Plants have evolved outstanding capacities to adapt their development and physiology to changes in the environment. The availability, distribution, and activity of endogenous signals, plant hormones, underlie and coordinate these responses (Santner and Estelle, 2009). Auxin, one such signaling molecule, affects a multitude of developmental processes (Woodward and Bartel, 2005; Leyser, 2006; Vanneste and Friml, 2009). The differential distribution of auxin within tissues is essential for many adaptive responses, including embryo and leaf patterning, root and stem elongation, lateral root initiation, leaf expansion, tropisms, regenerative growth, and vascular tissue formation (Vanneste and Friml, 2009). The best characterized auxin signaling pathway operates in the nucleus (Mockaitis and Estelle, 2008). In this pathway, auxin is perceived by and promotes the interaction of TIR1/AFB and Aux/IAA coreceptor proteins (Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Tan et al., 2007), and downstream signaling is mediated by ubiquitination and proteosome-mediated degradation of Aux/IAA transcriptional repressors (Gray et al., 2001; dos Santos Maraschin et al., 2009), which themselves regulate the activity of ARF transcription factors (Tiwari et al., 2004; Guilfoyle and Hagen, 2007). Ultimately, this mechanism leads to complex transcriptional reprogramming in response to auxin. The large number of different Aux/IAA and ARF proteins means that different combinations can be expressed in different cell types, allowing the pathway to produce a multitude of different developmental and physiological responses to auxin (Mockaitis and Estelle, 2008; Vanneste and Friml, 2009). In addition to this nuclear signaling pathway, the observation of rapid cellular responses to auxin suggests the existence of another, presumably nontranscriptional, signaling pathway that might be related to the action of auxin-binding protein1 (ABP1). ABP1 was identified on the basis of its ability to bind to natural and synthetic auxins with high affinity, and has been found in various monocot and dicot species (Napier et al., 2002). However, despite decades of studies, its mode of action remains unclear (Badescu and Napier, 2006; Schenck et al., 2010). The majority of ABP1 is detected in the endoplasmic reticulum (ER), but its physiological effects, mainly relating to cell expansion and division, have been attributed to the smaller apoplastic pool of the protein (Napier et al., 2002; Perrot-Rechenmann, 2010). In theory, auxin could be perceived at several different subcellular destinations, including the nucleus, ER, and extracellular compartment. The final output of auxin signaling is therefore likely to be influenced not only by the overall cellular auxin content, but also by the relative distribution of auxin among different subcellular compartments. Thus subcellular auxin distribution may represent an important regulatory level in the complex action of auxin.
AUXIN TRANSPORT SYSTEMS AT THE PLASMA MEMBRANE
Conceptually, cellular levels of auxins, such as naturally occurring indole-3-acetic acid (IAA), are determined by metabolic processes and transport across the plasma membrane (PM). IAA is produced by several biosynthetic pathways and through the regulated release of IAA from various storage forms, including amide- or ester-linked amino acid conjugates, peptides, and sugars as well as from indole-3-butyric acid, which serves as an important precursor to IAA (Woodward and Bartel, 2005). Conversely, excess IAA can be converted to conjugated forms for storage and degradation (Normanly, 2010). The transport of IAA across the PM depends on specialized influx and efflux carriers, making it a highly regulated process (Robert and Friml, 2009). IAA transporters include amino acid permease-like AUXIN RESISTANT1/LIKE AUX (AUX1/LAX) proteins, which mediate auxin influx (Bennett et al., 1996; Yang et al., 2006), the PIN-FORMED (PIN) efflux carriers (Luschnig et al., 1998; Petrášek et al., 2006), and the MULTIDRUG RESISTANCE/P-GLYCOPROTEIN ABCB class of ATP-binding cassette auxin transporters (Noh et al., 2001; Geisler et al., 2005; Bandyopadhyay et al., 2007). The fact that indole-3-butyric acid and presumably other auxin metabolites can also be selectively transported across the PM in a polar manner adds another level of complexity to the regulation of cellular auxin levels (Strader and Bartel, 2009; Łangowski et al., 2010; Růžička et al., 2010). In principle, all of the PM-based auxin transport systems can regulate the quantity of auxin in different cell types. Distinctively, the polar localization of PIN auxin efflux carrier proteins to one side of the cell confers directionality to the flow of auxin between cells (Friml et al., 2004; Wiśniewska et al., 2006). The Arabidopsis (Arabidopsis thaliana) genome encodes eight PIN-related sequences, five of which have been characterized in terms of their functions in auxin efflux at the PM and their roles in regulating different aspects of plant development (Křeček et al., 2009). These results demonstrate that the polar localization patterns of PIN proteins in different cells and developmental contexts mediate directional auxin fluxes during a wide variety of developmental processes, including embryogenesis, organogenesis, vascular differentiation, and regeneration and root meristem patterning (Petrášek and Friml, 2009). Many studies dealing with auxin transport-mediated development therefore focus on the mechanisms and regulation of polar PIN localization at the PM (Kleine-Vehn and Friml, 2008).
