Simulations for each mutation have been performed in both the reactant and activated precursor state, and are compared with the wild type simulation results. The activated precursor state is described in more detail below, and is distinguished by deprotonation of the 2′OH nucleophile of C17. This change in charge state facilitates the migration of a divalent metal ion into a position that bridges the A9 and scissile phosphates in the present simulation models,25–29 although, to date, this binding mode has not been directly observed experimentally. The U7 mutation is benign, and is used as a control simulation. Single mutations at both the C3 and G8 positions (C3U, G8A, G8I and G8D) are explored, where ”D” indicates 2,6-diaminopurine, and ”I” indicates inosine. In addition, double mutants that exhibit a partial rescue effect have been examined, including the isosteric C3U/G8D and hydrogen bond-preserving C3G/G8C switch mutations. A simulation of a C3U/G8D mutation, for which there currently exists, to our knowledge, no experimental measurement, predicts an almost complete rescue effect. Finally, a series of single mutations at the G5 position have been studied, including G5I, G5A and G5D.
The general structure of the hammerhead ribozyme active site, including identification of indexes used to characterize key hydrogen bond networks and base stacking interactions involving conserved residues are shown in Fig. 1. Representative hydrogen bond patterns observed in the simulations for the C3 and G8 mutants are shown in Fig. 2, and for the G5 mutants are shown in Fig. 3. Averages and fluctuations for key indexes used to characterize the active site are listed in Tables 1 and 2 for the reactant state and Tables 3 and 4 for the activated precursor state. Table 5 lists indexes used to characterize the base stacking interactions between G8 and C1.1 in the wild type, benign U7C and G8I single mutants, and double mutant simulations. A summary of the overall mutational effects inferred from the MD simulations are provided in Table 6 and compared to experimental values for the relative catalytic rates.
Table 1.
Characterization of the active site structure and fluctuations for the C3 and G8 mutants in reactant states.
| WT | U7C | C3U | G8A | C3U/G8A | G8I | G8D | C3U/G8D | C3G/G8C | |
|---|---|---|---|---|---|---|---|---|---|
| rNu | 4.07(0.25) | 3.78(0.43) | 3.88(0.42) | 3.2(0.1) | 3.35(0.36) | 3.96(0.35) | 4.1(0.17) | 3.63(0.45) | 4.2(0.13) |
| θinl | 124.3(8.6) | 133.3(15.1) | 132(12.8) | 155.5(7.6) | 150.2(14.1) | 125.6(12) | 123.7(8.6) | 140.4(16.3) | 122(4.2) |
| F* | 0.25(0.11) | 0.38(0.2) | 0.34(0.18) | 0.68(0.09) | 0.6(0.19) | 0.29(0.15) | 0.23(0.06) | 0.46(0.22) | 0.21(0.03) |
| d0 | 3.97(0.39) | 4.27(0.63) | 6.64(1.59) | 4.26(0.34) | 4.12(0.45) | 4.17(0.57) | 5.86(1.65) | 4.68(0.63) | 4.26(0.44) |
| rHB‡ | 2.07(0.25) | 2.1(0.28) | 2.88(0.85) | 2(0.15) | 2.44(0.57) | 2.05(0.26) | 2.54(1.00) | 2.11(0.37) | 2.5(0.72) |
| θHB‡ | 152.7(13.4) | 153.6(13.4) | 136.9(22.5) | 163.9(8.5) | 150(15.7) | 155.7(12.1) | 149.5(18.4) | 158.8(12.8) | 145.2(19) |
| %¶ | 98 | 97 | 52 | 100 | 82 | 98 | 77 | 95 | 76 |
| %|| | 62 | 64 | 34 | 93 | 51 | 71 | 55 | 78 | 46 |
| rHA§ | 2.75(0.45) | 3.41(1.02) | 5.88(2.4) | 2.97(0.34) | 3.61(0.6) | 2.85(0.71) | 5.74(1.81) | 4.3(1.38) | 2.84(0.5) |
| θHA§ | 115.7(18.6) | 107.4(36.3) | 68.9(47.5) | 118.1(14.9) | 136.7(23.8) | 115.9(26.2) | 58.3(38.3) | 83.7(48.7) | 108.5(16.2) |
| %¶ | 28 | 19 | 19 | 33 | 6 | 32 | 5 | 15 | 17 |
| %|| | 4 | 2 | 10 | 1 | 1 | 7 | 0 | 1 | 2 |
| rNN† | 2.98(0.09) | 2.96(0.09) | 3.65(0.22) | 5.39(0.57) | 2.94(0.14) | 2.96(0.12) | 3.69(0.28) | 3.1(0.18) | 2.93(0.09) |
| r1 | 2.01(0.17) | 2.02(0.18) | - | - | - | - | 2.58(0.74) | 2.32(0.4) | 1.9(0.13) |
| θ1 | 162.9(9) | 163.2(8.8) | - | - | - | - | 156.7(15.4) | 157(13.4) | 163.5(8.8) |
| r2 | 2.01(0.1) | 2(0.1) | 2.17(0.27) | - | 1.97(0.16) | 1.99(0.13) | 2.30(0.67) | 2.14(0.22) | 1.98(0.11) |
| θ2 | 162.1(8.7) | 161(8.8) | 156.4(12.8) | - | 161.9(9.8) | 163.1(8.4) | 151.3(17.0) | 160.7(11.6) | 158.9(11.8) |
| r3 | 1.89(0.13) | 1.88(0.12) | 1.89(0.13) | 2.08(0.31) | 2.1(0.22) | 2.01(0.21) | - | 1.91(0.14) | 2.05(0.2) |
| θ3 | 163.5(8.8) | 164.4(8.3) | 162.1(9.5) | 149.3(14.8) | 162.7(9.6) | 163.8(8.9) | - | 155.9(10.4) | 161.3(10.2) |
This table lists key structural indexes fluctuations for the C3 and G8 mutants, along with the control mutant U7C in reactant states. Data analysis was performed over the last 65 ns of each simulation with a 10 ps sampling frequency. Distance and angles (Figure 1) are in Å and degrees, respectively. Standard deviations (SD) are listed in parenthesis. Boldface font is used to highlight key quantities that are significantly altered with respect to the wild type (WT) simulation upon mutation and that are discussed in the text.
In-line fitness index.66
The N3 ···N1 distance between nucleobases in the 3 and 8 position.
rHB and θHB are the hydrogen bond length and angle for the general base step; defined by G12:N1.…C17:HO2′..C17:O2′.
rHA and θHA are the hydrogen bond length and angle for the general acid step; defined by C1.1:O5′ …G8:HO2′..G8:O2′.
The hydrogen bond contact percentage for the above entries, defined as the percentage of the snapshots in which r ≤ 3.0 Å and θ ≥120 °.
The hydrogen bond contact percentage for the above entries, defined as the percentage of the snapshots in which r ≤ 2.5 Å and θ ≥150 °.
Table 2.
