Abstract
Given the substantial changes in mitochondrial gene expression, the mitochondrial proteome, and respiratory function during rice (Oryza sativa) germination under anaerobic and aerobic conditions, we have attempted to identify changes in mitochondrial membrane transport capacity during these processes. We have assembled a preliminary rice mitochondrial carrier gene family of 50 members, defined its orthology to carriers of known function, and observed significant changes in microarray expression data for these rice genes during germination under aerobic and anaerobic conditions and across rice development. To determine if these transcript changes reflect alteration of the carrier profile itself and to determine which members of the family encode the major mitochondrial carrier proteins, we analyzed mitochondrial integral membrane protein preparations using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and peptide mass spectrometry, identifying seven distinct carrier proteins. We have used mass spectrometry-based quantitative approaches to compare the abundance of these carriers between mitochondria from dry seeds and those from aerobic- or anaerobic-germinated seeds. We highlight an anaerobic-enhanced basic amino acid carrier and show concomitant increases in mitochondrial arginase and the abundance of arginine and ornithine in anaerobic-germinated seeds, consistent with an anaerobic role of this mitochondria carrier. The potential role of this carrier in facilitating mitochondrial involvement in arginine metabolism and the plant urea cycle during the growth of rice coleoptiles and early seed nitrate assimilation under anaerobic conditions are discussed.
The number and activity of mitochondria vary between cells, tissues, and species and can change depending on the metabolic and/or environmental conditions that the cell or organism is experiencing. In mammals, changes in mitochondrial numbers and activity have been extensively studied during muscle development and remodeling (Moyes and Hood, 2003), and comparative analyses of mitochondria from different mammalian organs indicate that only half of the organelle proteome sets were shared between organs such as brain, heart, liver, and muscle (Mootha et al., 2003). A central factor for specialization in mammalian mitochondria is the substrates most commonly consumed by organelles in different tissues, ranging from end products of glycolysis to intermediates of β-oxidation of fatty acids and the urea cycle. In plants, differences in mitochondrial numbers, biochemical activities, and proteomes are readily observed between mitochondria isolated from leaves, roots, seeds, and cell cultures (Humphrey-Smith et al., 1992; Douce et al., 2001; Bardel et al., 2002; Lee et al., 2008). Mitochondrial number and respiratory capacity also vary markedly in plants exposed to elevated CO2 levels (Robertson et al., 1995; Gonzalez-Meler et al., 1996; Davey et al., 2004; Griffin et al., 2004), low oxygen (Gupta et al., 2009), or oxidative stress (Tiwari et al., 2002), indicating an organelle biogenesis response to environmental conditions.
Plants offer an attractive system to study mitochondrial biogenesis, as the seed stage of the life cycle is a natural and extreme quiescent state and induction to an active state occurs upon water uptake, a process termed imbibition (Bewley, 1997). Microscopic analyses of dormant seeds indicate that mitochondria are present but lack a typical cristae structure. Furthermore, the matrix appears less electron dense compared with mature mitochondria, indicating a lower protein content (Vartapetian et al., 2003). Studies examining embryo mitochondria during maize (Zea mays) and rice (Oryza sativa) germination provide evidence for a maturation process and the presence of mature and immature mitochondrial structures (Logan et al., 2001; Howell et al., 2006). Another feature of mitochondrial biogenesis is the role of oxygen as a regulator of nuclear gene expression for mitochondrial components. This phenomenon has been investigated in both yeast (Hon et al., 2003) and rice (Howell et al., 2007), as both organisms can survive extensive periods of growth without oxygen, allowing mitochondria to be isolated from long-term anoxic tissues. We have also noted that the oxygen dependence for biogenesis is not consistent for all pathways involved in biogenesis. For example, while import of many proteins via the general import pathway, which is dependent on the mitochondrial processing peptidase incorporated in the cytochrome bc1 complex, is decreased under anaerobic conditions, import of metabolite transporters via the mitochondrial carrier (MC) import pathway shows no significant alteration in rate between aerobic and anaerobic conditions (Howell et al., 2007).
The transport of metabolites and other solutes across the mitochondrial inner membrane is critical for the provision of substrates for oxidation, such as organic acids, the transport of amino acids, and the release of products such as ATP to the cytosol. These transport steps are catalyzed by a series of specific carriers that operate as exchangers/cotransporters. A superfamily of related MCs has been described in eukaryotes that contain a tripartite structure of 100-amino acid segments each consisting of two membrane-spanning α-helices separated by an extramembrane hydrophilic loop (Walker and Runswick, 1993). For many years, these carriers were thought to operate as homodimers with a 12-transmembrane domain structure (Saraste and Walker, 1982); however, the yeast ADP/ATP carrier 2 has been shown to act as a monomer with a six-transmembrane domain structure (Bamber et al., 2007), and a recent review has proposed that most if not all carriers in this class are functional monomers (Kunji and Crichton, 2010). They are responsible for the transport of a wide variety of metabolites between mitochondria and the cytosol. Studies have considered the structure, function, and import of MCs into mammalian and yeast mitochondria (Palmieri et al., 1996). There are 35 genes for MCs in the yeast genome, and approximately two-thirds of the members of this family have been assigned a function, to date, based on biochemical evidence or genetic inference (Belenkiy et al., 2000; Palmieri et al., 2001, 2006a; Prohl et al., 2001; Arco and Satrustegui, 2005). Expression profiling has revealed that many yeast MCs of unknown function are differentially expressed in response to growth conditions (Belenkiy et al., 2000).
Biochemical characterization of plant mitochondrial membrane transport over the last 30 years has revealed the operation of carriers for phosphate, adenine nucleotides, monocarboxylates, dicarboxylates, and tricarboxylates, amino acids, and cofactors such as NAD+ and CoA (Wiskich, 1977; Day and Wiskich, 1984; Douce et al., 1997). Analysis of the Arabidopsis (Arabidopsis thaliana) genome reveals as many as 58 MC genes based on homology with yeast and animal counterparts (Millar and Heazlewood, 2003; Picault et al., 2004). A combination of overexpression and studies of liposome transport, proteomics of mitochondrial membranes, and investigations of genetically modified plants has identified specific MCs on the inner mitochondrial membrane: adenine nucleotide transporters (ANT), succinate-fumarate carrier, phosphate carriers (PiC), a dicarboxylate/tricarboxylate transporter (DTC), uncoupling proteins (UCP), a dicarboxylate carrier, basic amino acid carriers (BAC), S-adenosyl Met carrier, and NAD+ transporter (Haferkamp, 2007; Palmieri et al., 2008, 2009). While transcript studies in plants indicate that the MCs are dynamically regulated at the transcriptional level (Zimmermann et al., 2004), there is virtually no information on relative changes in the abundance of carrier proteins of plant mitochondria, apart from the changes observed in the abundance of UCPs during stress (Douette and Sluse, 2006; Sluse et al., 2006). Here, we analyze the rice MC genes, the carriers located in mitochondria, and the changes in the carrier profile during rice germination under aerobic and anaerobic conditions.
RESULTS
Defining the Rice MC Family
The MC family protein sequences typically have a predicted alkaline pI, a molecular mass between 30 and 40 kD, a higher than average grand average of hydropathicity score, six transmembrane domains, and multiple copies of a functional domain currently defined in Prosite as SOLCAR (for solute carrier; Palmieri, 1994; Palmieri et al., 1996; Millar and Heazlewood, 2003). To define the genes of this family of proteins in the rice predicted proteome, we conducted a BLASTp analysis using each of the 58 MC proteins defined in Arabidopsis, yielding a set of 194 rice proteins identified as putative MC homologs. Of these 194 proteins, 29 had significant homology to all 58 Arabidopsis MC protein (AtMC) sequences. In addition, eight were homology hits to 57 AtMCs, seven to 56 AtMCs, eight to 55 AtMCs, two to 53 AtMCs, and four to 52 AtMCs. Beyond this, only one rice protein was found that hit 49, 42, and 30 AtMC proteins. Collectively, these 61 proteins constituted the rice proteins with a significant number of homology hits to suggest high degrees of similarity to the Arabidopsis MC superfamily. Based on analysis of the predicted number of transmembrane domains and the presence of the Prosite SOLCAR motif, a further 11 proteins were excluded from this set, yielding a current set of 50 putative OsMC proteins. Using the gene locus numbers for rice (Osxxgxxxxx), which place each gene in its genomic context, it is clear that the OsMCs are relatively randomly distributed across the first 11 chromosomes of the rice genome. Most OsMC members are physically separated by hundreds of genes, indicating that there is little evidence of physical clusters caused by recent duplication of genes, which is often observed in other large gene families in plants (Sappl et al., 2004). This approach could miss some rice carriers that lack the SOLCAR motifs or substantial homology with Arabidopsis MCs; hence, it is a preliminary carrier set that could be added to based on further analysis.