PIN-DEPENDENT AUXIN TRANSPORT AT THE ER
A recent report shows that Arabidopsis PIN5, an atypical member of the PIN protein family, is not present at the PM, but at the ER (Mravec et al., 2009). When heterologously produced in the yeast Saccharomyces cerevisiae, however, PIN5 localizes to a significant extent at the PM. This useful artifact made it possible to directly assess the auxin transport capacity of PIN5 with radioactively labeled auxin. Indeed, PIN5 appears to be a functional auxin transporter, which, when present at the PM (in yeast), mediates auxin export from the cell. This transport orientation implies that the endogenous function of PIN5 is to transport auxin from the cytosol into the lumen of the ER. In planta auxin accumulation experiments revealed that plants carrying the pin5 loss-of-function allele accumulated auxin at a higher concentration than that of the wild type, whereas gain-of-function alleles reduced the capacity of cells to export auxin through the PM. These results are consistent with the assumption that PIN5 mediates auxin flow into the lumen of the ER, thus limiting the availability of cytosolic auxin for PM-based transport (Mravec et al., 2009). The challenge for future studies will be in demonstrating the transport function at the ER more directly, preferentially at isolated ER membranes.
The Arabidopsis axr4 mutation leads to retention of the auxin influx carrier AUX1 at the ER membrane, thus blocking its activity (Dharmasiri et al., 2006). It is therefore interesting to speculate that the activity of multiple auxin transporters might be controlled in a similar manner, by a mechanism that regulates whether they are retained at the ER or secreted to the PM. Although it has been demonstrated that the controlled movement of PIN proteins between the PM and endosomal compartments is an important regulatory process in many adaptive growth responses (Kleine-Vehn and Friml, 2008), the intriguing possibility that PIN proteins or other auxin transporters might shuttle between the ER and PM in response to endogenous or external signals awaits demonstration.
ER: MORE THAN JUST THE TRASH BIN FOR AUXIN?
The observation that auxin is transported across the ER membrane was unexpected because no previous models of auxin transport or auxin action had proposed regulation at this level. So the key question remains, what is the function of PIN proteins at the ER? PIN5 gain-of-function plants show strong, in some cases lethal, auxin-related phenotypes, indicating that PIN5 action is relevant for regulating plant physiology and development (Mravec et al., 2009). pin5 loss-of-function mutants show relatively weak phenotypes in different auxin-related processes, but these observations can be explained by functional redundancy with other ER-localized auxin transporters (Mravec et al., 2009). Furthermore, multiple lines of evidence suggest that PIN5 function at the ER membrane is required for the regulation of intracellular auxin homeostasis and metabolism. PIN5 gain-of-function plants show reduced levels of free IAA, whereas IAA levels are increased in pin5 loss-of-function plants. Accordingly, loss of PIN5 function enhances the phenotypes of the auxin-overexpressing yucca allele (Mravec et al., 2009). Cultured tobacco (Nicotiana tabacum) cells in which PIN5 is expressed inducibly show dramatic changes in their IAA metabolic profile after induction: free IAA is quickly metabolized, IAA-sugar conjugates decrease, and the capacity to conjugate IAA to amino acids, such as Asp or Glu, increases. These relatively rapid and major changes in the metabolic fate of IAA in response to manipulations of the PIN5 activity at the ER membrane suggest that the auxin metabolic pathways are compartmentalized to the ER. This notion is supported by the demonstrated presence of several auxin metabolic enzymes and regulatory proteins in the ER (Bartel and Fink, 1995; Woodward and Bartel, 2005). As the subcellular localization of most other IAA metabolic enzymes is currently unknown, it is possible that more will also be found to reside selectively in the ER. According to this scenario, free IAA transported into the ER is inactivated by different metabolic processes.
