The initiation of flowering is a critical life-history trait; plants have presumably evolved to flower at a time of year that ensures maximal reproductive success in a given region. Decades of physiological studies have revealed that flowering is initiated in response to both environmental cues and endogenous pathways. Commonly studied environmental cues include changes in temperature and daylength. Endogenous pathways function independently of environmental signals and are related to the developmental state of the plant; such pathways are sometimes referred to as “autonomous” to indicate the lack of environmental influence. The relative contributions of autonomous and environmental inputs to the flowering “decision” vary among, and even within, species. For example, flowering is considered entirely due to autonomous pathways in a variety of tobacco (Nicotiana tabacum) that forms a fixed number of nodes before flowering regardless of the environment in which it is grown (McDaniel and Hsu, 1976). Yet, a single-gene change can cause tobacco to require short days to flower (Allard, 1919), which indicates that the underlying biochemical differences between environment-sensing and endogenous pathways can be minimal. Also, endogenous and environmental pathways can interact. For example, some plants pass through a juvenile phase in which they are not responsive to environmental cues that promote flowering (Poethig, 1990); that is, the transition from the juvenile to adult phase is a type of endogenous pathway that is necessary to provide competence for environmental pathways to promote flowering. The recent addition of molecular genetics to the range of approaches used to study the initiation of flowering has provided some molecular insights into these endogenous and environment-sensing pathways and has revealed how inputs from multiple pathways are integrated into the flowering decision.
(Due to the sustained efforts of a multitude of scientists working in many species, we have learned much about the timing of flowering that is worth celebrating. Unfortunately, only a small part of this extensive body of work can be covered in this article because of length and reference limits. Accordingly, we frequently refer readers to recent review articles for more in-depth discussions, and we apologize to our colleagues whose work was not cited due to these constraints.)
PHOTOPERIODISM AND FLORIGEN: AN ANCIENT PATHWAY
The annual fluctuations in daylength that occur over much of the surface of our planet provide a reliable environmental cue regarding the time of year. It is not surprising, therefore, that the pathways that detect and promote flowering in response to photoperiod are among the most ancient and conserved. Physiological experiments first done in the 1930s (Knott, 1934) demonstrated that inductive photoperiods are sensed by leaves. This raised two fundamental questions: how do leaves measure daylength, and what is the nature of the flowering signal (known as florigen) that must travel from the leaves to the shoot apical meristem? After another seven decades of research, we now have relatively clear and satisfying answers to these questions, especially in Arabidopsis (Arabidopsis thaliana).
Arabidopsis flowers more rapidly in long days than in short days and is thus a facultative long-day plant. The regulation of the floral promoter CONSTANS (CO) is key in the perception of inductive long days (Turck et al., 2008). The circadian clock regulates CO transcription such that peak expression occurs late in the day in long days but after dusk in short days (Suarez-Lopez et al., 2001). CO protein, in turn, is stabilized by light and rapidly degraded in darkness (Valverde et al., 2004). As a result, CO protein can only accumulate during inductive long days. CO is expressed in the vasculature of leaves, and its role in flowering is to activate the expression of FLOWERING LOCUS T (FT), which encodes a small protein that is florigen (Fig. 1). In both rice (Oryza sativa) and Arabidopsis, FT is a strong promoter of flowering that is translocated from the vasculature of leaves to the shoot apical meristem (Corbesier et al., 2007; Tamaki et al., 2007). In the meristem, FT forms a complex with the bZIP transcription factor FD and initiates flowering by activating floral meristem-identity genes such as APETALA1 and other floral promoters such as SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1; Michaels, 2009). Thus, FT up-regulation lies at the end of an environment-sensing pathway and initiates flower development. In addition to the photoperiod pathway, FT and SOC1 are also regulated by other flowering pathways (e.g. vernalization; see below) and therefore are referred to as floral integrators.
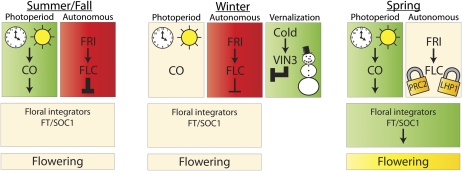
Figure 1.