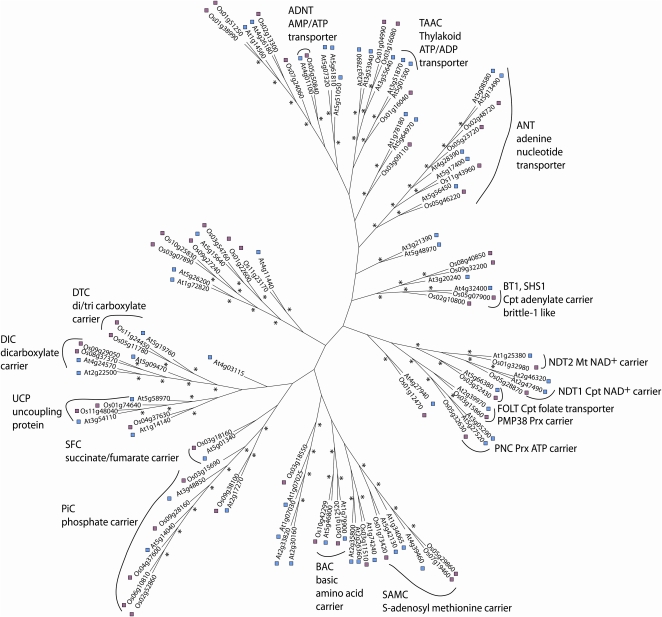
ClustalX was used to generate a bootstrapped (10,000 reiterations) simple unrooted phylogenetic tree of the OsMC and AtMC sets, based on sequence alignments using the neighbor-joining algorithm. Bootstrap values greater than 70% at branch points were considered as a cutoff for a significant relationship. A series of subfamilies were revealed that provide putative annotation for OsMCs based on clustering with AtMCs of known function (Fig. 1). These include DTC, dicarboxylate carrier, UCP, succinate/fumarate carrier, PiC, BAC, ANT, AMP/ATP carrier, and the S-adenosyl Met carrier. A series of AtMCs have been shown not to be targeted to mitochondria but to reside in the chloroplast or the peroxisome membranes. Clear OsMC homologs of these proteins are evident, for example, the chloroplast folate carrier (Os03g52430), thylakoid ATP/ADP transporter (Os01g16040), and adenylate carriers related to brittle-1 like carriers (Os08g40850, Os09g32200, Os05g07900, Os02g10800) and, in addition, the peroxisomal ATP carrier (Os05g32630) and PMP38 carrier (Os03g15860).
Figure 1.
Phylogenetic unrooted tree of rice and the Arabidopsis MC family proteins. In total, 58 Arabidopsis (Atxgxxxxx) and 50 rice (Osxxgxxxxx) protein sequences were used in the analysis. Asterisks indicate greater than 70% branch confidence (10,000 reiterations of bootstrapping). Naming of carrier function is based on the Arabidopsis carriers in each case. Cpt, Chloroplastic; Mt, mitochondrial; Prx, peroxisomal.
Expression Profile of the Rice MCs
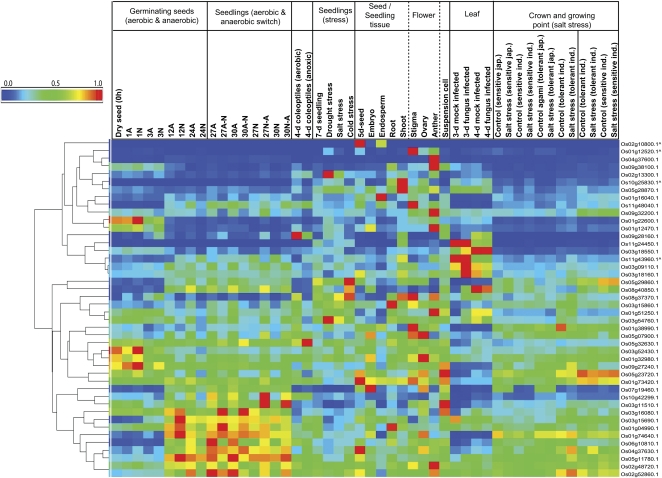
Rice Affymetrix microarrays contain probe sets for 44 of the set of 50 OsMCs. Using data we have already obtained from previous rice transcriptome analyses (Howell et al., 2009; Narsai et al., 2009) in combination with publicly available data, clustering of transcript data for this set of 44 was performed, revealing a wide variety of expression patterns across germination, during plant development, and in response to biotic and abiotic stresses (Fig. 2). Several shoot- and flower-specific carriers were identified, and specific members of the ATP/ADP transport family, the succinate/fumarate carrier, and a separate DTC were selectively expressed in leaves and had enhanced expression during fungal infection. Several were highly expressed in dry seeds or during the first 1 h post imbibition, which included the rice homolog of the chloroplast folate carrier (Os03g52430) and two carriers of unknown function (Os01g22600, Os09g27240). In contrast, some carriers including an unknown carrier (Os10g25830) and an ADP/ATP carrier protein (Os11g43960) displayed highly specific expression patterns (i.e. were expressed in seedlings and under certain biotic and abiotic stresses but were not expressed [absent] in dry seeds, during early germination, and in at least four other tissues/stresses). The majority of the carriers with substantial expression during germination were maximally expressed at 12 to 24 h post imbibition; some were more expressed under anaerobic conditions (e.g. Os03g16080), some others under aerobic conditions (e.g. Os08g40850), and some were relatively unaffected by oxygen availability (Fig. 2). Many of these germination-expressed genes are homologs of the AtMCs known to be mitochondria-localized carriers for phosphate, ATP/ADP, and carboxylic acids.
Figure 2.
Hierarchical clustering of transcripts for genes encoding MC proteins from rice. The Affymetrix rice genome microarray data were derived from the Gene Expression Omnibus within the National Center of Biotechnology Information database. The normalized data were hierarchically clustered using average linkage based on Euclidean distance. Genes annotated with carets indicate that these genes were absent in dry seeds, during early germination, and in more than four other tissues/stresses. A, Air; N, nitrogen.
Analysis of Rice Mitochondrial Respiration during Germination
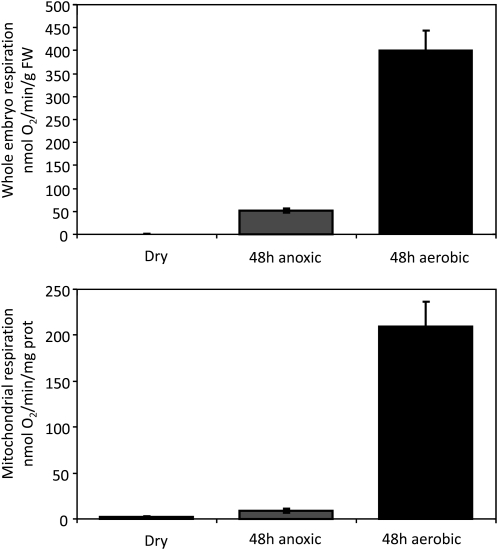
To investigate the respiratory competence of rice mitochondria in more detail during germination and their requirement for carrier proteins, Figure 3 shows functional studies of rice mitochondria during the germination process. Embryos excised from dry rice seeds respire at an extremely slow rate, as do mitochondria isolated from this tissue (Fig. 3). However, after 48 h post imbibition of the intact seed under aerobic conditions, the respiratory rate of the excised embryos increased several hundredfold and the specific respiratory rate of the mitochondria isolated from these embryos also increased markedly to rates on a protein basis that resemble those of mitochondria isolated from many plant tissues (Fig. 3). In contrast, if seeds were imbibed for the same period of time under anaerobic conditions, the respiratory capacity of the tissue, and of the mitochondria isolated from it, was only 3% to 10% of aerobically grown samples.
Figure 3.
Respiratory function of mitochondria during rice embryo germination. Whole embryo and mitochondrial respiration rates were obtained from dry seeds, 48-h-imbibed anaerobic-treated seeds, and 48-h-imbibed aerobic-treated seeds. Data presented are averages ± se (n = 3). FW, Fresh weight.
Enrichment of Rice MCs
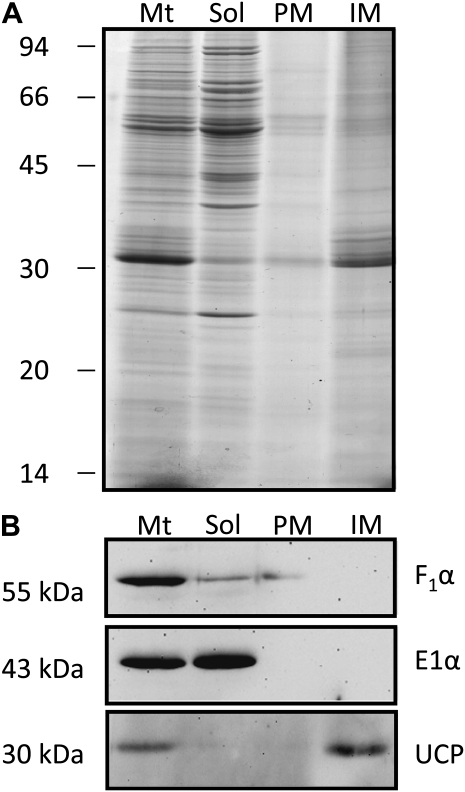
To produce protein fractions from rice embryos enriched in the mitochondrial membrane transporters, we isolated membrane proteins from mitochondrial preparations and extracted the integral membrane fraction by carbonate stripping of the isolated membranes. The first two lanes of Figure 4A show the protein profiles of the whole mitochondrial samples and the soluble proteins. A major difference is the loss of the approximately 30-kD band in the soluble protein lane, which represents hydrophobic membrane proteins in the mitochondrial membranes. Carbonate treatment of the membrane fraction led to two fractions, a peripheral membrane fraction containing proteins stripped from the surface of the membranes and an integral membrane fraction containing proteins embedded in the membrane. This integral membrane fraction is enriched for approximately 30-kD proteins, which is the size of the majority of the predicted plant MCs (Millar and Heazlewood, 2003). Antibodies to a peripheral membrane protein, the F1α-subunit of the ATP synthase, a soluble marker protein, the E1 α-subunit of pyruvate dehydrogenase, and an integral membrane protein, the UCP, confirmed that the fractionation of rice mitochondria had been successful (Fig. 4B).