Characterization of the active site structure and fluctuations for the G5 mutants in reactant states.
| WT | U7C | G5I | G5A | G5D | |
|---|---|---|---|---|---|
| rNu | 4.07(0.25) | 3.78(0.43) | 4.18(0.14) | 3.84(0.46) | 3.94(0.39) |
| θinl | 124.3(8.6) | 133.3(15.1) | 118.3(9.2) | 129.8(15.2) | 128.1(14.1) |
| F* | 0.25(0.11) | 0.38(0.2) | 0.2(0.04) | 0.35(0.19) | 0.31(0.17) |
| d0 | 3.97(0.39) | 4.27(0.63) | 4.64(0.56) | 4.95(0.8) | 5.2(0.43) |
| rHB‡ | 2.07(0.25) | 2.1(0.28) | 2.22(0.76) | 2.08(0.33) | 2.31(0.64) |
| θHB‡ | 152.7(13.4) | 153.6(13.4) | 155.5(14.4) | 156.9(12.7) | 149.5(18) |
| %¶ | 98 | 97 | 91 | 97 | 84 |
| %|| | 62 | 64 | 72 | 75 | 56 |
| rHA§ | 2.75(0.45) | 3.41(1.02) | 5.1(1.1) | 3.48(1.35) | 5.22(0.63) |
| θHA§ | 115.7(18.6) | 107.4(36.3) | 40(38) | 93.8(45.7) | 43.9(25.7) |
| %¶ | 28 | 19 | 5 | 33 | 1 |
| %|| | 4 | 2 | 1 | 11 | 0 |
| r4 | 2.11(0.21) | 2.17(0.27) | 4.99(0.64) | 2.04(0.28) | 2.18(0.58) |
| θ4 | 153.0(12.9) | 154.2(12.5) | 159.1(10.7) | 159.5(11.0) | 155.0(13.4) |
| r5 | 2.98(0.58) | 2.77(0.67) | - | - | 3.13(0.96) |
| θ5 | 119.1(16.8) | 130.2(20.2) | - | - | 129.4(31.3) |
| r6 | 2.08(0.22) | 2.07(0.19) | - | - | 2.31(0.79) |
| θ6 | 153.4(10.9) | 150.1(11.3) | - | - | 149.4(16.0) |
This table lists key structural indexes fluctuations for the G5 mutants, along with the control mutant U7C in reactant states. Data analysis was performed over the last 65 ns of each simulation with a 10 ps sampling frequency. Distance and angles (Figure 1) are in Å and degrees, respectively. Standard deviations (SD) are listed in parenthesis. Boldface font is used to highlight key quantities that are significantly altered with respect to the wild type (WT) simulation upon mutation and that are discussed in the text.
In-line fitness index.66
The N3 ···N1 distance between nucleobases in the 3 and 8 position.
rHB and θHB are the hydrogen bond length and angle for the general base step; defined by G12:N1 …C17:HO2′..C17:O2′.
rHA and θHA are the hydrogen bond length and angle for the general acid step; defined by C1.1:O5′ …G8:HO2′..G8:O2′.
The hydrogen bond contact percentage for the above entries, defined as the percentage of the snapshots in which r ≤ 3.0 Å and θ ≥120 °.
The hydrogen bond contact percentage for the above entries, defined as the percentage of the snapshots in which r ≤ 2.5 Å and θ ≥150 °.
Table 3.
Characterization of the active site structure and fluctuations for the C3 and G8 mutants in activated precursor states.
| d-WT¶ | d-U7C | d-C3U | d-G8A | d-C3U/G8A | d-G8I | d-G8D | d-C3U/G8D | d-C3G/G8C | |
|---|---|---|---|---|---|---|---|---|---|
| rNu | 3.59(0.17) | 3.64(0.17) | 3.65(0.16) | 3.73(0.2) | 3.6(0.16) | 3.69(0.23) | 3.64(0.21) | 3.34(0.13) | 3.61(0.17) |
| θinl | 156.8(7.9) | 153.6(8.7) | 154.9(7.5) | 148.1(8.9) | 155.7(7.6) | 149(12.7) | 152.5(10.4) | 164.1(7.4) | 155.4(8.2) |
| F* | 0.49(0.09) | 0.46(0.09) | 0.46(0.08) | 0.41(0.1) | 0.48(0.08) | 0.43(0.12) | 0.46(0.11) | 0.65(0.1) | 0.48(0.09) |
| d0 | 2.94(0.13) | 2.93(0.12) | 2.93(0.13) | 2.95(0.12) | 2.93(0.12) | 2.93(0.13) | 3(0.13) | 2.97(0.13) | 2.95(0.13) |
| rHB‡ | 2.03(0.37) | 1.94(0.14) | 1.93(0.14) | 1.93(0.2) | 1.95(0.14) | 1.95(0.18) | 1.91(0.13) | 1.86(0.1) | 1.96(0.15) |
| θHB‡ | 154.6(11.9) | 155.8(9.4) | 155.6(9.4) | 158.4(9.2) | 155.5(9.3) | 157.2(9.8) | 158.1(9.3) | 161.2(8.4) | 156(9.2) |
| %¶ | 95 | 100 | 100 | 99 | 100 | 100 | 100 | 100 | 100 |
| %|| | 72 | 73 | 73 | 82 | 74 | 77 | 81 | 91 | 75 |
| rHA§ | 2.61(0.77) | 2.5(0.79) | 2.63(0.76) | 4.13(0.63) | 2.6(0.9) | 2.44(0.52) | 2.46(0.44) | 3.53(0.74) | 2.42(0.57) |
| θHA§ | 131.8(34.5) | 135.5(35.2) | 122.3(34.3) | 82.6(21.4) | 130.5(38) | 137.3(27.7) | 139.6(24.6) | 117.2(31.2) | 138(29.3) |
| %¶ | 69 | 75 | 57 | 1 | 69 | 74 | 81 | 20 | 77 |
| %|| | 35 | 44 | 24 | 0 | 41 | 37 | 34 | 1 | 41 |
| rNN† | 2.96(0.1) | 2.96(0.09) | 3.78(0.21) | 6.68(0.42) | 3.04(0.15) | 2.92(0.11) | 3.57(0.19) | 3.03(0.12) | 2.97(0.09) |
| r1 | 1.92(0.15) | 2.02(0.17) | - | - | - | - | 1.98(0.11) | 2.23(0.28) | 1.9(0.13) |
| θ1 | 160.9(10) | 163.8(8.5) | - | - | - | - | 163.1(9.1) | 157.9(13.1) | 163.7(8.3) |
| r2 | 1.98(0.1) | 1.99(0.1) | 1.91(0.15) | - | 2.08(0.17) | 1.94(0.11) | 2.06(0.16) | 2.06(0.14) | 2.01(0.1) |
| θ2 | 163.2(8.3) | 162.3(8.8) | 160.6(10.7) | - | 160.8(10.1) | 163.9(8.3) | 160.7(10.8) | 163.1(9.8) | 160.1(8.6) |
| r3 | 2(0.19) | 1.88(0.11) | 1.96(0.17) | 2.77(0.4) | 1.96(0.17) | 2.13(0.24) | - | 1.9(0.13) | 1.94(0.15) |
| θ3 | 163.7(9) | 164(8.5) | 161.1(9.7) | 106.8(11) | 160.3(10.5) | 162.8(9.6) | - | 156.1(10.4) | 162.2(9.9) |
This table lists key structural indexes fluctuations for the C3 and G8 mutants, along with the control mutant U7C in activated precursor states. Data analysis was performed over the last 65 ns of each simulation with a 10 ps sampling frequency. Distance and angles (Figure 1) are in Å and degrees, respectively. Standard deviations (SD) are listed in parenthesis. Boldface font is used to highlight key quantities that are significantly altered with respect to the wild type (WT) simulation upon mutation and that are discussed in the text.