Seasonal regulation of flowering in winter-annual Arabidopsis. The flowering pathways that are active in each season are indicated by green or red boxes; green is promotive and red is repressive. Beige indicates inactive. In the summer/fall establishment phase, FLC prevents flowering by repressing floral integrators that would otherwise be induced by CO in response to long days (left). During the short days of winter, the photoperiod pathway is not active and vernalization leads to the induction of VIN3 and epigenetic repression of FLC (center). By the spring season, FLC repression is complete and is maintained by PRC2 and LHP1 (and VRN1; not pictured); in the lengthening days of spring, CO activates floral integrators free of competition from FLC and flowering is initiated (right).
The coupling of CO and FT appears to be an ancient and evolutionarily adaptable module. Unlike the situation in Arabidopsis, in which long days lead to CO activation and FT induction, the rice CO/FT homologs (HEADING DATE1/HEADING DATE3A) have evolved different circuitry that triggers flowering in response to short days (Turck et al., 2008). It will be interesting to explore the possible role of CO and FT in more complex photoperiod response types, such as various species of Bryophyllum, which require long days followed by short days for flowering to occur (i.e. plants maintained under constant long or short days do not flower). There is also evidence that the role of CO and FT extends beyond flowering. In poplar (Populus spp.) trees, CO and FT are involved in the initiation of photoperiod-dependent dormancy (Turck et al., 2008). There is also intriguing data demonstrating that the use of CO as a daylength indicator may predate flowering plants. Chlamydomonas reinhardtii lacks FT but does contain a CO-like gene (CrCO) that is an output of the circadian clock; remarkably, CrCO can partially rescue co mutants in Arabidopsis (Serrano et al., 2009). Given that CO exists in a relatively large gene family (17 CO-like genes in Arabidopsis), it is possible that CO-related genes play additional yet-to-be-discovered roles in plant responses to daylength.
VERNALIZATION
Vernalization is defined as the process by which exposure to the cold of winter renders plants competent to flower (Kim et al., 2009). The passage of winter is an environmental cue that, when coupled to photoperiod sensing, provides clear seasonal information that distinguishes the spring and fall seasons. For cold to be a reliable cue for winter, plants need to be able to distinguish the long cold exposure characteristic of winter from short fluctuations in temperature that might occur, for example, in the fall. Thus, it is not surprising that vernalization (and in many species the breaking of bud dormancy) requires exposure to prolonged cold. A vernalization requirement is often found in winter-annual and biennial plants that flower early in the spring; these plants typically become established in the fall, and a vernalization requirement ensures that premature flowering does not occur during the fall establishment phase.
In winter-annual Arabidopsis, the vernalization-responsive block to flowering requires the interaction of two genes, FLOWERING LOCUS C (FLC) and FRIGIDA (FRI; Michaels and Amasino, 1999; Sheldon et al., 1999; Johanson et al., 2000). FLC is a MADS domain-containing transcription factor that acts as a floral repressor, and FRI is a plant-specific gene of unknown biochemical function that is required for high levels of FLC expression. FLC inhibits flowering by directly repressing the key promoters of flowering, FT, SOC1, and FD (Michaels, 2009; Fig. 1). Vernalization permits plants to flower rapidly in the lengthening days of spring through repression of FLC (Fig. 1). FRI and FLC were first identified genetically in crosses between winter-annual and rapid-cycling accessions (Napp-Zinn, 1979; Burn et al., 1993; Lee et al., 1993; Clarke and Dean, 1994; Gazzani et al., 2003; Michaels et al., 2003); winter annuals contain functional alleles of both genes, whereas rapid-cycling accessions contain loss-/reduction-of-function mutations in either FRI or FLC (Kim et al., 2009). Thus, rapid-cycling accessions evolved from winter annuals by shedding the vernalization requirement conferred by the interaction of FRI and FLC.