Figure 4.
Isolation of integral membrane proteins from rice mitochondria. A, SDS-PAGE analysis of whole mitochondria (Mt), carbonated extracted soluble fraction (Sol), peripheral membrane proteins (PM), and integral membrane proteins (IM) stained with colloidal Coomassie Brilliant Blue G250. B, Western blot of a matrix protein (E1α), peripheral membrane protein (F1α), and integral membrane protein (UCP). Numbers on the left indicate apparent molecular mass.
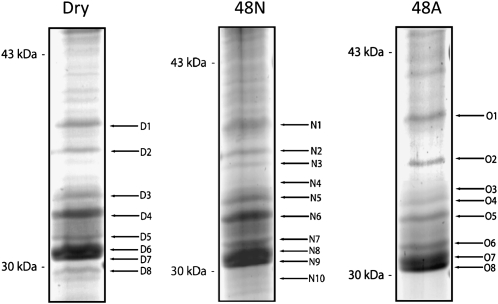
Integral membrane fractions from mitochondria prepared from rice embryos from dry seeds and from seeds imbibed for 48 h under aerobic or anaerobic conditions were separated on gels and stained with colloidal Coomassie Brilliant Blue (Fig. 5). Analysis focused on the region between the 30- and 43-kD markers, the expected size of the OsMCs in mitochondria. Relatively similar patterns of bands were observed for each sample. To determine the identity of the major bands, gel bands were excised and in-gel digested with trypsin, and extracted peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) and matched against the predicted database of rice proteins. In each case, this led to the identification of five MC family proteins: two ANTs (Os02g48720, Os05g23720), redundant matches to two PiCs (Os02g52860, Os06g10810), and a DTC (OsOs5g11780). Two carriers were found in some samples but not in others: these were a UCP (Os11g48040) and a BAC (Os10g42299; Table I).
Figure 5.
Integral membrane proteins from mitochondria isolated from rice embryos dissected from dry seeds and germinated under aerobic and anaerobic conditions. Representative SDS-PAGE gels are shown from carbonate-extracted mitochondria isolated from embryos of dry seeds, 48-h-imbibed anaerobic-treated seeds (48 N), and 48-h-imbibed aerobic-treated seeds (48A) revealing the integral membrane proteome. Numbers on the left indicate apparent molecular mass, and alphanumerics on the right indicate the protein bands extracted for analysis by MS (Table I).
Table I. MS-based identification of members of the OsMC family.
Analysis of protein bands extracted from gels shown in Figure 5. Details of the six gene products and their annotations are shown with their experimental [MM (Gel)] and theoretical [MM (Match)] molecular masses. The Mascot scores, sequence coverage, and number of peptides shown here are the combined values from all MS/MS-analyzed bands containing peptides matching carrier gene products.
| Sample | Band No. | Os Locus | Description | MM (Gel) | MM (Match) | Score | Coverage | No. of Peptides |
| D | D | % | ||||||
| Dry seeds | D6, D7 | Os02g48720 | Adenine nucleotide translocator | 33,000 | 48,650 | 472 | 32 | 13 |
| D3 | Os02g52860/Os06g10810 | Mitochondrial phosphate transporter | 35,500 | 38,934 | 130 | 11 | 4 | |
| D6 | Os05g11780 | Dicarboxylate/tricarboxylate carrier | 34,500 | 37,518 | 116 | 7 | 2 | |
| D6, D7 | Os05g23720 | Adenine nucleotide translocator | 32,000 | 40,951 | 585 | 42 | 13 | |
| Aerobic-germinated seeds | O5, O6, O7, O8 | Os02g48720 | Adenine nucleotide translocator | 33,000 | 48,650 | 461 | 30 | 12 |
| O3 | Os02g52860/Os06g10810 | Mitochondrial phosphate transporter | 35,500 | 38,934 | 55 | 10 | 4 | |
| O5, O6 | Os05g11780 | Dicarboxylate/tricarboxylate carrier | 34,500 | 37,518 | 303 | 19 | 6 | |
| O6 | Os05g23720 | Adenine nucleotide translocator | 32,000 | 40,951 | 164 | 22 | 7 | |
| O8 | Os10g42299 | Basic amino acid carrier | 32,500 | 31,534 | 250 | 29 | 8 | |
| Anaerobic-germinated seeds | N7, N8, N9 | Os02g48720 | Adenine nucleotide translocator | 33,000 | 48,650 | 483 | 29 | 12 |
| N4, N5 | Os02g52860/Os06g10810 | Mitochondrial phosphate transporter | 35,500 | 38,934 | 56 | 7 | 3 | |
| N4, N5 | Os02g52860/Os06g10810 | Mitochondrial phosphate transporter | 35,500 | 38,934 | 56 | 7 | 3 | |
| N6, N7 | Os05g11780 | Dicarboxylate/tricarboxylate carrier | 34,500 | 37,518 | 460 | 30 | 9 | |
| N7, N8 | Os05g23720 | Adenine nucleotide translocator | 32,000 | 40,951 | 567 | 44 | 14 | |
| N9 | Os10g42299 | Basic amino acid carrier | 32,500 | 31534 | 251 | 29 | 8 | |
| N9 | Os11g48040 | Putative uncoupling protein | 30,500 | 32,071 | 85 | 6 | 2 |
Identification and Analysis of Rice MCs by LC-MS/MS
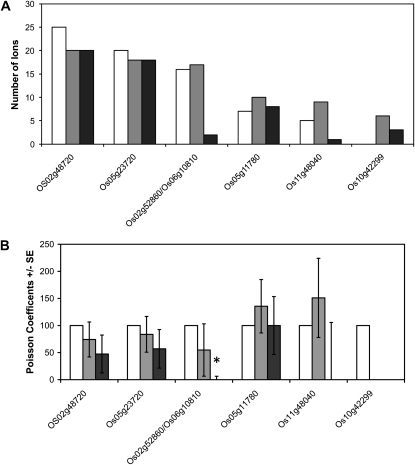
In order to gain information on the relative abundance of these carriers in the three different samples, we undertook direct analysis of carbonate-stripped membrane samples by trypsin digestion and LC-MS/MS analysis of triplicate biological sample sets. Gene-specific peptides were again found for all the carrier proteins listed above. Total spectral counts across the three experiments are shown in Figure 6A. Based on the noncontinuous nature of the data generated by a counting procedure, statistical analysis of the number of spectral matches to each carrier was undertaken using Poisson regression analysis. This revealed no significant change in the number of times peptides were observed for the DTC, UCP, and both ANTs (Fig. 6B). The only single significant change was for the phosphate carriers (P < 0.05), which had a significantly lower peptide count in aerobically germinated samples compared with the dry seeds and the anaerobically germinated samples (Fig. 6; Supplemental Table S1).
Figure 6.
The relative abundance of members of the MC family in rice. Samples were prepared from carbonate-extracted mitochondrial membranes isolated from embryos of dry seeds (white bars), 48-h-imbibed anaerobic-treated seeds (gray bars), and 48-h-imbibed aerobic-treated seeds (black bars). A, Average spectral counts for each OsMC member. B, Analysis of spectral counts by Poisson regression analysis displayed as relative Poisson coefficients ± se (n = 3). The statistical analysis results can be seen in Supplemental Table S1. * P < 0.05.