It seems, however, that the ER might be more than a “trash bin” for free auxin and serve rather as a recycling station. The predicted ER localization of the IAA-amino acid hydrolases and the slight contribution of these enzymes to increasing IAA levels (Rampey et al., 2004) indicate that the ER can in some instances function also as a source of IAA. Regardless of what the outcome of experiments clarifying this issue will be, it seems clear that the subcellular transport-mediated auxin compartmentalization into ER provides a so-far unappreciated level for regulating auxin homeostasis.
Beyond metabolism, auxin in the ER may also participate more directly in signaling. It is well known that the majority of the ABP1 in the cell is retained at the ER as demonstrated by both localization studies and the ER retention signal in the ABP1 sequence (Tian et al., 1995). It remains unclear what the role of ABP1 in the ER would be. It might merely bind and aid auxin retention at the ER or perform a receptor and signaling function there. It has been regularly noted that ABP1 can only bind auxin poorly in the ER because of the unfavorable pH in the ER lumen (Tian et al., 1995). However, the data on pH in the ER are not conclusive and the optimum pH at which auxin binds to ABP1 might be strongly influenced by as yet unidentified accessory proteins. Indeed, some observations link ABP1 function to the regulation of auxin metabolism (Chen et al., 2006), which, in the light of the PIN5 results, supports the hypothesis that ABP1 might play an active role in the ER. Also unclear at the moment is the relationship between auxin transport into the ER and auxin availability for nuclear-located TIR1-based transcriptional auxin signaling. These and other interesting questions remain open for future investigation.
EVOLUTIONARY ASPECTS OF AUXIN TRANSPORT AT THE ER
PIN5 belongs to an otherwise functionally uncharacterized subclade of PIN proteins that is characterized by a hydrophilic domain that is much shorter than that in all “classical” PM-localized PIN proteins. In Arabidopsis, this group also encompasses PIN6 and PIN8, in which the hydrophilic loop is reduced or completely lacking, respectively (Křeček et al., 2009). Expression of these proteins in cultured tobacco cells suggests that they are ER localized similarly to PIN5 (Mravec et al., 2009). Based on the sequence and localization data obtained by studying Arabidopsis PIN proteins, all of the available PIN sequences from other plant species can be divided into two major subclasses: the ER-localized “short” and the PM-localized “long” PIN proteins that contain a large central hydrophilic loop. An obvious question is which of these localizations and, thus, functions is the ancestral one.
The exact origin of PIN proteins in evolutionary history is unclear; PIN-like sequences have been found in the genomes of land plants, but not in those of more primitive species, suggesting that they arose somewhere during the adaptation to life on land (Paponov et al., 2005; Křeček et al., 2009). This is after auxin signaling itself is predicted to have appeared (Cooke et al., 2002; Lau et al., 2009). The most ancient PIN sequences identified so far come from the moss Physcomitrella patens and the club moss Selaginella moellendorffii. These PIN proteins do not cluster clearly with any of the PIN subclasses of the seed plants, although analysis of Physcomitrella PIN proteins revealed that they are predominantly localized at the ER (Mravec et al., 2009). The ER localization of the moss PIN proteins suggests that mediating auxin homeostasis at the ER is the ancestral function of PIN proteins, and that diversification into a role in directional cell-to-cell auxin transport at the PM occurred at a later stage. Interestingly, ABP1 homologs have been found in the genomes of green algae, including Chlamydomonas reinhardtii, Ostreococcus tauri, and Ostreococcus lucimarinus, but homologs of TIR1/AFBs, Aux/IAAs, and ARFs have not (Lau et al., 2009). This adds further circumstantial evidence for a potentially non-nuclear ancestral auxin signaling pathway.