The notation ”d-” denotes the activated precursor state simulations having the C17:O2′ deprotonated.
In-line fitness index.66
The N3 ···N1 distance between nucleobases in the 3 and 8 position.
rHB and θHB are the hydrogen bond length and angle for the general base step; defined by G12:N1 …C17:HO2′..C17:O2′.
rHA and θHA are the hydrogen bond length and angle for the general acid step; defined by C1.1:O5′ …G8:HO2′..G8:O2′.
The hydrogen bond contact percentage for the above entries, defined as the percentage of the snapshots in which r ≤ 3.0 Å and θ ≥120 °.
The hydrogen bond contact percentage for the above entries, defined as the percentage of the snapshots in which r ≤ 2.5 Å and θ ≥150 °.
Table 4.
Characterization of the active site structure and fluctuations for the G5 mutants in activated precursor states.
| d-WT** | d-U7C | d-G5I | d-G5A | d-G5D | |
|---|---|---|---|---|---|
| rNu | 3.59(0.17) | 3.64(0.17) | 3.63(0.15) | 4.15(0.11) | 3.58(0.24) |
| θinl | 156.8(7.9) | 153.6(8.7) | 156.5(7.6) | 128.6(5.8) | 139.7(9.8) |
| F* | 0.49(0.09) | 0.46(0.09) | 0.47(0.08) | 0.24(0.03) | 0.43(0.11) |
| d0 | 2.94(0.13) | 2.93(0.12) | 3(0.13) | 2.9(0.12) | 2.88(0.12) |
| rHB‡ | 2.03(0.37) | 1.94(0.14) | 1.93(0.13) | 1.92(0.15) | 2.62(0.96) |
| θHB‡ | 154.6(11.9) | 155.8(9.4) | 155.2(9.4) | 153.6(8.9) | 152.5(11.9) |
| %¶ | 95 | 100 | 100 | 100 | 79 |
| %|| | 72 | 73 | 72 | 67 | 52 |
| rHA§ | 2.61(0.77) | 2.5(0.79) | 3.6(0.3) | 3.67(1.09) | 2.66(0.6) |
| θHA§ | 131.8(34.5) | 135.5(35.2) | 62.2(11.4) | 74.2(55.6) | 128.1(34) |
| %¶ | 69 | 75 | 0 | 30 | 62 |
| %|| | 35 | 44 | 0 | 12 | 24 |
| r4 | 1.97(0.14) | 1.97(0.15) | 1.99(0.16) | 2.11(0.30) | 2.66(0.97) |
| θ4 | 160.0(10.5) | 159.3(9.8) | 158.1(10.2) | 148.1(13.7) | 155.4(14.8) |
| r5 | 2.56(0.46) | 2.56(0.49) | - | - | 2.91(0.87) |
| θ5 | 135.3(12.7) | 135.6(13.4) | - | - | 130.7(18.0) |
| r6 | 2.12(0.20) | 2.09(0.17) | - | - | 2.09(0.21) |
| θ6 | 148.8(10.5) | 149.9(10.8) | - | - | 150.2(13.0) |
This table lists key structural indexes fluctuations for the G5 mutants, along with the control mutant U7C in activated precursor states. Data analysis was performed over the last 65 ns of each simulation with a 10 ps sampling frequency. Distance and angles (Figure 1) are in Å and degrees, respectively. Standard deviations (SD) are listed in parenthesis. Boldface font is used to highlight key quantities that are significantly altered with respect to the wild type (WT) simulation upon mutation and that are discussed in the text.
The notation ”d-” denotes the activated precursor state simulations having the C17:O2′ deprotonated.
In-line fitness index.66
The N3 ···N1 distance between nucleobases in the 3 and 8 position.
rHB and θHB are the hydrogen bond length and angle for the general base step; defined by G12:N1 …C17:HO2′..C17:O2′.
rHA and θHA are the hydrogen bond length and angle for the general acid step; defined by C1.1:O5′…G8:HO2′..G8:O2′.
The hydrogen bond contact percentage for the above entries, defined as the percentage of the snapshots in which r ≤ 3.0 Å and θ ≥120 °.
The hydrogen bond contact percentage for the above entries, defined as the percentage of the snapshots in which r ≤ 2.5 Å and θ ≥150 °.
Table 5.
Structural indexes characterizing the relative positions between C1.1 and G8.
| WT | U7C | G8I | C3U/G8A | C3U/G8D | C3G/G8C | |
|---|---|---|---|---|---|---|
| r7 | 4.17(0.38) | 4.15(0.35) | 4.19(0.36) | 4.70(0.69) | 4.89(0.49) | 4.24(0.33) |
| θ7 | 110.2(11.3) | 109.3(9.6) | 111.6(9.8) | 116.8(13.1) | 123.7(9.0) | 111.1(8.1) |
| r8 | 3.82(0.34) | 3.84(0.37) | 3.86(0.36) | 4.06(0.45) | 4.27(0.50) | 4.11(0.43) |
| φ8 | 47.6(8.5) | 45.4(8.3) | 43.8(7.3) | 61.1(20.8) | 61.4(11.9) | 32.5(9.0) |
| d-WT* | d-U7C | d-G8I | d-C3U/G8A | d-C3U/G8D | d-C3G/G8C | |
| r7 | 4.04(0.25) | 3.98(0.25) | 4.13(0.34) | 4.00(0.26) | 4.40(0.36) | 4.23(0.32) |
| θ7 | 111.6(7.1) | 105.7(7.9) | 111.8(8.7) | 108.8(7.9) | 108.6(10.5) | 115.1(9.1) |
| r8 | 3.76().31) | 3.62(0.29) | 3.65(0.34) | 3.51(0.27) | 4.13(0.38) | 3.56(0.27) |
| φ8 | 49.4(6.6) | 47.6(6.1) | 54.1(7.3) | 51.2(6.9) | 54.0(7.0) | 55.6(7.3) |
This table lists key structural indexes to characterize the base-stacking between G8 and C1.1 for different mutants. Data analysis was performed over the last 65 ns of each simulation with a 10 ps sampling frequency. Distance and angles (Figure 1) are in Å and degrees, respectively. r7 is r(C1.1:N1, G8:N9) ; θ7 is θ(C1.1:C2, C1.1:N1, G8:N9); r8 is r(C1.1:O4′, G8:C1′) ; and the torsion angle φ8 is φ(C1.1:C1′, C1.1:O4′,G8:C1′, G8:N9). Standard deviations (SD) are listed in parenthesis. Boldface font is used to highlight key quantities that are significantly altered with respect to the wild type (WT) simulation upon mutation and that are discussed in the text.
The notation ”d-” denotes the activated precursor state simulations having the C17:O2′ deprotonated.
When G8 is mutated to a C, G8:N9 is replaced by C8:N1.
Table 6.