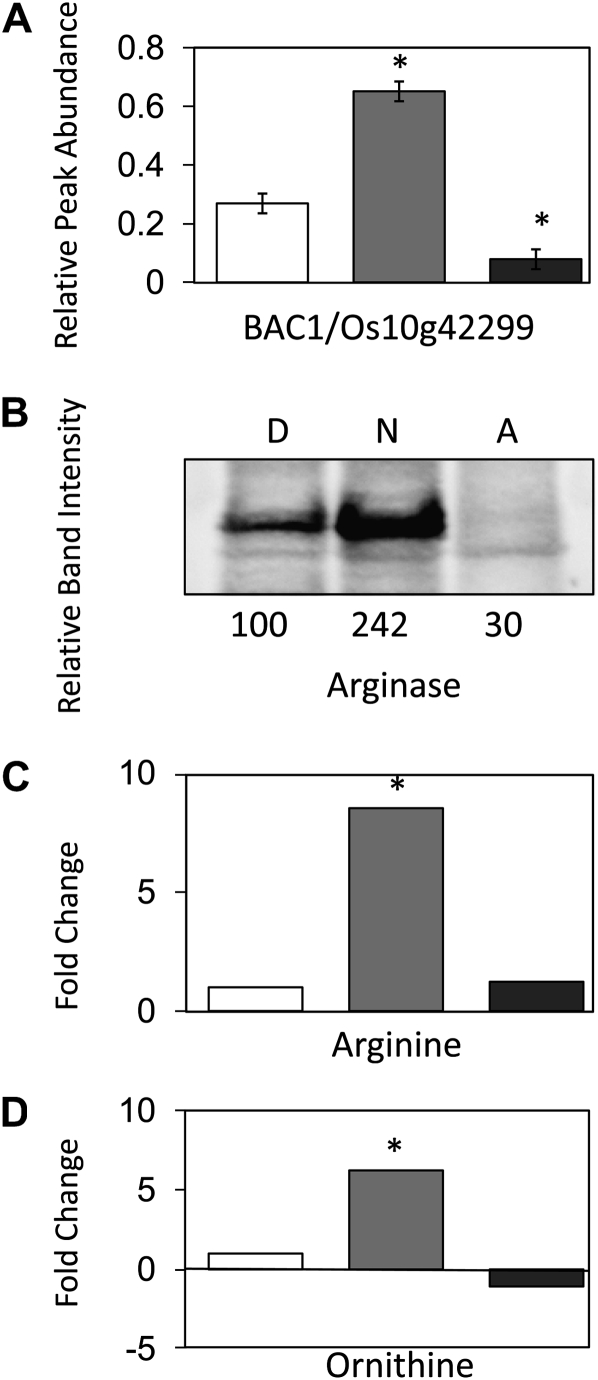
This Poisson regression analysis also revealed that the spectral counts for BAC/Os10g42299 were too low to provide any statistically reliable conclusion (Supplemental Table S1). For this reason, another MS approach was used to quantify the abundance of this protein in the three sets of samples. The MS/MS spectra for gene-specific peptide ions from BAC/Os10g42299 that were identified in our LC-MS/MS analyses were analyzed to design selected reaction monitoring (SRM) transitions. A triple quadrupole mass spectrometer was used based on the SRM transitions 862.90/769.39, 862.90/1,113.59, 862.90/1,184.59, 849.40/1,031.59, 849.40/735.40, and 849.40/1,130.59 to quantify the abundance of these peptides. This analysis showed a statistically significant (P < 0.05) 3-fold difference in abundance for these two peptides following imbibition for 48 h under anaerobic conditions when compared with dry seeds. Furthermore, a small but significantly (P < 0.05) lower abundance of BAC/Os10g42299 in mitochondrial membranes from 48-h aerobically germinated embryos was observed compared with dry seeds. The difference in the abundance of the peptides between the 48-h aerobic and anaerobic samples was greater than 7-fold (Fig. 7A; Supplemental Table S2).
Figure 7.
Changes in BAC, arginase, Arg, and Orn levels from dry, anoxically imbibed, and aerobically imbibed embryos. A, The relative abundance of BAC1 (Os10g42299) assessed by QqQ SRM MS. Mitochondria isolated from embryos of dry seeds (white bar), 48-h-imbibed anaerobic-treated seeds (gray bar), and 48-h-imbibed aerobic-treated seeds (dark gray bar) were carbonate extracted. The abundance from SRM transitions (862.90/769.39, 862.90/1,113.59, 862.90/1,184.59, 849.40/1,031.59, 849.40/735.40, 849.40/1,130.59) was then obtained. These abundance changes were then combined for each treatment triplicate, and the results are shown in relative abundance ± se (n = 3; * P < 0.05). B, The relative abundance of arginase assessed by western blot. Mitochondrial proteins isolated from embryos of dry seeds (D), 48-h-imbibed anaerobic-treated seeds (N), and 48-h-imbibed aerobic-treated seeds (A) were separated by SDS-PAGE. Numbers below represent relative band intensity. C and D, Ratio of metabolite abundance in embryos relative to dry seed embryos. Dry seeds (white bars), 48-h-imbibed anaerobic-treated seeds (gray bars), and 48-h-imbibed aerobic-treated seeds (dark gray bars) were used for Arg (C) and Orn (D). * P < 0.05.
Quantitative PCR Validation of Transcripts during Aerobic and Anaerobic Germination
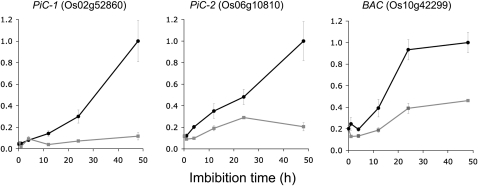
The apparent higher abundance of the phosphate carrier (Os02g52860/Os06g10810) and the basic amino acid carrier (Os10g42299) in membranes from anaerobic-germinated compared with aerobic-germinated embryos (Figs. 6 and 7A) appeared to negatively correlate with the microarray data (Fig. 2). Greater transcript abundance was reported for all three carriers in aerobic samples, an increase of transcript abundance was observed on transfer of anaerobic samples to air, and a decrease in transcript abundance was observed following transfer of aerobic samples to anaerobic conditions (Fig. 2). To confirm this negative correlation, we needed to ensure that incorrect microarray annotation or some cross-hybridization issue was not responsible and that no major changes in transcript abundance were occurring between the last microarray data point at 30 h (Fig. 2) and the 48-h time point used for the protein analysis (Figs. 6 and 7A). Quantitative reverse transcription (RT)-PCR analysis for the three transcripts using gene-specific primer pairs across the germination time line up to 48 h is presented in Figure 8. This clearly confirmed the microarray data, in that all three genes are more highly expressed in aerobic compared with anaerobic conditions, and shows that this difference in transcript abundance is apparent at all but the earliest time point during the first 48 h of the germination process.
Figure 8.
Quantitative transcript analysis of PiC and BAC in rice embryos. Gene-specific quantitative RT-PCR was used to determine transcript levels over the first 48 h of germination under aerobic (black circles) and anaerobic (gray circles) conditions. Data are given relative to total RNA, with average values ± se shown (n = 3).
Analysis of Arginase, Arg, and Orn Levels in Mitochondrial Samples
To place the much higher abundance of BAC in anaerobic samples into a biological context, we investigated the only mitochondrial enzyme known to use Arg, namely arginase, and its substrate and products, Arg and Orn. An antibody raised against Pinus taeda arginase (Todd et al., 2001) was used to assess the protein abundance in dry, 48-h aerobic, and 48-h anaerobic mitochondrial samples (Fig. 7B). The abundance of arginase very closely followed the changes we had seen in the BAC: a large increase was seen in embryos imbibed under anaerobic conditions, while a slight decrease in abundance was seen in embryos imbibed under aerobic conditions. We also reanalyzed gas chromatography-MS data that we recently reported (Narsai et al., 2009) to determine metabolite abundances in aerobic and anaerobic embryos compared with dry seed embryos. The abundance of Arg and Orn in rice embryo extracts followed a very similar pattern to BAC and arginase, with 6- to 8-fold increases in their abundance observed under anaerobic conditions (Fig. 7, C and D).
DISCUSSION
Sequence similarity searches have identified that the rice genome contains at least 50 MC-like genes, similar to the 58 identified in Arabidopsis (Picault et al., 2004), 46 in human (Haitina et al., 2006), and 35 in yeast (Palmieri et al., 2006a). Close pairing of orthologs, based on amino acid sequence, is apparent between carriers in Arabidopsis and rice (Fig. 1), indicating that discrete genes, and likely the functions of the proteins they encode, are maintained between these two model plants. The MS analysis of rice mitochondrial membranes (Table I) has identified close sequence orthologs to the carrier proteins identified by MS in Arabidopsis mitochondrial extracts (Millar and Heazlewood, 2003). This indicates that the expression of carriers in these divergent plant species is conserved, leading to the same set of carriers dominating the protein profile.
Oxygen-dependent increases in transcripts encoding mitochondrial proteins during seed germination have largely been interpreted as evidence for increased mitochondrial biogenesis and function under aerobic conditions (Howell et al., 2007). Additionally, the low respiratory capacity of mitochondria from anaerobic tissues (Fig. 3) suggests that the need for transport of substrates and cofactors for respiratory metabolism is probably low under anaerobic conditions. Conversely, carrier import is one of only a few processes in mitochondria that is reported to be unaffected by anaerobic conditions (Howell et al., 2007). Furthermore, to date, it is not known whether there are specific roles for rice MCs under anaerobic conditions that would require transport functions. As specific membrane carriers are not separable on two-dimensional gels, previous studies of the rice proteome during anaerobic conditions have not identified any OsMCs or changes in their abundance (Millar et al., 2004; Howell et al., 2006). Here, we found, using quantitative peptide-level analysis, that in some cases transcript pool size was not a good indicator of the changes in OsMC composition in mitochondrial membranes and that some OsMC members are more abundant under anaerobic than aerobic conditions (Figs. 6 and 7). We have previously observed similar phenomena with other rice mitochondrial proteins, where large differences in protein abundance between aerobic and anaerobic mitochondria are evident but are not reflected in transcript abundance (Howell et al., 2007). This could reflect the regulation of rice protein levels at the posttranscriptional level under anaerobic conditions by altering the rate or extent of transcript degradation (Narsai et al., 2007, 2009), protein synthesis (Bailey-Serres, 1999; Branco-Price et al., 2008), or protein degradation (Howell et al., 2006, 2007).