From an evolutionary perspective of auxin metabolism and action, it makes sense that the ancestral function of PIN proteins should be in the regulation of auxin homeostasis rather than transport between cells. The presence of IAA, its function as signaling molecule, and the capacity for IAA metabolism have all been detected in various algal species (Cooke et al., 2002; Lau et al., 2009). In contrast, transport-mediated asymmetric auxin distributions are typically required to pattern and coordinate the differentiation of multicellular organisms and are particularly important during developmental programs involved in adaptations specific to life on land, such as tropisms (Petrášek and Friml, 2009), and would therefore be expected to have evolved later. Notably, polar auxin transport activity has been detected in moss sporophytes (Fujita et al., 2008). If the observation that all Physcomitrella PIN proteins localize to the ER is confirmed, it will be interesting to uncover the mechanism underlying polar auxin transport in moss sporophytes. Another exciting issue concerns the identification of the point in the evolution of land plants at which the PIN proteins acquired their new function at the PM and the developmental innovations connected to this event. Moreover, were the various polar localizations exhibited by different PIN proteins in diverse cell types acquired from the very beginning or did they evolve gradually? Which preexisting cellular mechanism was adapted to regulate PIN localization? In general, given the lack of direct data from algae or more ancestral land plants, the evolutionary interpretations of the PIN action are still largely speculative and more surprising findings can be expected in the coming years.
ER: ARE THERE ANY OTHERS COMING TO THE PARTY?
The discovery of ancestral auxin transport at the ER membrane has identified a previously unappreciated mechanism for regulating auxin homeostasis and, presumably, also signaling through compartmentalization. In the case of auxin, several important compartments have been characterized. Metabolism is likely to be divided between the cytosol, ER and peroxisomes, ER-derived organelles, as indicated by the localization of enzymes, and corresponding genetic studies (Woodward and Bartel, 2005; Mravec et al., 2009). The TIR1-mediated signaling pathway for regulation of gene expression is located in the nucleus (Mockaitis and Estelle, 2008), while the ABP1-mediated signaling presumably operates in the ER and at the outer side of the PM (Tian et al., 1995). For other hormonal signaling pathways, the location of the hormone receptors are at least roughly known: All ubiquitination-based pathways including those for auxin, gibberellin, and jasmonate, are perceived in the nucleus; cytokinins, abscisic acid, and brassinosteroids are detected by PM receptors, but the possibility that some of these proteins also reside at the ER cannot be excluded, and another receptor for abscisic acid is found in the cytosol (Santner and Estelle, 2009). Interesting for our discussion is the presence of all five ethylene receptors at the ER membrane where they directly interact with EIN2, the central downstream regulator of ethylene signaling (Grefen et al., 2008; Bisson et al., 2009). It remains unclear why ethylene signaling is located specifically at the ER. It is possible that, because ethylene is a gaseous and, therefore, easily diffusible hormone, it does not matter where the perception occurs. Nevertheless, it is intriguing to speculate that the subcellular compartmentalization of both the ethylene and auxin signaling to the ER might play a role in their cross talk. Hormone cross talk often so decisively contributes to the final physiological and developmental output of hormone action (Benková and Hejátko, 2009) that understanding how the compartmentalization of hormone signaling pathways and hormones themselves contributes to their interactions will be an important focus for future work.
CONCLUSION
The recent identification of PIN-dependent auxin transport from the cytoplasm into the lumen of the ER revealed an unsuspected mode of regulation of auxin homeostasis through the subcellular compartmentalization of both auxin itself and the pathways that mediate its metabolism (Fig. 1). This observation puts forward a whole array of questions and new directions for research related, for example, to the compartmentalization of auxin metabolic enzymes; the regulation of ER-based transport; evolutionary aspects of ER versus PM auxin transport; connection of auxin in the lumen of the ER to the presumptive ER-localized auxin receptor ABP1; and the relevance of compartmentalization for signaling cross talk with other hormonal pathways, such as ER-localized ethylene perception. Whatever the answers to these questions, it is clear that compartmentalization of metabolism and signaling in conjunction with the ability to transport the signaling molecules between different cellular compartments has a largely underestimated potential to integrate different signals and provide an additional level of regulation that will influence the net output of the signaling pathways.
Figure 1.