Comparison of experimental evidence and simulation results for mutants studied.
| U7C | C3U | G8A | C3U/G8A | G8I | G8D | C3U/G8D | C3G/G8C | G5I | G5A | G5D | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Simulation | active site integraty | O | XX | O | O | O | XX | X | O | X | X | X |
| general base HB | O | XX | O | X | O | X | O | X | O | O | X | |
| in-line angle | O | O | O | O | O | O | O | O | O | XX | X | |
| general acid HB | O | O | XX | O | O | O | X | O | XX | X | O | |
| Experimental Krel | Ref. 33 | 0.68 | 0.006 | |||||||||
| Ref. 34 | 1.1 | |||||||||||
| Ref. 35 | 0.0003 | |||||||||||
| Ref. 30 | < 0.02 | < 0.004 | nd | |||||||||
| Ref. 36 | ~ 0.5 | ~ 10−3 | < 10−3 | ~ 10−4 | ||||||||
| Ref. 39 | 0.012 | 0.0068 | ||||||||||
| Ref. 43 | ~ 0.004 | |||||||||||
| Ref. 38 | ~ 0.5 | ~ 0.8 | ||||||||||
This table summarizes the mutational effects on different stages of HHR reaction, derived from simulations (upper rows), and the relative experimental cleavage reaction constants (lower rows, relative to the WT). For simulation entries, ”O” refers to benign effect; ”X” mild deleterious effect; ”XX” severe deleterious effect. For experimental entries, ”nd” means ”no catalysis activity detected”. The experimental rate constants are from various HHR sequences and may or may not be the same as the sequence in the simulations.
In the discussion that follows, we will apply certain mechanistic assumptions in our analysis, in particular, with regard to the role of G12 and G8 2′OH as the general base and acid, respectively. These roles are supported by structural data,43, 44 mutagenesis24 and biochemical59, 60 studies, but have not been definitively proven. Assuming this plausible mechanistic hypothesis, we then ask whether our simulation results can explain the origin of the mutational effects. It should be emphasized, however, that should the mechanistic assumptions regarding G12 and G8 be incorrect, so must be the interpretations of the simulation data that invoke this model. To resolve these details, further study is needed, for example, using molecular simulation of the catalytic chemical steps of the reaction with combined quantum mechanical/molecular mechanical methods.
Wild type and U7C control simulations
As a precursor to the discussion of the origin of mutation effects on reaction rate, a characterization of the key elements of the wild type simulation that affect catalysis is needed. Moreover, to lend credence to our simulation methodology and our mechanistic interpretation of simulation results, we perform a control simulation of a U7C mutation that has been observed experimentally to have no adverse effect on the relative rate of reaction as determined by the ratio of rate constants for wild type and mutant reactions, krel = kmut/kwt. The wild type and U7C control simulation results are included in all of the tables for reference and comparison.
Characterization of the active site structure and dynamics of the wild type simulation
The active site scaffold and hydrogen bond networks for the wild type simulation are depicted in Fig. 1. In the full length hammerhead ribozyme catalyzes the site-specific cleavage transesterification of the phosphodiester bond with rate enhancement up to 106-fold relative to the rate of non-catalysed cleavage.14 The rate for cleavage and ligation for the naturally occurring full length hammerhead motifs are around 1000 and 2000 times faster, respectively, than the corresponding rates for the so-called “minimal sequence” hammerhead ribozymes.32, 61, 62 Catalysis is generally believed to proceed by a general acid and base mechanism. In this mechanism, the endocyclic amine of G12 (G12:N1), in deprotonated form, acts as the general base to abstract a proton from the 2′OH of C17 (the nucleophile) to form an activated precursor. The activated precursor then proceeds in an in- line attack to the adjacent scissile phosphate to form a pentacovalent phos-phorane transition state. The 2′OH group of G8 (G8:O2′) acts as a general acid catalyst to donate a proton to the 5′ oxygen of C1.1 (the leaving group) to facilitate breakdown of the phosphorane and phosphodi-ester bond cleavage. The pKa of the general acid is believed to be shifted into the catalytic range through interaction of a divalent metal ion that bridges the phosphoryl oxygens of A9 and the scissile phosphate. These oxygens are positioned approximately 4.3 Å away from one another in the crystal structure, and both exhibit significant catalytic thio effects in the presence of Mg2+ ions that can be rescued by titration with thiophilic Cd2+ ions. Simulation results indicate that the divalent metal ion migrates from a distal binding site involving A9 and G10 in the reactant state to a bridging position between the A9 and scissile phosphates upon formation of the activated precursor.
From this mechanistic picture, several conditions for catalytic competency of the hammerhead ribozyme can be inferred. Firstly, the general base must be correctly positioned to abstract a proton from the nucleophile to form the activated precursor. Secondly, the structure of the active site must allow the activated nucleophile to be in-line with the scissile phosphate, and fluctuations must sample conformations that have a high degree of in-line fitness. Thirdly, the integrity of the active site, and in particular, the proximity of the A9 and scissile phosphates must be conducive to binding a bridging divalent metal ion. Fourthly, the general acid must be poised to donate a proton to the leaving group to facilitate cleavage. In order to satisfy these conditions, a specific network of hydrogen bonds and base stacking interactions must be in place. Indexes correlated with each of these conditions are depicted in Fig. 1. The average values and fluctuations of these indexes for the wild type simulation are listed in the tables for reference, and representative hydrogen bond networks involving conserved residues are shown in the figures.
U7C control simulation satisfies all of the conditions for wild-type catalysis
The U7C mutant has a catalytic rate virtually identical to that of the wild type (krel = 1.1).34 Comparison of the wild type and U7C control simulation show no major differences in the indexes likely to be key for catalysis (see tables). The general base forms a stable hydrogen bond with the nucleophile in the reactant state (rHB=2.1 Å), the distance between the A9 and scissile phosphates (d0) in the reactant state is around 4.3 Å as in the crystal structure, the activated precursor maintains in-line fitness comparable to the wild type simulation and significant hydrogen bonding between the general acid and the leaving group. Additionally, the base stacking interactions are very similar between the WT and U7C simulations (Table 5). Perhaps the most notable difference is that the U7C simulation of the reactant state does not exhibit a strong hydrogen bond between the general acid and leaving group (rHA=3.41 Å in the U7C simulation, whereas the corresponding value is 2.75 Å in the WT simulation). However, the general acid step occurs at a point farther along the reaction coordinate from the reactant state, and examination of the hydrogen bond of the general acid in the activated precursor state indicates a comparable, slightly stronger hydrogen bond interaction (rHA=2.50 and 2.61 Å in the U7C and WT activated precursor simulations, respectively). Overall, the U7C simulation results indicate very comparable integrity of the active site, in-line fitness, and positioning of the general base and acid that are conducive for catalysis.
Single mutations at the C3 and G8 positions
The C3 and G8 positions form a Watson-Crick base pair in the full length hammerhead structure,43, 44 and are important in stabilizing the active site structure, and positioning the 2′OH of G8 for acid catalysis. Here, we consider a series of single mutations (C3U, G8A, G8I and G8D), representative hydrogen bond patterns for which are shown in Fig. 2.
C3U mutation disrupts the active site in the reactant
The C3U mutation reduces the catalytic rate by a factor ~3 × 10−4.35 Simulation results indicate this mutation disrupts the normal Watson-Crick hydrogen bonding with G8 (Fig. 1), causing a base shift that disrupts the active site structure in the reactant state. The average distance between the A9 and scissile phosphate (d0) increases by 2.67 Å relative to the wild type simulation, and breaks key hydrogen bonds between the O2′ nucleophile of C17 and N1 of G12 (the implicated general base). These perturbations in the reactant state would prevent activation of the nucleophile and progress toward the transition state.