For the phosphate carriers, our data indicate the proteins are present in the embryos from dry seeds and are retained during anaerobic germination, but they are apparently depleted during aerobic germination (Fig. 6). This is similar to our published data on the general import pathway Tom40, Tom20, and Tim17/23 proteins, which decrease in abundance much faster during aerobic than anaerobic germination from their high level in dry seeds (Howell et al., 2006, 2007). Hence, the abundance of phosphate carriers most likely reflects that they are substrates for an aerobic-induced protein turnover pathway during mitochondrial maturation rather than them having a specific role under anaerobic conditions.
The only anaerobic-induced carrier from our analysis was BAC/Os10g42299 (Figs. 7 and 8). BAC was substantially greater in abundance in anaerobic samples than either dry seeds or aerobic-germinated samples, clearly indicating differential protein accumulation under anaerobic conditions and hence a probable role during anaerobic germination of rice. In Arabidopsis, two BACs have been identified through functional complementation of the knockout of the yeast translocator for Orn and Arg (Ort1p/Arg-11p; Catoni et al., 2003; Hoyos et al., 2003). Both proteins are members of the MC family, are localized to the mitochondria, and are called AtmBAC1 (At5g46800) and AtmBAC2 (At1g79900). AtmBAC1 is highly expressed during germination, while the expression of AtmBAC2 is relatively low and more ubiquitous, with maximal expression in flowers (Catoni et al., 2003; Hoyos et al., 2003). Both transporters have been shown to be efficient in the uniport/exchange of Arg, citrulline (Cit), Lys, Orn, and His, to be inactivated by the same inhibitors, and to exhibit very similar Km and Vmax values for Arg (Palmieri et al., 2006b). The only real differences identified are in pH optima, with AtmBAC1 having sustained activity over a wider range (pH 7–9) whereas AtmBAC2 has a distinct optimum at pH 8 (Palmieri et al., 2006b). It has been proposed that AtmBAC1 has a role in the uptake of Arg and in Arg/Orn exchange in conjunction with seed protein degradation (Palmieri et al., 2006b). Arg is the major store of nitrogen in the developing embryos of Arabidopsis, with at least 50% of the free amino acid nitrogen found in the Arg pool (Palmieri et al., 2006b). A specific role for AtmBAC2 in Orn transport has been proposed (Catoni et al., 2003), aligning its function with related MC carriers in human (Fiermonte et al., 2003) and the yeast translocator Ort1p/Arg-11p. Knockout and overexpression of BAC2 in Arabidopsis shows, on the one hand, that BAC2 is essential for the use of Arg as a nitrogen source and, on the other hand, that the level of BAC2 expression is a limiting factor for Arg transport in vivo (Toka et al., 2010).
To our knowledge, there is no report of a role of the Arabidopsis BACs in hypoxic or anaerobic metabolism in Arabidopsis and, notably, no polysomal enrichment of AtmBAC1 or AtmBAC2 during hypoxia (Branco-Price et al., 2008). Sequence alignments indicate, based on sequence similarity, that Os10g42299 is the rice BAC1 while Os10g12520 is the rice BAC2 (Fig. 1). This nomenclature is also consistent with expression profiles that show that Os10g42299 is highly expressed in seeds while Os10g12520 is lowly expressed in storage organs and highly expressed in the floral stigma (Fig. 2). However, explaining why BAC1 in rice might have an enhanced role under anaerobic conditions requires a broader appraisal of why plant mitochondria contain BACs as well as an understanding of the metabolic changes that occur under anoxia in rice, as outlined below.
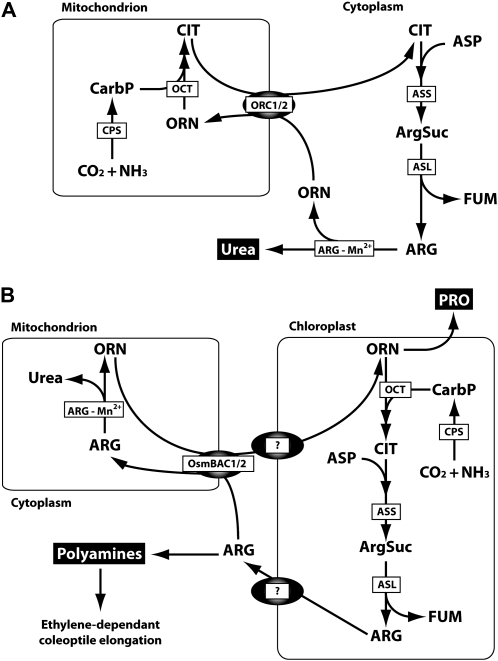
The BACs in animal mitochondria are critical to facilitate the Orn-urea cycle used to convert excess ammonia into urea for secretion from the cell. Animal mitochondria contain the first few enzymatic steps of this pathway (Fig. 9A), and the BACs facilitate Orn uptake and Cit export to maintain the cycle (Fiermonte et al., 2003). Much debate over several decades has surrounded the subcellular localization and role of the enzymes of the Orn-urea cycle in plants. Resolving this puzzle is essential to determine what substrate transport steps between plant organelle compartments are required for an Orn-urea cycle analogous to that in animals. By combining data collected from both enzymatic studies and recent protein localization studies in model plants (Supplemental Materials S1), we propose that the subcellular locations of these enzymes in plants have been resolved and that they are distinctly different from those of the Orn-urea cycle in animals. Plant mitochondria contain only one enzyme of the cycle, arginase (which is in the cytosol in mammals), and most of the enzymes distributed between the cytosol and the mitochondrion in animals are in the plastid in plants (Fig. 9). Assessment of the transcript levels for arginase and these plastidial enzymes in rice embryos through a reanalysis of our published data (Narsai et al., 2009) did not show any induction of transcripts under anaerobic conditions (Supplemental Fig. S1). This arrangement of the urea cycle enzymes in plants, with arginase in the mitochondrial matrix, necessitates Arg and Orn transport across the plant inner mitochondrial membrane rather than the transport of Orn and Cit as required in mammals. The reason for arginase being localized in mitochondria in plants is not clear, but it could be due to kinetic considerations of the plant arginase or perhaps the Mn2+ requirement of the enzyme and, thus, its placement in mitochondria alongside other prominent Mn2+-requiring enzymes in plants, including manganese-superoxide dismutase and NAD-dependent malic enzyme.
Figure 9.
The BAC and the Orn-urea cycle in animals (A) and plants (B). ARG-Mn2+, Arginase; ArgSuc, arginosuccinate; ASL, arginosuccinate lyase; ASS, arginosuccinate synthase; CarbP, carbamoyl phosphate; CPS, carbamoyl phosphate synthase; FUM, fumarate; OCT, Orn carbamoyltransferase; ORC1/2, human mitochondrial Orn carrier; OsmBAC1/2, rice mitochondrial BAC carrier. Question marks indicate currently unknown carriers for Arg and Orn in plastids.
Changes in the metabolome of the rice embryo during aerobic and anaerobic germination (Narsai et al., 2009; Fig. 7, C and D) show that Arg and Orn are six to eight times more abundant in anaerobic- than aerobic-germinated embryos, suggesting enhanced amino acid metabolism involving these compounds under anaerobic conditions. Reports have also noted that putrescine, synthesized from Arg, is much more abundant during anaerobic rice germination (Reggiani et al., 1989), and its synthesis is linked to the ethylene-dependent enhanced elongation of the rice coleoptile under anaerobic conditions (Lee and Chu, 1992). Also, Pro, which is synthesized from Orn, is more than 5-fold higher in abundance in anaerobic- than aerobic-germinated rice embryos (Narsai et al., 2009). Pro is commonly induced by environmental perturbations, with roles in desiccation tolerance, ion balance, and macromolecule stabilization. Reports have noted that Pro synthesis occurs very early in the germination of rice embryos under anaerobic conditions as a major product of seed nitrate assimilation (Reggiani et al., 1993, 2000), prior to the reliance of nitrogen metabolism on the breakdown and degradation of amino acids in storage proteins. Combining these data, we propose that Orn-Arg-Cit metabolism is enhanced during anaerobic rice germination and that the rice mitochondrial BAC1 plays a specific role in this metabolism through Arg and Orn transport. This transport facilitates the mitochondria-localized arginase reaction and is needed for specific aspects of basic amino acid metabolism of anaerobic rice, leading to putrescine-stimulated coleoptile elongation and early anaerobic Pro synthesis in the rice embryo.
MATERIALS AND METHODS
Rice Growth
Dehulled, sterilized rice (Oryza sativa ‘Amaroo’) seeds were grown under aerobic or anaerobic conditions in the dark at 30°C as described previously (Howell et al., 2007).
Mitochondrial Isolation
Mitochondria were isolated from rice embryos using a modified mitochondrial isolation protocol as described previously (Howell et al., 2006).