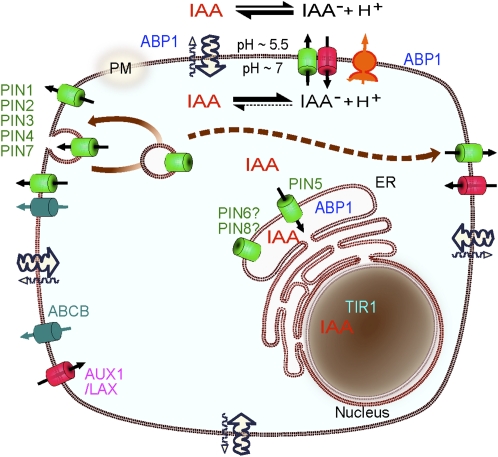
Schematic diagram of auxin fluxes and potential sites of auxin perception in the plant cell. The pH at the outer side of the PM is maintained at approximately 5.5 by the H+-ATPase activity of the PM (orange). As a consequence, a proportion of auxin (IAA) molecules in the apoplast remain protonated and can enter the cell via diffusion (wavy arrows). Auxin also enters cells through the action of specific uptake carriers of the AUX/LAX family (red). In the relatively higher pH of the cytoplasm, auxin molecules are deprotonated. The resulting ions cannot pass across the PM by diffusion and, instead, must be transported by efflux carriers including the PIN auxin transporters (green) and the MULTIDRUG RESISTANCE/P-GLYCOPROTEIN ABCB transporters (light blue). The coordinated polar localization of the auxin-efflux carriers from the long PIN subfamily at the PM determines the directionality of the auxin flow within the tissue. The long PIN proteins undergo constitutive endocytic recycling, which allows dynamic changes of PIN polarity by a transcytotic mechanism. PIN5, a member of the short PIN subfamily, is found at the membranes of the ER and leads to compartmentalization of auxin into the lumen of the ER. A number of auxin metabolic enzymes are also found in the lumen of the ER, and auxin metabolic profiling suggests that auxin entering the ER through PIN5-mediated transport rapidly undergoes metabolic conversion. The presumptive auxin receptor ABP1 (blue) is present both in the ER and in the apoplast. The receptor for the transcriptional auxin response pathway, TIR1 (light blue), is found in the nucleus. The different localizations of the long and short subfamilies of PIN efflux carriers, together with the spatial separation of the auxin receptors and localization of metabolic enzymes, implies that auxin signaling and metabolism, as well as auxin molecules themselves, are compartmentalized within the plant cell. Adapted from Křeček et al. (2009).
Acknowledgments
We thank Petr Skůpa for help with the graphics.
References
- Badescu GO, Napier RM. (2006) Receptors for auxin: will it all end in TIRs? Trends Plant Sci 11: 217–223 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, Blakeslee JJ, Lee OR, Mravec J, Sauer M, Titapiwatanakun B, Makam SN, Bouchard R, Geisler M, Martinoia E, et al. (2007) Interactions of PIN and PGP auxin transport mechanisms. Biochem Soc Trans 35: 137–141 [DOI] [PubMed] [Google Scholar]
- Bartel B, Fink GR. (1995) ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268: 1745–1748 [DOI] [PubMed] [Google Scholar]
- Benková E, Hejátko J. (2009) Hormone interactions at the root apical meristem. Plant Mol Biol 69: 383–396 [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950 [DOI] [PubMed] [Google Scholar]
- Bisson MM, Bleckmann A, Allekotte S, Groth G. (2009) EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem J 424: 1–6 [DOI] [PubMed] [Google Scholar]
- Chen JG, Wang S, Lazarus CM, Napier RM, Jones AM. (2006) Altered expression of auxin-binding protein 1 affects cell expansion and auxin pool size in tobacco cells. J Plant Growth Regul 25: 69–78 [Google Scholar]
- Cooke TJ, Poli D, Sztein AE, Cohen JD. (2002) Evolutionary patterns in auxin action. Plant Mol Biol 49: 319–338 [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri S, Swarup R, Mockaitis K, Dharmasiri N, Singh SK, Kowalchyk M, Marchant A, Mills S, Sandberg G, Bennett MJ, et al. (2006) AXR4 is required for localization of the auxin influx facilitator AUX1. Science 312: 1218–1220 [DOI] [PubMed] [Google Scholar]
- dos Santos Maraschin F, Memelink J, Offringa R. (2009) Auxin-induced SCFTIR1-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J 59: 100–109 [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PBF, Ljung K, Sandberg G, et al. (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Fujita T, Sakaguchi H, Hiwatashi Y, Wagstaff SJ, Ito M, Deguchi H, Sato T, Hasebe M. (2008) Convergent evolution of shoots in land plants: lack of auxin polar transport in moss shoots. Evol Dev 10: 176–186 [DOI] [PubMed] [Google Scholar]
- Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, Titapiwatanakun B, Peer WA, Bailly A, Richards EL, et al. (2005) Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J 44: 179–194 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. (2001) Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Grefen C, Städele K, Růžička K, Obrdlik P, Harter K, Horák J. (2008) Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Mol Plant 1: 308–320 [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. (2007) Auxin response factors. Curr Opin Plant Biol 10: 453–460 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Friml J. (2008) Polar targeting and endocytic recycling in auxin-dependent plant development. Annu Rev Cell Dev Biol 24: 447–473 [DOI] [PubMed] [Google Scholar]
- Křeček P, Skůpa P, Libus J, Naramoto S, Tejos R, Friml J, Zažímalová E. (2009) The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol 10: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łangowski Ł, Růžička K, Naramoto S, Kleine-Vehn J, Friml J. (2010) Trafficking to the outer polar domain defines the root-soil interface. Curr Biol 20: 904–908 [DOI] [PubMed] [Google Scholar]
- Lau S, Shao N, Bock R, Jürgens G, De Smet I. (2009) Auxin signaling in algal lineages: fact or myth? Trends Plant Sci 14: 182–188 [DOI] [PubMed] [Google Scholar]
- Leyser O. (2006) Dynamic integration of auxin transport and signalling. Curr Biol 16: R424–R433 [DOI] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR. (1998) EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev 12: 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M. (2008) Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24: 55–80 [DOI] [PubMed] [Google Scholar]
- Mravec J, Skůpa P, Bailly A, Hoyerová K, Křeček P, Bielach A, Petrášek J, Zhang J, Gaykova V, Stierhof YD, et al. (2009) Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1140 [DOI] [PubMed] [Google Scholar]
- Napier RM, David KM, Perrot-Rechenmann C. (2002) A short history of auxin-binding proteins. Plant Mol Biol 49: 339–348 [PubMed] [Google Scholar]
- Noh B, Murphy AS, Spalding EP. (2001) Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J. (2010) Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb Perspect Biol 2: a001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov IA, Teale WD, Trebar M, Blilou I, Palme K. (2005) The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci 10: 170–177 [DOI] [PubMed] [Google Scholar]
- Perrot-Rechenmann C. (2010) Cellular responses to auxin: division versus expansion. Cold Spring Harb Perspect Biol 2: a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrášek J, Friml J. (2009) Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Petrášek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertová D, Wiśniewska J, Tadele Z, Kubeš M, Čovanová M, et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Rampey RA, LeClere S, Kowalczyck M, Ljung K, Sandberg G, Bartel B. (2004) A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol 135: 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert HS, Friml J. (2009) Auxin and other signals on the move in plants. Nat Chem Biol 5: 325–332 [DOI] [PubMed] [Google Scholar]
- Růžička K, Strader LC, Bailly A, Yang H, Blakeslee J, Łangowski Ł, Nejedlá E, Fujita H, Itoh H, Syōno K, et al. (2010) Arabidopsis PIS1 encodes the ABCG37 transporter of the auxinic compounds including the auxin precursor indole-3-butyric acid. Proc Natl Acad Sci USA 107: 10749–10753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Estelle M. (2009) Recent advances and emerging trends in plant hormone signalling. Nature 459: 1071–1078 [DOI] [PubMed] [Google Scholar]
- Schenck D, Christian M, Jones A, Lüthen H. (2010) Rapid auxin-induced cell expansion and gene expression: a four-decade-old question revisited. Plant Physiol 152: 1183–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L, Bartel B. (2009) The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell 21: 1992–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobas LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Tian H, Klämbt D, Jones AM. (1995) Auxin-binding protein 1 does not bind auxin within the endoplasmic reticulum despite this being the predominant subcellular location for this hormone receptor. J Biol Chem 270: 26962–26969 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J. (2009) Auxin: a trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Wiśniewska J, Xu J, Seifertová D, Brewer PB, Růžička K, Blilou I, Rouquié D, Benková E, Scheres B, Friml J. (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E. (2006) High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol 16: 1123–1127 [DOI] [PubMed] [Google Scholar]