G8I mutation is relatively benign
The G8I mutation does not significantly alter catalytic activity, the measured rate reduction being less than a factor of 2.33, 36 The removal of the exocyclic amine at the C2 position weakens the hydrogen bonded base pair with C3, but does not alter the structure (Fig. 2). None of the structural features derived from the simulations in either the reactant state of activated precursor state show any marked differences from the wild type simulations. Overall, the marginal effect on catalysis is likely a consequence of modest weakening, but not disruption, of the base pair between C3 and G8.
G8A mutation blocks the general acid step in the activated precursor
The G8A mutation reduces the catalytic rate by a factor ≤ 0.004.30 Simulation results indicate the G8A mutation considerably weakens the base pair with C3 with only one weak hydrogen bond that remains intact (Fig. 1). In the reactant state simulation, G8A does not appear to dramatically alter the active site contacts relative to the wild-type simulation, with the exception of the A8:N1···C3:N3 distance which increases due to a shift in the hydrogen bond pattern (Fig. 2). In the activated precursor state, however, the hydrogen bond between G8:H2′ and C1.1:O5′ is significantly disrupted relative to the wild-type simulation. The breaking of this key hydrogen bond (rHA=4.13 Å) in the G8A mutation and movement of the general acid away from the leaving group effectively blocks the general acid step of the reaction. The G8A mutation has the largest effect on catalysis of all the single mutations at the G8 position considered here.
G8D mutation alters the active site structure and weakens the interaction of the nucleophile and general base
Mutation of G8 to 2-aminopurine24, 36 or to 2,6-diaminopurine (G8D),36 reduces the reaction rate by approximately three orders of magnitude. The affect of this mutation in the reactant state is to weaken the hydrogen bonding between G8D and C3 relative to the wild type simulation. This results in corruption of the active site structure (d0 increases by 2.89 Å relative to the wild type simulation) and impairs hydrogen bonding of the 2′OH nucleophile with the the general base.
Double mutations at the C3/G8 positions
In order to better understand the role played by individual residues on ribozyme structure and function from mutagenesis data, it is useful to consider the effects of double mutations that are able to restore, at least in part, activity caused by deleterious single mutations. Here, we consider a series of double (C3G/G8C, C3U/G8A and C3U/G8D) mutations, representative hydrogen bond patterns for which are shown in Fig. 2.
C3G/G8C switch mutation has a partial rescue effect
The C3G/G8C switch mutation preserves strong hydrogen bond interactions between the 3 and 8 positions, and has a partial rescue effect relative to the C3G and G8C single mutations, but nonetheless still reduces activity 150–200-fold relative to the wild type.39, 40 The purine/pyrimidine substitutions alter the local stacking environment and lead to subtle structural changes in the active site. As a consequence, the average distance between the A9 and scissile phosphates increases by 0.29 Å relative to the wild type simulation, and the hydrogen bond between the 2′OH nucleophile and general base is weakened (rHB decreases from 2.07 Å in the wild type to 2.50 Å in the C3G/G8C mutant).
C3U/G8A mutation is relatively benign
The C3U/G8A mutation is an isosteric substitution that slightly weakens the basepair hydrogen bonding, and results in a relatively benign mutation with krel of around 0.5–0.012.38, 39 The simulation results for C3U/G8A are similar to that of the relatively benign G8I single mutation in both hydrogen bond pattern and the isosteric pyrimidine/purine base pair (Fig. 2). The C3U/G8A mutation is a slightly larger perturbation than G8I, and the C3U/G8A simulation results indicate correspondingly more significant deviations from the wild type. The C3U/G8A double mutation is overall similar to the C3G/G8C switch mutation, which although the pyrimidine/purine pattern is reversed in C3G/G8C, it does preserve the number of base pair hydrogen bonds in the wild type. The average distance between the A9 and scissile phosphates increases by 0.15 Å in the C3U/G8A mutant relative to the wild type simulation, and the average hydrogen bond distance between the 2′OH nucleophile and general base (rHB) is increased by 0.37 Å. This is expected to only moderately retard the efficiency of the general base step of the reaction. No other notable effects on the structure or dynamics were observed as a result of this mutation.
C3U/G8D mutation is predicted to have a rescue effect
The results of the hydrogen bond preserving C3G/G8C switch mutation, and isosteric C3U/G8A mutations both had partial to strong rescue effects on hammerhead activity. Computationally, we performed simulations of a C3U/G8D mutation that would be an isosteric mutation that also preserved the number of native basepair hydrogen bonds. Despite the corresponding single mutations having significant deleterious effects on activity, the double mutation is predicted by our simulations to have a significant rescue effect. The only significant deviation from the wild type was in d0 in the reactant state simulation, which increased by around 0.7 Å, but did not lead to disruption of any of the key hydrogen bonds or in- line fitness indexes. A closer examination, however, reveals some differences in the base stacking of C3U/G8D relative to the wild type and control simulations (Table 5). The alteration of the relative position between C1.1 and G8 (Fig. 1), in particular the increase in the r7 stacking distance, shifts the position of the A9 phosphate leading to the modest increase in d0. Consequently, the simulations results predict this double mutation to have a significant rescue effect, but not necessarily fully rescue activity to that of the wild type.
Single mutations at the G5 position
The G5 position forms an important element in the structural scaffold of the active site that helps to hold the scissile phosphate in position via hydrogen bond interactions with the sugar and base functional groups of C17, and with the nucleobase of A14 (Fig. 1). These include hydrogen bonds between the G5:N1 endocyclic amine and C17:O2 (r4), and the G5:N2 exocyclic amine and C17:O4′ (r5) and A14:N1 (r6). Here, we consider a series of single mutations (G5I, G5A and G5D), representative hydrogen bond patterns for which are shown in Fig. 3.
G5I mutation retards the general acid step
Experimental rate measurements for the G5I mutation suggest a reduction of the catalytic rate by 0.006 to <10−3 in references 33 and 36, respectively. This mutation eliminates the exocyclic amine of guanine, and hence the hydrogen bond with C17:O6 and A14:N1. As a result, in the reactant state, the integrity of the active site is somewhat disrupted (d0 increases by 0.67 Å). In the activated precursor state, the hydrogen bond between G8:H2′ and C1.1:O5′ is lost (rHA=3.6 Å), which would severely impair the general acid step of catalysis.
G5A mutation severely disrupts the active site integrity and in-line fitness
The G5A mutation leads to no detectable activity in the hammerhead ribozyme.30 This mutation eliminates both the endocyclic and exocyclic amines at the 1 and 2 positions of guanine that hydrogen bond with C17 and A14, and replaces the O6 carbonyl group with a new exocyclic amine at the 6 position. The result is that the nucleobase shifts such that the exocyclic amine at the 6 position of inosine hydrogen bonds with C17:O4′, previously hydrogen bonded to the endocyclic amine at the 1 position. The other hydrogen bonds to C17 and A14 are eliminated. This results in considerable degradation of the active site (d0 increases by 0.98 Å in the reactant state), poor in-line fitness in the activated precursor state (F decreases by 50%), and greatly reduces hydrogen bonding interaction between G8:H2′ and C1.1:O5′ (rHA increases by 1.1 Å). Overall, the active site structure and positioning of the general acid in the activated precursor are not conducive for catalysis.