Respiratory Assays
Oxygen uptake by whole rice embryos was measured using 10 embryos in 2 mL of fresh coleoptile culture medium at 30°C in a Clarke-type oxygen electrode (Rank Bros). Cyanide-sensitive oxygen consumption was calculated by adding KCN to a final concentration of 1 mm and subtracting this residual rate of oxygen consumption. Oxygen consumption by isolated mitochondria was measured by adding 80 to 110 μg of mitochondrial protein to 1 mL of mitochondria reaction medium at 25°C in a Clarke-type oxygen electrode. The following reagents and inhibitors were added to the reaction medium to examine mitochondrial cytochrome c oxidase activity: ascorbate (10 mm), cytochrome c (50 mm), Triton X-100 (0.05%, w/v), and KCN (1 mm). Cytochrome c oxidase activity was calculated by subtracting the rate (y) of cytochrome c oxidase KCN-sensitive activity after the addition of 1 mm KCN from the rate (x) of cytochrome c oxidase activity, prior to the addition of 1 mm KCN.
Carbonate Extraction of Mitochondria
Carbonate extraction was performed to separate the soluble, peripheral, and integral proteins of mitochondria samples. Approximately 400 μg of mitochondrial protein was used for each extraction. The samples were freeze thawed three times using liquid N2 to disrupt the mitochondrial membranes, releasing the soluble proteins, and then spun in a centrifuge at 20,000g for 10 min at 4°C to pellet the membrane proteins. The supernatant (soluble protein fraction) was retained and stored at −20°C. The pellet was resuspended in 1 mL of double deionized water, and the remaining soluble proteins were removed by repeated freeze-thaw cycles and centrifugation steps. Peripheral membrane proteins were stripped and solubilized by resuspending the pellet in 1 mL of 100 mm sodium carbonate. The sample was spun at 20,000g for 10 min at 4°C, and the supernatant (peripheral membrane protein fraction) was stored at −20°C. The pellet was resuspended in 1 mL of 100 mm sodium carbonate and spun again in a centrifuge at 20,000g for 10 min at 4°C, and the supernatant was discarded. The resultant pellet (integral membrane protein fraction) was stored at −20°C. Prior to further analysis, peripheral proteins were concentrated using a 3-kD NMWL Ultra 0.5-mL centrifugal filter (Millipore), while the soluble protein fraction was precipitated with acetone at −20°C.
One-Dimensional SDS-PAGE and Immunodetection
Approximately 30 μg of concentrated soluble, peripheral, and integral mitochondrial fractions was loaded onto a 20-cm × 20-cm × 1-mm polyacrylamide gel. Electrophoresis was performed at room temperature at a constant 20 mA. The gel was stained overnight with colloidal Coomassie Brilliant Blue G250 and destained in 0.5% (v/v) orthophosphoric acid. For immunodetection, 20 μg of mitochondrial proteins was resolved by SDS-PAGE and transferred to a Hybond-C+ nitrocellulose membrane using a semidry blotting apparatus (Millipore). Immunodetection of the proteins was performed using the BM Chemiluminescence Blotting Substrate (POD) Kit (GE Healthcare), and chemiluminescence was detected on a LAS 100 instrument (Fuji). Antibodies used were as follows: E1α (matrix protein marker) from Dr. T. Elton (University of Nebraska, Lincoln), 1:5,000; F1α (peripheral protein marker) from Dr. T. Elton, 1:10,000; Glycine max UCP1a (integral membrane protein marker [AAL68562]; Considine et al., 2001). For arginase western blot, anti-arginase antibodies were kindly provided by Christopher Todd (Todd et al., 2001); 30 μg of mitochondrial proteins was separated via SDS-PAGE using a Criterion mini gel (10%–20% Tris-HCl; Bio-Rad) following the manufacturer’s instructions and transferred to a nitrocellulose membrane (Hybond C+ Extra; GE Healthcare) by standard procedures. Following overnight blocking with 1% (w/v) blocking solution (Amersham ECL Advance; GE Healthcare), membranes were incubated with primary antibodies (anti-arginase, 1:5,000) at room temperature for 1 h, followed by an appropriate secondary antibody (anti-Rabbit IgG POD; 1:20,000) at room temperature for 1 h. Chemiluminescence was visualized using an Amersham ImageQuant RT-ECL camera (GE Healthcare), and band intensity was quantified using Amersham Image-Quant TL software (GE Healthcare).
MS Analysis of In-Gel Trypsin-Digested Samples
Colloidal Coomassie Brilliant Blue bands around the 30-kD region were excised from the integral protein fractions and processed by the method described by Taylor et al. (2004). Digested samples were analyzed on a QStar Pulsar i MS/MS system (Applied Biosystems). Each sample was loaded onto a reverse-phase column (C18) in 5% acetonitrile and 0.1% formic acid using an Agilent 1100 LC system (Agilent Technologies). Bound peptides were differentially eluted into the mass spectrometer with increasing concentrations of acetonitrile (5%–80% in 0.1% formic acid). Eluted peptides were ionized through an electrospray ionization source and automatically selected for fragmentation using independent data acquisition with collision-induced disassociation undertaken with nitrogen gas (Millar and Heazlewood, 2003). Data produced on the QStar Pulsar i MS/MS system were exported as .mgf files from Analyst QS version 1.1 using a purpose-built script freely available from Matrix Sciences (version 1.6b23). The script was set up to centroid the survey scan ions (time of flight [TOF] MS) at a height percentage of 50% and a merge distance of 0.1 atomic mass unit (for charge state determination), centroid MS/MS data at a height percentage of 50% and a merge distance of 2 atomic mass units, reject a collision-induced disassociation if less than 10 peaks, and discard ions with charge equal and greater than 5+.
MS Analysis from Non-Gel LC-MS/MS of Integral Membrane Digests
Whole organelle protein extracts were digested overnight at 37°C in the presence of trypsin, and insoluble components were removed by centrifugation at 20,000g for 5 min (Eubel et al., 2008). Samples were analyzed on an Agilent 6510 Q-TOF mass spectrometer with an HPLC Chip Cube source (Agilent Technologies). The chip consisted of a 40-nL enrichment column (Zorbax 300SB-C18, 5-μm pore size) and a 150-mm separation column (Zorbax 300SB-C18, 5-μm pore size) driven by the Agilent Technologies 1100 series nano/capillary LC system. Both systems were controlled by MassHunter Workstation Data Acquisition for Q-TOF (version B.01.03, B1045; Agilent Technologies). Peptides were loaded onto the trapping column at 4 μL min−1 in 5% (v/v) acetonitrile and 0.1% (v/v) formic acid with the chip switched to enrichment and using the capillary pump. The chip was then switched to separation, and peptides were eluted during a 1-h gradient (5% [v/v] acetonitrile to 60% [v/v] acetonitrile) directly into the mass spectrometer. The mass spectrometer was run in positive ion mode, and MS scans were run over a mass-to-charge ratio range of 275 to 1,500 and at four spectra s−1. Precursor ions were selected for auto MS/MS at an absolute threshold of 500 and a relative threshold of 0.01, with maximum three precursors per cycle, and active exclusion set at two spectra and released after 1 min. Precursor charge-state selection and preference were set to 2+ and then 3+, and precursors were selected by charge and then abundance. Resulting MS/MS spectra were opened in MassHunter Workstation Qualitative Analysis (version B.01.03, build 1.3.157.0, patch 2; Agilent Technologies), and MS/MS compounds were detected by Find Auto MS/MS using default settings. The resulting compounds were then exported as .mzdata files.
Protein Identification Searches
Gel Spots
Output files were analyzed by the MS interrogator program Mascot (http://www.matrixscience.com). Searches were undertaken against the in-house database, which comprises The Institute for Genomic Research (TIGR) rice proteome (Osa3.all.pep: TIGR) set, at error tolerances of MS ± 0.15 D and MS/MS ± 0.05 D. Protein sequences matching peptides with a Mascot score greater than 37 (P < 0.05) and annotated as MC proteins were used for further analysis.
Complex Mixture Analysis
Output files from Q-TOF data were searched using the Mascot search engine version 2.1.04 (Matrix Science) and a database that comprises the TIGR rice proteome (Osa5.all.pep: TIGR) utilizing error tolerances of ±100 ppm for MS and ±0.5 D for MS/MS, Max Missed Cleavages set to 1, the Oxidation (M), Carboxymethyl (C), variable modifications, and Instrument set to ESI-Q-TOF, and Peptide charge set at 2+ and 3+. Results were filtered using MUDPIT scoring, with Maximum Number of Hits set to 20, Significance Threshold at P < 0.05, and Ion Score Cutoff at 33. Protein matches were only claimed if at least two distinct peptides were detected per protein, resulting in Mascot scores typically higher than 70 (P < 0.05 significance level is score > 33).
SRM of Os10g42299 Peptides
Protein extracts from non-gel LC-MS/MS of integral membrane digests were analyzed on an Agilent 6410 QqQ mass spectrometer with an HPLC Chip Cube source (Agilent Technologies). The chip consisted of a 40-nL enrichment column (Zorbax 300SB-C18, 5-μm pore size) and a 150-mm separation column (Zorbax 300SB-C18, 5-μm pore size) driven by Agilent Technologies 1200 series nano/capillary LC system. Both systems were controlled by MassHunter Workstation Data Acquisition for QqQ (version B.01.04, build 84; Agilent Technologies). Peptides were loaded onto the trapping column at 4 μL min−1 in 5% (v/v) acetonitrile and 0.1% (v/v) formic acid with the chip switched to enrichment and using the capillary pump. The chip was then switched to separation, and peptides were eluted during a 20-min gradient (5% [v/v] acetonitrile to 80% [v/v] acetonitrile) directly into the mass spectrometer. Parent ions and transitions were selected based on previous Q-TOF experiments and were 862.90/769.39, 862.90/1,113.59, 862.9/1,148.59, 849.40/735.4, 849.40/1,031.5, and 849.40/1,130.59. The mass spectrometer was run in positive ion mode, and for each parent/transition the fragmentor was set to 160, collision energy was 26, and dwell time was 100 ms. Resulting total ion chromatograms were opened in MassHunter Workstation Qualitative Analysis (version B.01.04, build 1.4.126.0; Agilent Technologies), and SRM chromatograms were obtained using the Extract Chromatogram feature. Each SRM chromatogram was then integrated, and the area under the peak within ±30 s of the expected retention time was calculated. The data for each transition was then combined and expressed as relative abundance (calculated on total areas in all samples).