After winter has passed, there is a permanent “memory” of winter in many plant species (i.e. the vernalized state is stable during subsequent growth and mitotic cell division). Mitotic stability in the absence of the inducing signal (cold) is a classic definition of an epigenetic change of state (Amasino, 2004). In Arabidopsis, the epigenetic nature of the vernalized state results from a series of modifications to FLC chromatin that result in mitotically stable repression. Specifically, the levels of two repressive modifications, trimethylation of histone H3 at Lys-9 (H3K9) and Lys-27 (H3K27), increase at FLC chromatin during and after cold exposure (Bastow et al., 2004; Sung and Amasino, 2004). H3K27 methylation at FLC results from the activity of Polycomb Repressive Complex2 (PRC2), which was first identified in animals and is conserved in eukaryotes (Kim et al., 2009). During cold exposure, VERNALIZATION INSENSITIVE3 (VIN3), a gene encoding a plant-specific component of the PRC2 complex that is essential for FLC repression, is induced (Wood et al., 2006; De Lucia et al., 2008). The PRC2 complex in plants and animals is involved in the repression of a large number of genes, but in Arabidopsis, the cold-induced VIN3 is a component specific for the vernalization process; thus, the VIN3-containing version of PRC2 is likely to target a vernalization-specific subset of genes. There is a family of VIN3-like genes in Arabidopsis (Kim et al., 2009), and why VIN3 is specifically critical for vernalization-mediated silencing of FLC is an intriguing issue to resolve. It is also intriguing that during cold exposure, there is a transient increase in expression of a noncoding RNA complementary to FLC known as COOLAIR (Swiezewski et al., 2009), but it remains to be determined what role, if any, this RNA plays in vernalization-mediated FLC silencing.
Polycomb repression in animals does not typically involve H3K9 methylation, whereas repression of FLC involves both H3K9 and H3K27 methylation. The methylase involved in vernalization-mediated H3K9 methylation has not been identified, but the plant-specific VERNALIZATION1 (VRN1) protein and a plant relative of a protein first identified in animals that binds methylated H3K9 (LIKE HETERCHROMATIN PROTEIN1 [LHP1]) are required to maintain H3K9 methylation and FLC repression (Kim et al., 2009). It is interesting that in animals, a Polycomb complex called PRC1 is involved in maintaining Polycomb-mediated repression, but plants do not possess PRC1 components; thus, VRN1 and LHP1 may play a PRC1-like role.
AUTONOMOUS CONTROL OF FLOWERING
In Arabidopsis, two antagonistic autonomous pathways regulate prevernalization levels of FLC expression. FLC is positively regulated by the FRI pathway and negatively regulated by a group of genes known collectively as the autonomous floral-promotion pathway. In winter annuals, the FRI pathway acts epistatically to the autonomous pathway and activates FLC expression to create vernalization-responsive late flowering. In rapid-cycling accessions, which typically lack functional alleles of FRI, the autonomous pathway represses FLC; thus, recessive autonomous-pathway mutants are late flowering due to high levels of FLC expression and are vernalization responsive. It is important to note that vernalization represses FLC expression without impacting the expression of FRI or autonomous-pathway genes. This, as well as other data, indicates that the effect of FRI and the autonomous pathway on FLC does not appear to be regulatory per se; rather, they are involved in setting basal levels of FLC expression via constitutive activation/repression.
Although the genetic circuitry by which FRI and the autonomous pathway control FLC expression is well established, our knowledge of molecular mechanism remains limited. The predicted biochemical functions of many autonomous-pathway proteins suggest that FLC repression may involve a coupling of RNA-binding/processing and chromatin-remodeling events (Kim et al., 2009; Michaels, 2009). Three proteins, FCA, FPA, and FLOWERING LOCUS K, contain RNA-binding domains, and a fourth, FY, shows homology to RNA-processing factors. In addition, dicer-like1 dicer-like3 double mutants have elevated levels of FLC, suggesting that small RNA processing may play a role in FLC repression. Other autonomous-pathway proteins act, or are predicted to act, as histone methyltransferases (e.g. several members of the PRMT family) or histone demethylases (e.g. FLOWERING LOCUS D and RELATIVE OF EARLY FLOWERING6). One of the remaining major challenges in Arabidopsis is to determine how these RNA- and chromatin-related elements function together to set the level of FLC expression. One thing that has recently become clear is that the function of many of the so-called autonomous-pathway genes is not restricted to the regulation of flowering time. Although the phenotypes of most autonomous-pathway single mutants are largely limited to delayed flowering, some autonomous-pathway double mutants show strong pleiotropic phenotypes and many autonomous-pathway mutants show defects in gene silencing (Baurle et al., 2007; Veley and Michaels, 2008). Interestingly, these loss-of-gene-silencing phenotypes are correlated with changes in DNA methylation at the affected loci. The fact that DNA methylation is not observed at the FLC locus suggests that proteins of the autonomous pathway may participate in multiple repressive pathways.