G5D mutation severely disrupts the active site integrity and in-line fitness
The G5D mutation leads to reduction of catalytic rate by 4 orders of magnitude (krel = 10−4).36 The hydrogen bond pattern for this mutation is not as severely perturbed as for the G5A mutation. Nonetheless, in the reactant state, the average distance between A9 and the scissile phosphates (d0) increases to 5.2 Å, and hydrogen bond between the 2′OH nucleophile and G12:N1 (the implicated general base) is less pronounced. In the activated precursor state, the general base hydrogen bond is dramatically weakened (rHB=2.62 Å), and the average in-line attack angle, θinl, is reduced to 139.7°. These structural deviations are expected to hinder the attack of the nucleophile to the scissile phosphate.
Discussion
In the reactant state, shifted residue positions stabilized by hydrogen bond networks disrupt the active site integrity
Simulation results (Table 1) show that, among the C3 and G8 mutants, d0 shows the greatest deviation for C3U (d0 = 6.64 Å) and G8D (d0 = 5.86 Å), compared to the wild-type (d0 = 3.97 Å) and U7C control simulation (d0 = 4.27 Å). In these two mutants, the C3/G8 base pairs are held together by two hydrogen bonds and the bases are shearer relative to the WT (Fig. 2). In other mutants, including C3U/G8A, G8I, and C3G/G8C, all d0 values are similar to WT and U7C. In the case of the C3U/G8D mutation, subtle changes in base stacking produce a slight shift in the relative positions of C.1. and G8 (Table 5), and this deviation is stabilized by the strong hydrogen bond network, leading to a modest increase in d0 (d0 = 4.68 Å). In the case of mutations at G5, the G5D mutant has the largest value for d0 (d0 = 5.2 Å). The G5D mutant also displays a significant shift in the orientation of D5 with respect to C17 (Fig. 3) that is stabilized by a fairly strong hydrogen bond network. Thus, the mutants that exhibit large shifts in the relative positions of key residues in the active site scaffold and that are locked by hydrogen bonding networks (i.e., C3U, G8D, G5D), generally display the largest disruptions of the active site architecture. Alternately, mutants that do not exhibit shifts of the relative positions of residues (i.e., G8I, C3U/G8A and C3G/G8C) or that have weakened hydrogen bond networks (G8A, G5A, G5I) have d0 values more similar to the Wt and control simulations. Figure 4 shows the cluster-average structures of the active sites from the reactant states of WT, C3U, and G8D.
In the activated precursor state, strong hydrogen bond networks stabilize the active site architecture
In all of the activated precursor simulations, the relative positions of the A9 and scissile phosphates are very similar (d0 values of around 3.0 Å). This is a result of the bridging Mg2+ ion that locks the A9:O2P and C1.1 O2P positions. The general base indexes are also similar between the different simulations due to the strong hydrogen bond that forms between G12:N1 and the deprotonated C17:O2′ nucleophile. The one exception is for d-G5D, where the loss of a hydrogen bond between D5 and the C17:O4′ causes the position of the sugar to shift and the hydrogen bond between the activated nucleophile and G12 to be less populated.
The orientation of the implicated general acid, G8:O2′, so as to be poised to donate a proton to the C1.1:O5′ leaving group is important for catalytic competency. Alterations in the hydrogen bond network that disrupt the position of G8:O2′, particularly the base pair hydrogen bonds between C3 and G8, will have an adverse effect of the general acid step, and can be tracked by monitoring the hydrogen bond distance rHA. Table 3 and Table 4 indicate that activated precursor simulations generally have similar general acid indexes (rHA and θHA), with a few notable exceptions. In the d-G8A simulation, the C3-G8 hydrogen bond network is lost. In the d-C3U/G8D simulation the C1.1-G8 base stacking is significantly different from other mutants. In the d-G5I and d-G5A simulations, the hydrogen bond network between G5 and C17 is disrupted.
Hence, in the activated precursor state, the two strands are brought together by the bridging divalent metal ion and a strong C3-G8 hydrogen bond network is necessary to keep G8 in position so that the general acid (G8:O2′) is poised for catalysis. The interaction between G5 and C17 is important since G5 stabilizes the orientation of both the nucleobase and ribose of C17, and helps to position the nucleophile in-line with the scissile phosphate. Disruption of G5-C17 hydrogen bonds will either prevent in-line attack (d-G5A and d-G5D) or the alter the position of the general acid (d-G5I). For the double mutant d-C3U/G8D, although all U3-D8 hydrogen bonds are maintained (Table 3), the C1.1-G8 base stacking distance is slightly elongated as indicated by larger r7 and r8 in Table 5, and as a result, the general acid, G12:O2′ is shifted away from the leaving group C1.1:O5′ (rHA in Table 3).
Figures 5 and 6 demonstrates the significant mutational effects on the activated precursor states due to C3U/G8D and G8A (Figure 5) and G5I, G5A, and G5D (Figure 6) by showing the corresponding cluster-average structures.
C3/G8 mutations may induce structural changes that have remote consequences at the general base step
There is evidence that minimal sequence hammerhead motifs are conformationally variable, and that there is an equilibrium between active and inactive conformational states.32, 39, 63 It has been hypothesized that minimal and extended (full length) hammerheads may utilize a similar dynamic reaction mechanism for catalysis, and that observed correlations between mutations between C17 and C3/G8 may be explained by transient pairing between residues 3 and 17 and 8 and 13 arising from interactions in the inactive conformation.39 The supposition was based, in part, on the observation that C3/G8 are distant from C17 in the extended hammerhead structures, making it difficult to rationalize the observed correlations between mutations between C17 and C3/G8 in an active conformational state. In the inactive minimal sequence structures, C3 pairs with C17 and G8 pairs with A13.40, 42, 64 Our simulations results of the active full-length hammerhead suggest that mutations at C3/G8, in some cases, may induce remote changes in the structure and dynamics at C17 that affect the general base step of the reaction. Recent kinetics studies have indicated that the native full length hammerhead constructs exhibit less conformational variability than the minimal sequence motifs, and maintain a higher population of active conformations.32, 63 The present simulations results afford an alternate interpretation of the experimentally observed correlations between mutations between C17 and C3/G8 positions that does not rely, at least as heavily, on the assumption that mutants of the full length hammerhead are in a dynamic equilibrium that populate an inactive conformation with contacts resembling that of the minimal-sequence.
Variation of base stacking interactions of G8 and C3 with C1.1 and G2.1 may lead to new HHR motifs with modified activity
Our results suggest that the identities of nucleotides at C3 and G8, including some double-mutants that preserve base pairing, can adversely effect HHR catalytic activity. This implies that the environmental effect due to the base stacking interactions between G8/C1.1 and C3/G2.1 may be important to hold G8/C1.1 in the proper relative position for the general acid step of HHR catalysis (Table 5). In recent studies on various HHRs, it has been shown that the tolerance of mutations at C3 and G8 is dependent on the identities of surrounding nucleotides, including C1.1 and G2.1. Residues C1.1 and G2.1 are not conserved,17, 30, 65 and hence it may be possible to exploit variations in C1.1 and G2.1, along with correlated mutations at G8 and C3, to design new HHR motifs with modified activity.