Statistical Analysis of MS-Derived Data
The noncontinuous spectral counting data sets were analyzed by Poisson regression analysis and Fisher exact tests using the R statistics package (2.9.0). Data obtained from QqQ MS was analyzed in Excel (Microsoft) for Student’s t test.
Identifying Putative MC Protein Family Members in Rice
The Arabidopsis (Arabidopsis thaliana) MC superfamily was obtained from Picault et al. (2004). Sequences were analyzed using BLASTp against release 3 of the TIGR Rice Genome Annotation Database (Osa.3.all.pep: TIGR). Each query sequence was filtered using a low-complexity filter, expect value of 10, and a BLOSUM62 substitution matrix to give the top 200 most similar sequences in rice. The results were compiled, and sequences with the highest number of hits were extracted. The number of SOLCAR domains within the rice sequences indicating carrier protein function was retrieved from Prosite (http://au.expasy.org/prosite/).
RNA Isolation, cDNA Synthesis, and Quantitative RT-PCR
Total RNA was isolated from rice embryos as described previously (Howell et al., 2006). Three independent RNA preparations were performed for each developmental stage/growth condition, and the concentration of RNA was determined spectrophotometrically. cDNA was prepared from 1 μg of total RNA using the iScript cDNA synthesis kit (Bio-Rad). Quantitative RT-PCR analysis was carried out using an iCycler (Bio-Rad) or LightCycler 480 instrument (Roche) and iQ SYBR Green Supermix (Bio-Rad) or LightCycler 480 SYBR Green I Master (Roche) under conditions optimized to maximize the amplification efficiency and minimize primer-dimer formation. For every transcript, each cDNA sample was analyzed in duplicate, and relative transcript abundance was calculated by normalizing to the maximum level during the 48-h time course. Primers used for quantitative PCR (PiC-1, Os02g52860; PiC-2, Os06g10810; BAC, Os10g42299) were as follows: PiC-1-FWD, 5′-GCTGTGAAGGTTCGTGTGC-3′; PiC-1-REV, 5′-CAATGGTCTCAAAGGAGGC-3′; PiC-2-FWD, 5′-CAAATCTACAAGCATGCTG-3′; PiC-2-REV, 5′-CACAGAAGACACCGGCAAT-3′; BAC-FWD, 5′-CATGAAGGGTTTGTTCAAGG-3′; BAC-REV, 5′-CTGCCGAGGTTTGATGTGTC-3′.
Microarray Data Analyses
In order to compare the transcript abundance changes of genes encoding MCs across different tissues and conditions, rice array data were retrieved from the Gene Expression Omnibus within the National Center of Biotechnology Information database; the data sets were E-MEXP-1766 and E-MEXP-2267 undertaken by the authors during rice germination and GSE6908, GSE6893, GSE6901, and GSE7965 from other researchers. All data were MAS5.0 normalized and normalized against average ubiquitin expression, as carried out previously (Huang et al., 2009). For each probe set, the maxximum expression was set to 1.0 with all other data shown relative to this. This facilitates cross-comparison of arrays from all of the different studies at once. Hierarchical clustering (average linkage) based on Euclidian distance was carried out using Partek Genomic suite software (version 6.4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Changes in transcript abundance for genes of components of the urea cycle from rice.
Supplemental Table S1. Poisson regression analysis of spectra counts for each identified member of the rice MCF family.
Supplemental Table S2. Student’s t test of QqQ MS data from Os10g42299.
Supplemental Materials S1. Summarized data on the subcellular localization of components of the urea cycle in Arabidopsis and rice.
Supplementary Material
Acknowledgments
Kristen Feher (University of Western Australia) is thanked for helpful discussions in relation to statistical analysis, and Damien Callahan and the staff at Metabolomics Australia are thanked for help with QqQ analysis. Dr. Christopher D. Todd (University of Saskatchewan) is thanked for the gift of anti-arginase antibodies.
References
- Arco AD, Satrustegui J. (2005) New mitochondrial carriers: an overview. Cell Mol Life Sci 62: 2204–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J. (1999) Selective translation of cytoplasmic mRNAs in plants. Trends Plant Sci 4: 142–148 [DOI] [PubMed] [Google Scholar]
- Bamber L, Harding M, Monne M, Slotboom DJ, Kunji ER. (2007) The yeast mitochondrial ADP/ATP carrier functions as a monomer in mitochondrial membranes. Proc Natl Acad Sci USA 104: 10830–10834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardel J, Louwagie M, Jaquinod M, Jourdain A, Luche S, Rabilloud T, Macherel D, Garin J, Bourguignon J. (2002) A survey of the plant mitochondrial proteome in relation to development. Proteomics 2: 880–898 [DOI] [PubMed] [Google Scholar]
- Belenkiy R, Haefele A, Eisen MB, Wohlrab H. (2000) The yeast mitochondrial transport proteins: new sequences and consensus residues, lack of direct relation between consensus residues and transmembrane helices, expression patterns of the transport protein genes, and protein-protein interactions with other proteins. Biochim Biophys Acta 1467: 207–218 [DOI] [PubMed] [Google Scholar]
- Bewley JD. (1997) Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJ, Larive CK, Bailey-Serres J. (2008) Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 56: 743–755 [DOI] [PubMed] [Google Scholar]
- Catoni E, Desimone M, Hilpert M, Wipf D, Kunze R, Schneider A, Flugge UI, Schumacher K, Frommer WB. (2003) Expression pattern of a nuclear encoded mitochondrial arginine-ornithine translocator gene from Arabidopsis. BMC Plant Biol 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Considine MJ, Daley DO, Whelan J. (2001) The expression of alternative oxidase and uncoupling protein during fruit ripening in mango. Plant Physiol 126: 1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey PA, Hunt S, Hymus GJ, DeLucia EH, Drake BG, Karnosky DF, Long SP. (2004) Respiratory oxygen uptake is not decreased by an instantaneous elevation of [CO2], but is increased with long-term growth in the field at elevated CO2. Plant Physiol 134: 520–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DA, Wiskich J. (1984) Transport processes of isolated plant mitochondria. Physiol Veg 22: 241–261 [Google Scholar]
- Douce R, Aubert S, Neuburger M. (1997) Metabolic exchange between the mitochondrion and the cytosol. Dennis DT, Turpin DH, Lefebvre DD, Layzell DB, , Plant Metabolism. Addison Wesley Longman, Essex, UK, pp 234–251 [Google Scholar]
- Douce R, Bourguignon J, Neuburger M, Rebeille F. (2001) The glycine decarboxylase system: a fascinating complex. Trends Plant Sci 6: 167–176 [DOI] [PubMed] [Google Scholar]
- Douette P, Sluse FE. (2006) Mitochondrial uncoupling proteins: new insights from functional and proteomic studies. Free Radic Biol Med 40: 1097–1107 [DOI] [PubMed] [Google Scholar]
- Eubel H, Meyer EH, Taylor NL, Bussell JD, O’Toole N, Heazlewood JL, Castleden I, Small ID, Smith SM, Millar AH. (2008) Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol 148: 1809–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiermonte G, Dolce V, David L, Santorelli FM, Dionisi-Vici C, Palmieri F, Walker JE. (2003) The mitochondrial ornithine transporter: bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J Biol Chem 278: 32778–32783 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Meler MA, Ribas-Carbo M, Siedow JN, Drake BG. (1996) Direct inhibition of plant mitochondrial respiration by elevated CO2. Plant Physiol 112: 1349–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin KL, Anderson OR, Tissue DT, Turnbull MH, Whitehead D. (2004) Variations in dark respiration and mitochondrial numbers within needles of Pinus radiata grown in ambient or elevated CO2 partial pressure. Tree Physiol 24: 347–353 [DOI] [PubMed] [Google Scholar]
- Gupta KA, Zablaza A, van Dongen JT. (2009) Regulation of respiration when the oxygen availability changes. Physiol Plant 137: 383–389 [DOI] [PubMed] [Google Scholar]
- Haferkamp I. (2007) The diverse members of the mitochondrial carrier family in plants. FEBS Lett 581: 2375–2379 [DOI] [PubMed] [Google Scholar]
- Haitina T, Lindblom J, Renstrom T, Fredriksson R. (2006) Fourteen novel human members of mitochondrial solute carrier family 25 (SLC25) widely expressed in the central nervous system. Genomics 88: 779–790 [DOI] [PubMed] [Google Scholar]
- Hon T, Dodd A, Dirmeier R, Gorman N, Sinclair PR, Zhang L, Poyton RO. (2003) A mechanism of oxygen sensing in yeast: multiple oxygen-responsive steps in the heme biosynthetic pathway affect Hap1 activity. J Biol Chem 278: 50771–50780 [DOI] [PubMed] [Google Scholar]