Genetic screens conducted in FRI-containing or autonomous-pathway mutant backgrounds have also identified many genes required for high levels of FLC expression (Kim et al., 2009; Michaels, 2009). Perhaps not surprisingly, many of these proteins are associated with activating histone modifications. For example, such screens have identified components of a RNA polymerase II-associated factor 1 complex, which promotes activating H3K4 and H3K36 methylation, as well as other complexes that promote histone 2B monoubiquitination and deposition of the histone variant H2A.Z. Although the majority of these genes are required for high levels of FLC expression in FRI-containing or autonomous-pathway mutant backgrounds, it is interesting that some genes, such as FRI-LIKE1, SUPPRESSOR OF FRI4, and FRI ESSENTIAL1, are only required for the up-regulation of FLC by FRI (Michaels et al., 2004; Schmitz et al., 2005; Kim et al., 2006; Kim and Michaels, 2006). Thus, in terms of molecular mechanism, the activation of FLC in an autonomous-pathway mutant is not exactly the same as the activation of FLC by FRI. It will be quite interesting to discover the molecular relationships between the common and pathway-specific components that positively regulate FLC.
Whether FLC-like genes repress flowering outside of the crucifers is an open question. It is possible that the FLC-based pathways described above are crucifer specific; however, other autonomous pathways may be more widespread. There is a microRNA, miR156, involved in the timing of the juvenile-to-adult transition in both maize (Zea mays) and Arabidopsis, and expression of this microRNA delays flowering, whereas expression of another microRNA, miR172, promotes flowering in part by relieving FT repression (Fornara and Coupland, 2009; for more on miRNAs and phase transitions, see Poethig, 2010). Expression of these microRNAs is in part under developmental control; thus, this system could be considered a more conserved autonomous pathway.
INTEGRATION OF PHOTOPERIOD AND VERNALIZATION
As discussed above, the basic photoperiod pathway appears to be conserved in flowering plants, and as illustrated in Figure 1, in Arabidopsis the circuitry of how the vernalization and photoperiod pathways interact is clear: FLC represses expression of flowering promoters (integrators) until this repression is removed through the silencing of FLC by vernalization. However, in cereals, the vernalization pathway is distinct from that in Arabidopsis. In the cereal pathway, there is a flowering repressor, VRN2, that, like FLC, is turned off during cold exposure. However, FLC is a MADS box protein, whereas VRN2 is a zinc-finger protein that does not have a homolog in the Arabidopsis genome. FLC expression is repressed solely by cold, whereas in cereals, VRN2 expression is repressed by cold, short days, and induction of the meristem-identity gene VERNALIZATION1 (note: in cereals, VERNALIZATION1 is a MADS box gene unrelated in amino acid sequence to Arabidopsis VRN1).
Despite their differences, there is a common feature of the interface between the vernalization and photoperiod pathways in Arabidopsis and cereals: both FLC and VRN2 repress the key photoperiod pathway gene FT (VRN3 in cereals). This example of convergent evolution in how the pathways interface is perhaps not surprising. In contrast to the ancient photoperiod pathway, vernalization pathways arose after the divergence of major groups of flowering plants, as an adaptation to the new environments created by climate change and continental drift (Amasino, 2010). As vernalization pathways evolved, FT presented a prime regulation point for floral repression. It will be interesting to determine how vernalization pathways have been “constructed” in other groups of plants and how often these pathways target FT expression.
How plants sense and measure the prolonged cold of winter and transduce this into a vernalization response is not understood. An output of this cold-sensing system in Arabidopsis is the induction of VIN3 expression and increased expression of the COOLAIR RNA. However, at present, genetic variation (i.e. mutants or natural variation) in the cold-sensing system has not been identified, and there are not any biochemical clues to how this system operates. It will be quite interesting, as well as a challenge, to understand the molecular basis of how plants sense prolonged cold for both vernalization and the breaking of bud dormancy, and whether such a system is conserved or has independently evolved multiple times, as appears to be the case for downstream parts of the vernalization pathway in cereals and Arabidopsis.