Long time simulations are required to understand some mutational effects
The current study reports 100 ns simulations for a series of HHR mutants in the reactant and activated precursor states. As pointed out previously,27 long time simulations are necessary to relax the mutations and sufficiently sample the conformational space required to observe the mutational effects. Shown in Figure 7, the simulation of the activated precursor state of G8A reaches a stable relaxed state after 30 ns. Figure 8 shows the backbone heavy atom traces from the first 100 ns of the reactant state trajectory of the native sequence (WT), and suggests that the system undergoes a significant conformational motion in the ns time-scale. However, on this time scale, for the extended hammerhead ribozyme, we do not observe an equilibrium between proposed active and inactive states that involve changes in stem contacts, as has been inferred from biochemical studies.39 Recent time-resolved NMR spectroscopic investigations have indicated that an ensemble of conformations, sensitive to the presence of Mg2+, are accessible to the minimal hammerhead ribozyme, and that dynamic transitions, localized around the catalytic core, occur on the ms time scale.63 This time scale extends beyond that which has been probed in the current work for the full length hammerhead, but nonetheless, the question of dynamic conformational equilibrium and it role is catalysis remains an important one to address in future work.
Figure 7.
The time series of general acid hydrogen bond indexes, rHA and θHA, as well as the hydrogen bond distance (r3, see Figure 1 and Figure 2), from WT and G8A activated precursor state simulations. The distances are in Å and the angles are in degrees. These results indicate that the time scale for equilibration and stable fluctuation in some cases exceed 30 ns (shown by vertical dashed red line).
Figure 8.

The phosphate backbone heavy atom traces from the reactant state simulations of the native sequence (WT). In total, 100 structures are used here, extracting from the first 100 ns of the trajectory, with one structure for every one ns. The yellow colored trace is the starting structure. Two different angle views are shown (A and B).
Conclusions
The present molecular simulation results provide insight into the origin of mutational effects in the full length hammerhead ribozyme not easily derivable from available experimental structural data. We report results of molecular dynamics simulations of the native and mutated full length hammerhead ribozymes in the reactant state and in an activated precursor state. A key result from this work is that in performing computational mutagenesis simulations of ribozymes, one must consider time scales greater than 30 ns for relaxation of mutant structures, and in some instances look beyond the reactant state along the catalytic reaction coordinate in order to reconcile the origin of mutational effects. The present study makes predictions, and offers new insights into the understanding of the hammerhead mechanism as interpreted through mutational data.
Mutant simulations at the C3, G8, and G5 positions were performed and the simulation results are consistent with a mechanistic model where G12 acts as the general base while the 2′OH of G8 acts as a general acid, although it does not definitively exclude other mechanisms. Simulations suggest that the Watson-Crick base pair interaction between G8 and C3, the hydrogen bond network between C17 and G5, and the base stacking interactions between G8 and C1.1, collectively contribute to the stabilization of the active site scaffold that is required for catalytic activity. Disruption of any of these components can affect the catalysis at different stages of the reaction. Analysis of the simulations indicate that C3/G8 mutations may induce structural changes that have remote consequences at the general base step, and afford a possible alternative, or partial, explanation of experimentally observed correlations between mutations between C17 and C3/G8 positions. Simulation results suggest that base stacking interactions of G8 and C3 with non-conserved residues C1.1 and G2.1 can affect activity, and provide insights into the design of new HHR motifs with modified activity. Finally, we predict that the C3U/G8D double mutation is predicted to have a rescue effect.
The results reported here offer molecular-level details into the origin of mutational effects on full length hammerhead ribozyme catalysis. It should be emphasized that the interpretations made here do impose particular assumptions about the mechanism, such as the roles of G12 and G8 2′OH as general base and acid, respectively. It is possible that other mechanistic scenarios whereby these roles are played by different residues, including metal ions, might prove equally consistent with the data. To fully resolve these questions, combined quantum mechanical/molecular mechanical simulations of the chemical steps of the reaction must be performed. Nonetheless, the present computational mutagenesis study provides a detailed characterization of the structure and dynamics of a series of important hammerhead mutations, and makes predictions about compensatory mutations that can be tested experimentally.
Supplementary Material
Acknowledgments
The authors are grateful for financial support provided by the National Institutes of Health (GM62248 to DY). Computational resources from The Minnesota Supercomputing Institute for Advanced Computational Research (MSI) were utilized in this work. This work was also supported in part by a generous allocation on an IBM Blue Gene BG/P with 4,096 850 Mhz CPUs at the IBM Advanced Client Technology Center in Rochester, Minnesota with further thanks to Cindy Mestad, Steven Westerbeck, and Geoffrey Costigan for technical assistance.
References
- 1.Scott WG. Curr Opin Struct Biol. 1998;8:720–726. doi: 10.1016/s0959-440x(98)80091-2. [DOI] [PubMed] [Google Scholar]
- 2.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 3.Takagi Y, Ikeda Y, Taira K. Top Curr Chem. 2004;232:213–251. [Google Scholar]
- 4.Scott WG. Curr Opin Struct Biol. 2007;17:280–286. doi: 10.1016/j.sbi.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert W. Nature. 1918;319:618. [Google Scholar]
- 6.Scott WG. Biochem Soc Trans. 1996;24:604–608. doi: 10.1042/bst0240604. [DOI] [PubMed] [Google Scholar]
- 7.Gesteland RF, Cech TR, Atkins JF. The RNA World: The Nature of Modern RNA Suggests a Prebiotic RNA. 2. Cold Spring Harbor Laboratory Press; New York: 1999. [Google Scholar]
- 8.Yarus M. Curr Opin Chem Biol. 1999;3:260–267. doi: 10.1016/S1367-5931(99)80041-6. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Li N, Ellington AD. Chem Biodivers. 2007;4:633–655. doi: 10.1002/cbdv.200790055. [DOI] [PubMed] [Google Scholar]
- 10.Rubenstein M, Tsui R, Guinan P. Drugs of the Future. 2004;29:893–909. [Google Scholar]
- 11.Vaish NK, Dong F, Andrews L, Schweppe RE, Ahn NG, Blatt L, Seiwert SD. Nature Biotech. 2002;20:810–815. doi: 10.1038/nbt719. [DOI] [PubMed] [Google Scholar]