- Howell KA, Cheng K, Murcha MW, Jenkin LE, Millar AH, Whelan J. (2007) Oxygen initiation of respiration and mitochondrial biogenesis in rice. J Biol Chem 282: 15619–15631 [DOI] [PubMed] [Google Scholar]
- Howell KA, Millar AH, Whelan J. (2006) Ordered assembly of mitochondria during rice germination begins with pro-mitochondrial structures rich in components of the protein import apparatus. Plant Mol Biol 60: 201–223 [DOI] [PubMed] [Google Scholar]
- Howell KA, Narsai R, Carroll A, Ivanova A, Lohse M, Usadel B, Millar AH, Whelan J. (2009) Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol 149: 961–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyos ME, Palmieri L, Wertin T, Arrigoni R, Polacco JC, Palmieri F. (2003) Identification of a mitochondrial transporter for basic amino acids in Arabidopsis thaliana by functional reconstitution into liposomes and complementation in yeast. Plant J 33: 1027–1035 [DOI] [PubMed] [Google Scholar]
- Huang S, Taylor NL, Narsai R, Eubel H, Whelan J, Millar AH. (2009) Experimental analysis of the rice mitochondrial proteome, its biogenesis, and heterogeneity. Plant Physiol 149: 719–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey-Smith I, Colas des Francs-Small C, Ambart-Bretteville F, Remy R. (1992) Tissue-specific variation of pea mitochondrial polypeptides detected by computerized image analysis of two-dimensional electrophoresis gels. Electrophoresis 13: 168–172 [DOI] [PubMed] [Google Scholar]
- Kunji ER, Crichton PG. (2010) Mitochondrial carriers function as monomers. Biochim Biophys Acta 1797: 817–831 [DOI] [PubMed] [Google Scholar]
- Lee CP, Eubel H, O’Toole N, Millar AH. (2008) Heterogeneity of the mitochondrial proteome for photosynthetic and non-photosynthetic Arabidopsis metabolism. Mol Cell Proteomics 7: 1297–1316 [DOI] [PubMed] [Google Scholar]
- Lee TM, Chu C. (1992) Ethylene-induced polyamine accumulation in rice (Oryza sativa L.) coleoptiles. Plant Physiol 100: 238–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DC, Millar AH, Sweetlove LJ, Hill SA, Leaver CJ. (2001) Mitochondrial biogenesis during germination in maize embryos. Plant Physiol 125: 662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Heazlewood JL. (2003) Genomic and proteomic analysis of mitochondrial carrier proteins in Arabidopsis. Plant Physiol 131: 443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Trend AE, Heazlewood JL. (2004) Changes in the mitochondrial proteome during the anoxia to air transition in rice focus around cytochrome-containing respiratory complexes. J Biol Chem 279: 39471–39478 [DOI] [PubMed] [Google Scholar]
- Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, et al. (2003) Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115: 629–640 [DOI] [PubMed] [Google Scholar]
- Moyes CD, Hood DA. (2003) Origins and consequences of mitochondrial variation in vertebrate muscle. Annu Rev Physiol 65: 177–201 [DOI] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Carroll A, Ivanova A, Millar AH, Whelan J. (2009) Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol 151: 306–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Millar AH, O’Toole N, Small I, Whelan J. (2007) Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19: 3418–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri F. (1994) Mitochondrial carrier proteins. FEBS Lett 346: 48–54 [DOI] [PubMed] [Google Scholar]
- Palmieri F, Agrimi G, Blanco E, Castegna A, Di Noia MA, Iacobazzi V, Lasorsa FM, Marobbio CM, Palmieri L, Scarcia P, et al. (2006a) Identification of mitochondrial carriers in Saccharomyces cerevisiae by transport assay of reconstituted recombinant proteins. Biochim Biophys Acta 1757: 1249–1262 [DOI] [PubMed] [Google Scholar]
- Palmieri L, Agrimi G, Runswick MJ, Fearnley IM, Palmieri F, Walker JE. (2001) Identification in Saccharomyces cerevisiae of two isoforms of a novel mitochondrial transporter for 2-oxoadipate and 2-oxoglutarate. J Biol Chem 276: 1916–1922 [DOI] [PubMed] [Google Scholar]
- Palmieri F, Bisaccia F, Capobianco L, Dolce V, Fiermonte G, Iacobazzi V, Indiveri C, Palmieri L. (1996) Mitochondrial metabolite transporters. Biochim Biophys Acta 1275: 127–132 [DOI] [PubMed] [Google Scholar]
- Palmieri F, Rieder B, Ventrella A, Blanco E, Do PT, Nunes-Nesi A, Trauth AU, Fiermonte G, Tjaden J, Agrimi G, et al. (2009) Molecular identification and functional characterisation of Arabidopsis thaliana mitochondrial and chloroplastic NAD+ carrier proteins. J Biol Chem 284: 31249–31259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L, Santoro A, Carrari F, Blanco E, Nunes-Nesi A, Arrigoni R, Genchi F, Fernie AR, Palmieri F. (2008) Identification and characterization of ADNT1, a novel mitochondrial adenine nucleotide transporter from Arabidopsis. Plant Physiol 148: 1797–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L, Todd CD, Arrigoni R, Hoyos ME, Santoro A, Polacco JC, Palmieri F. (2006b) Arabidopsis mitochondria have two basic amino acid transporters with partially overlapping specificities and differential expression in seedling development. Biochim Biophys Acta 1757: 1277–1283 [DOI] [PubMed] [Google Scholar]
- Picault N, Hodges M, Palmieri L, Palmieri F. (2004) The growing family of mitochondrial carriers in Arabidopsis. Trends Plant Sci 9: 138–146 [DOI] [PubMed] [Google Scholar]
- Prohl C, Pelzer W, Diekert K, Kmita H, Bedekovics T, Kispal G, Lill R. (2001) The yeast mitochondrial carrier Leu5p and its human homologue Graves’ disease protein are required for accumulation of coenzyme A in the matrix. Mol Cell Biol 21: 1089–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiani R, Hochkoeppler A, Bertani A. (1989) Polyamines in rice seedlings under oxygen-deficit stress. Plant Physiol 91: 1197–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiani R, Mattana M, Aurisano N, Bertani A. (1993) Utilization of stored nitrate during the anaerobic germination of rice seeds. Plant Cell Physiol 34: 379–383 [Google Scholar]
- Reggiani R, Nebuloni M, Mattana M, Brambilla I. (2000) Anaerobic accumulation of amino acids in rice roots: role of the glutamine synthetase/glutamate synthase cycle. Amino Acids 18: 207–217 [DOI] [PubMed] [Google Scholar]
- Robertson EJ, Williams M, Harwood JL, Lindsay JG, Leaver CJ, Leech RM. (1995) Mitochondria increase three-fold and mitochondrial proteins and lipid change dramatically in postmeristematic cells in young wheat leaves grown in elevated CO2. Plant Physiol 108: 469–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sappl PG, Heazlewood JL, Millar AH. (2004) Untangling multi-gene families in plants by integrating proteomics into functional genomics. Phytochemistry 65: 1517–1530 [DOI] [PubMed] [Google Scholar]
- Saraste M, Walker JE. (1982) Internal sequence repeats and the path of polypeptide in mitochondrial ADP/ATP translocase. FEBS Lett 144: 250–254 [DOI] [PubMed] [Google Scholar]
- Sluse FE, Jarmuszkiewicz W, Navet R, Douette P, Mathy G, Sluse-Goffart CM. (2006) Mitochondrial UCPs: new insights into regulation and impact. Biochim Biophys Acta 1757: 480–485 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Day DA, Millar AH. (2004) Lipoic acid-dependent oxidative catabolism of alpha-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol 134: 838–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari BS, Belenghi B, Levine A. (2002) Oxidative stress increased respiration and generation of reactive oxygen species, resulting in ATP depletion, opening of mitochondrial permeability transition, and programmed cell death. Plant Physiol 128: 1271–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd CD, Cooke JEK, Gifford DJ. (2001) Purification and properties of Pinus taeda arginase from germinated seedlings. Plant Physiol Biochem 39: 1037–1045 [Google Scholar]
- Toka I, Planchais S, Cabassa C, Justin AM, De Vos D, Richard L, Savoure A, Carol P. (2010) Mutations in the hyperosmotic stress-responsive mitochondrial BASIC AMINO ACID CARRIER2 enhance proline accumulation in Arabidopsis. Plant Physiol 152: 1851–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartapetian BB, Andreeva IN, Generozova IP, Polyakova LI, Maslova IP, Dolgikh YI, Stepanova AY. (2003) Functional electron microscopy in studies of plant response and adaptation to anaerobic stress. Ann Bot (Lond) 91: 155–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Runswick MJ. (1993) The mitochondrial transport protein superfamily. J Bioenerg Biomembr 25: 435–446 [DOI] [PubMed] [Google Scholar]
- Wiskich J. (1977) Mitochondrial metabolite transport. Annu Rev Plant Physiol 28: 45–69 [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.