References
- Allard HA. (1919) Gigantism in Nicotiana tabacum and its alternate inheritance. Am Nat 53: 218–233 [Google Scholar]
- Amasino R. (2004) Vernalization, competence, and the epigenetic memory of winter. Plant Cell 16: 2553–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amasino R. (2010) Seasonal and developmental timing of flowering. Plant J 61: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Baurle I, Smith L, Baulcombe DC, Dean C. (2007) Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science 318: 109–112 [DOI] [PubMed] [Google Scholar]
- Burn JE, Smyth DR, Peacock WJ, Dennis ES. (1993) Genes conferring late flowering in Arabidopsis thaliana. Genetica 90: 147–155 [Google Scholar]
- Clarke JH, Dean C. (1994) Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Mol Gen Genet 242: 81–89 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AM, Greb T, Dean C. (2008) A PHD-Polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci USA 105: 16831–16836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Coupland G. (2009) Plant phase transitions make a SPLash. Cell 138: 625–627 [DOI] [PubMed] [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C. (2003) Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol 132: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Kim DH, Doyle MR, Sung S, Amasino RM. (2009) Vernalization: winter and the timing of flowering in plants. Annu Rev Cell Dev Biol 25: 277–299 [DOI] [PubMed] [Google Scholar]
- Kim S, Choi K, Park C, Hwang HJ, Lee I. (2006) SUPPRESSOR OF FRIGIDA4, encoding a C2H2-type zinc finger protein, represses flowering by transcriptional activation of Arabidopsis FLOWERING LOCUS C. Plant Cell 18: 2985–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Michaels SD. (2006) SUPPRESSOR OF FRI 4 encodes a nuclear-localized protein that is required for delayed flowering in winter-annual Arabidopsis. Development 133: 4699–4707 [DOI] [PubMed] [Google Scholar]
- Knott JE. (1934) Effect of a localized photoperiod on spinach. Proc Am Soc Hortic Sci 31: 152–154 [Google Scholar]
- Lee I, Bleecker A, Amasino R. (1993) Analysis of naturally occurring late flowering in Arabidopsis-thaliana. Mol Gen Genet 237: 171–176 [DOI] [PubMed] [Google Scholar]
- McDaniel CN, Hsu HF. (1976) Position-dependent development of tobacco meristems. Nature 259: 564–566 [Google Scholar]
- Michaels S, Amasino R. (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD. (2009) Flowering time regulation produces much fruit. Curr Opin Plant Biol 12: 75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Bezerra IC, Amasino RM. (2004) FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc Natl Acad Sci USA 101: 3281–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, He Y, Scortecci KC, Amasino RM. (2003) Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci USA 100: 10102–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napp-Zinn K. (1979) On the genetical basis of vernalization requirement in Arabidopsis thaliana (L.) Heynh. Champagnat P, Jaques R, , La Physiologie de la Floraison. Collogues Internationaux Centre National de la Recherche Scientifique, Paris, pp 217–220 [Google Scholar]
- Poethig RS. (1990) Phase change and the regulation of shoot morphogenesis in plants. Science 250: 923–930 [DOI] [PubMed] [Google Scholar]
- Poethig RS. (2010) The past, present, and future of vegetative phase change. Plant Physiol 154: 541–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Hong L, Michaels S, Amasino RM. (2005) FRIGIDA-ESSENTIAL 1 interacts genetically with FRIGIDA and FRIGIDA-LIKE 1 to promote the winter-annual habit of Arabidopsis thaliana. Development 132: 5471–5478 [DOI] [PubMed] [Google Scholar]
- Serrano G, Herrera-Palau R, Romero JM, Serrano A, Coupland G, Valverde F. (2009) Chlamydomonas CONSTANS and the evolution of plant photoperiodic signaling. Curr Biol 19: 359–368 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES. (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM. (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164 [DOI] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C. (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. (2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Veley KM, Michaels SD. (2008) Functional redundancy and new roles for genes of the autonomous floral-promotion pathway. Plant Physiol 147: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA. (2006) The Arabidopsis thaliana vernalization response requires a Polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA 103: 14631–14636 [DOI] [PMC free article] [PubMed] [Google Scholar]