- 12.Breaker RR. Curr Opin Biotechnol. 2002;13:31–39. doi: 10.1016/s0958-1669(02)00281-1. [DOI] [PubMed] [Google Scholar]
- 13.Scott WG. Q Rev Biophys. 1999;32:241–294. doi: 10.1017/s003358350000353x. [DOI] [PubMed] [Google Scholar]
- 14.Blount KF, Uhlenbeck OC. Biochem Soc Trans. 2002;30:1119–1122. doi: 10.1042/bst0301119. [DOI] [PubMed] [Google Scholar]
- 15.Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 16.Forster AC, Symons RH. Cell. 1987;50:9–16. doi: 10.1016/0092-8674(87)90657-x. [DOI] [PubMed] [Google Scholar]
- 17.Uhlenbeck O. Nature. 1987;328:596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- 18.Link KH, Guo L, Ames TD, Yen L, Mulligan RC, Breaker RR. Biol Chem. 2007;388:779–786. doi: 10.1515/BC.2007.105. [DOI] [PubMed] [Google Scholar]
- 19.Wieland M, Gfell M, Hartig JS. RNA. 2009;15:968–976. doi: 10.1261/rna.1220309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sano M, Taira K. Methods Mol Biol. 2007;360:143–153. doi: 10.1385/1-59745-165-7:143. [DOI] [PubMed] [Google Scholar]
- 21.Sarver N, Cantin EM, Chang PS, Zaia JA, Ladne PA, Stephens DA, Rossi JJ. Science. 1990;247:1222–1225. doi: 10.1126/science.2107573. [DOI] [PubMed] [Google Scholar]
- 22.Michienzi A, Cagnon L, Bahner I, Rossi JJ. Proc Natl Acad Sci U S A. 2000;97:8955–8960. doi: 10.1073/pnas.97.16.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alvarez-Salas LM, Benítez-Hess ML, DiPaolo JA. Antivir Ther. 2003;8:265–278. [PubMed] [Google Scholar]
- 24.Blount KF, Uhlenbeck OC. Annu Rev Biophys Biomol Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- 25.Lee TS, Silva-Lopez C, Martick M, Scott WG, York DM. J Chem Theory Comput. 2007;3:325–327. doi: 10.1021/ct6003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee T-S, Silva Lopez C, Giambasu GM, Martick M, Scott WG, York DM. J Am Chem Soc. 2008;130:3053–3064. doi: 10.1021/ja076529e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee TS, York DM. J Am Chem Soc. 2008;130:7168–7169. doi: 10.1021/ja711242b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee TS, Giambau GM, Sosa CP, Martick M, Scott WG, York DM. J Mol Biol. 2009;388:195–206. doi: 10.1016/j.jmb.2009.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee T-S, Giambaşu GM, Nam K, Guerra F, Giese TJ, York DM. Unraveling the mechanisms of ribozyme catalysis with multi-scale simulations. In: York DM, Lee T-S, editors. Multi-scale Quantum Models for Biocatalysis: Modern Techniques and Applications. CHALLENGES AND ADVANCES IN COMPUTATIONAL CHEMISTRY AND PHYSICS Springer Verlag; 2009. [Google Scholar]
- 30.Ruffner DE, Stormo GD, Uhlenbeck OC. Biochemistry. 1990;29:10695–10712. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- 31.la Peña MD, Gago S, Flores R. EMBO Journal. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Nature Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 33.Tuschl T, Ng MM, Pieken W, Benseler F, Eckstein F. Biochemistry. 1993;32:11658–11668. doi: 10.1021/bi00094a023. [DOI] [PubMed] [Google Scholar]
- 34.Burgin AB, Jr, Gonzalez C, Matulic-Adamic J, Karpeisky AM, Usman N, McSwiggen JA, Beigelman L. Biochemistry. 1996;35:14090–14097. doi: 10.1021/bi961264u. [DOI] [PubMed] [Google Scholar]
- 35.Baidya N, Uhlenbeck OC. Biochemistry. 1997;36:1108–1114. doi: 10.1021/bi962165j. [DOI] [PubMed] [Google Scholar]
- 36.Han J, Burke JM. Biochemistry. 2005;44:7864–7870. doi: 10.1021/bi047941z. [DOI] [PubMed] [Google Scholar]
- 37.Lambert D, Heckman JE, Burke JM. Biochemistry. 2006;45:7140–7147. doi: 10.1021/bi052457x. [DOI] [PubMed] [Google Scholar]
- 38.Przybilski R, Hammann C. RNA. 2007;13:1625–1630. doi: 10.1261/rna.631207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson JA, Uhlenbeck OC. RNA. 2008;14:43–54. doi: 10.1261/rna.717908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott WG, Murray JB, Arnold JRP, Stoddard BL, Klug A. Science. 1996;274:2065–2069. doi: 10.1126/science.274.5295.2065. [DOI] [PubMed] [Google Scholar]
- 41.Murray JB, Terwey DP, Maloney L, Karpeisky A, Usman N, Beigelman L, Scott WG. Cell. 1998;92:665–673. doi: 10.1016/s0092-8674(00)81134-4. [DOI] [PubMed] [Google Scholar]
- 42.Murray JB, Szöke H, Szöke A, Scott WG. Mol Cell. 2000;5:279–287. doi: 10.1016/s1097-2765(00)80423-2. [DOI] [PubMed] [Google Scholar]
- 43.Martick M, Scott WG. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martick M, Lee TS, York DM, Scott WG. Chem Biol. 2008;15:332–342. doi: 10.1016/j.chembiol.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humphrey W, Dalke A, Schulten K. J Mol Grapics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 46.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 47.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kaleé L, Schulten K. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cornell WD, Cieplak P, Bayly CI, Gould IR, Ferguson DM, Spellmeyer DC, Fox T, Cald-well JW, Kollman PA. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 49.Case DA, et al. AMBER 7. University of California San Francisco; San Francisco: 2002. [Google Scholar]
- 50.Pearlman DA, Case DA, Caldwell JW, Ross WR, Cheatham T, III, DeBolt S, Ferguson D, Seibel G, Kollman P. Comput Phys Commun. 1995;91:1–41. [Google Scholar]
- 51.Andersen HC. J Chem Phys. 1980;72:2384–2393. [Google Scholar]
- 52.Nosé S, Klein ML. Mol Phys. 1983;50:1055–1076. [Google Scholar]
- 53.Hoover WG. Phys Rev A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 54.Essmann U, Perera L, Berkowitz ML, Darden T, Hsing L, Pedersen LG. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 55.Sagui C, Darden TA. Annu Rev Biophys Biomol Struct. 1999;28:155–179. doi: 10.1146/annurev.biophys.28.1.155. [DOI] [PubMed] [Google Scholar]
- 56.Allen M, Tildesley D. Computer Simulation of Liquids. Oxford University Press; Oxford: 1987. [Google Scholar]
- 57.Ryckaert JP, Ciccotti G, Berendsen HJC. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 58.Ponomarev SY, Thayer KM, Beveridge DL. Proc Natl Acad Sci USA. 2004;101:14771–14775. doi: 10.1073/pnas.0406435101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas JM, Perrin DM. J Am Chem Soc. 2008;130:15467–15475. doi: 10.1021/ja804496z. [DOI] [PubMed] [Google Scholar]
- 60.Thomas JM, Perrin DM. J Am Chem Soc. 2009;131:1135–1143. doi: 10.1021/ja807790e. [DOI] [PubMed] [Google Scholar]
- 61.Canny MD, Jucker FM, Kellogg E, Khorova A, Jayasena SD, Pardi A. J Am Chem Soc. 2004;126:10848–10849. doi: 10.1021/ja046848v. [DOI] [PubMed] [Google Scholar]
- 62.Canny MD, Jucker FM, Pardi A. Biochemistry. 2007;46:3826–3834. doi: 10.1021/bi062077r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fürtig B, Richter C, Schell P, Wenter P, Pitsch S, Schwalbe H. RNA Biol. 2008;5:41–48. doi: 10.4161/rna.5.1.5704. [DOI] [PubMed] [Google Scholar]
- 64.Pley HW, Flaherty KM, McKay DB. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 65.Sheldon CC, Symons RH. Nucleic Acids Res. 1989;17:5679–5685. doi: 10.1093/nar/17.14.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soukup GA, Breaker RR. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.