Abstract
VERNALIZATION INSENSITIVE3 (VIN3) induction by vernalization is one of the earliest events in the vernalization response of Arabidopsis (Arabidopsis thaliana). However, the mechanism responsible for vernalization-mediated VIN3 induction is poorly understood. Here, we show that the constitutive repression of VIN3 in the absence of the cold is due to multiple repressive components, including a transposable element-derived sequence, LIKE-HETEROCHROMATIN PROTEIN1 and POLYCOMB REPRESSION COMPLEX2. Furthermore, the full extent of VIN3 induction by vernalization requires activating complex components, including EARLY FLOWERING7 and EARLY FLOWERING IN SHORT DAYS. In addition, we observed dynamic changes in the histone modifications present at VIN3 chromatin during the course of vernalization. Our results show that the induction of VIN3 includes dynamic changes at the level of chromatin triggered by long-term cold exposure.
The transition from vegetative growth to reproductive growth is one of major developmental transitions in the life cycle of plants. Flowering plants have evolved to maximize the reproductive success by optimizing the timing of flowering. The onset of floral transition in flowering plants is affected by various environmental cues, including changing daylength and temperature. Plants use such environment cues to monitor seasonal changes and determine the timing of flowering. In temperate climates, the winter season imposes a prolonged period of cold to plants. In many plant species, exposure to prolonged period of cold provides competence to flower in the following spring through the process known as vernalization (for review, see Sung and Amasino, 2005; Dennis and Peacock, 2007).
While most lab strains of Arabidopsis (Arabidopsis thaliana) do not require vernalization treatment to flower rapidly, many naturally occurring accessions of Arabidopsis flower very late unless vernalized (Clarke and Dean, 1994; Lee and Amasino, 1995; Michaels and Amasino, 1999; Gazzani et al., 2003). In Arabidopsis, the vernalization requirement is conferred by two dominant genes, FRIGIDA (FRI) and FLOWERING LOCUS C (FLC; Lee et al., 1993; Clarke and Dean, 1994; Michaels and Amasino, 1999; Sheldon et al., 1999; Johanson et al., 2000). FLC encodes a MADS box DNA binding protein that functions as a repressor of the floral integrators, FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) (Michaels and Amasino, 1999; Sheldon et al., 1999; Lee et al., 2000; Samach et al., 2000; Hepworth et al., 2002; Helliwell et al., 2006; Searle et al., 2006). FLC antagonizes the effect of CONSTANS (CO) by directly binding to regulatory regions within FT and SOC1. It appears that FRI contributes to the vernalization requirement solely by activating FLC. FRI encodes a protein with unknown biochemical function (Johanson et al., 2000). Vernalization results in the stable repression of FLC (Michaels and Amasino, 1999; Bastow et al., 2004; Sung and Amasino, 2004) so that floral integrators can be activated when the photoperiod pathway activates CO (Parcy, 2005). Thus, vernalization renders plants to be competent to flower upon exposure to inductive photoperiods in winter annuals and biennials.
Other than FRI, another group of genes involved in FLC activation have been identified from screens for early flowering in certain genotypes or photoperiod conditions. They include EARLY FLOWERING5 (ELF5), ELF7, ELF8, VERNALIZATION INDEPENDENCE3 (VIP3), VIP4, EARLY FLOWERING IN SHORT DAYS4 (ESD4), PHOTOPERIOD INDEPENDENT EARLY1 (PIE1), EARLY FLOWERING IN SHORT DAYS (EFS), and ARABIDOPSIS HOMOLOG OF TRITHORAX1 (ATX1)/ATX2/ARABIDOPSIS TRITHORAX-RELATED7 (ATXR7; Reeves et al., 2002; Zhang and van Nocker, 2002; Noh and Amasino, 2003; He et al., 2004; Noh et al., 2004; Oh et al., 2004; He and Amasino, 2005; Kim et al., 2005; Zhao et al., 2005; Choi et al., 2007; Saleh et al., 2008; Tamada et al., 2009). Some of these genes encode proteins with chromatin modification functions, including components of RNA Polymerase II-associated factor 1 (PAF1) complex (VIP3, VIP4, ELF7, and ELF8), a Histone H3 Lys-36 methyltransferase (EFS), a Histone H3 Lys-4 methyltransferase (ATX1, ATX2, and ATXR7), and a SWR1-related nucleosome remodeling factor (PIE1).
Mitotically stable repression of FLC by vernalization is also achieved by chromatin modifications (Michaels and Amasino, 1999; Bastow et al., 2004; Sung and Amasino, 2004). FLC mRNA expression is repressed during the course of cold exposure, and several repressive histone marks accumulate at FLC chromatin, including methylations at Histone H3 Lys-9 (H3K9) and Histone H3 Lys-27 (H3K27). The accumulation of histone modifications at FLC chromatin depends on the activity of chromatin remodeling complexes. During the course of cold exposure, POLYCOMB REPRESSION COMPLEX2 (PRC2), which has H3K27 methyltransferase activity, is enriched at FLC chromatin (Wood et al., 2006; De Lucia et al., 2008) and establishes the stable repression of FLC through H3K27 methylation. PRC2 biochemically copurifies with members of the VERNALIZATION INSENSITIVE3 (VIN3) family of proteins, including VIN3, VIN3-LIKE1 (VIL1)/VERNALIZATION5 (VRN5), and VIL2/VERNALIZATION LIKE1 (VEL1; Wood et al., 2006; De Lucia et al., 2008).
The vernalization response involves two phases. The first is a cold perception that measures the cumulative time of exposure to cold. Vernalization requires cold exposure over the course of weeks rather than minutes or hours. The second phase is essentially the output of the cold perception. When a sufficient duration of cold has been perceived, a series of changes of gene expression ensue, ultimately leading to the epigenetic repression of FLC. VIN3, which is a repressive chromatin-remodeling component, is induced only after a sufficient duration of cold has been perceived. One of the early molecular events in the vernalization response is the induction of VIN3 by prolonged cold exposure. Upstream of VIN3, there must be a biochemical mechanism to sense cold. However, nothing is known about the upstream event. The induction of VIN3 by cold is unique in that VIN3 induction takes several days of cold, unlike many cold-induced genes, which are induced within hours of cold exposure (Thomashow, 2001). Furthermore, VIN3 mRNA expression is quickly rerepressed once plants are moved to warm temperature.
Interestingly, the induction of VIN3 also involves changes in active histone marks at VIN3 chromatin, including Histone H3 acetylation, Histone H4 acetylation, and Histone H3 Lys-4 trimethylation (H3K4me3; Finnegan et al., 2005; Bond et al., 2009). However, no chromatin remodeling complexes have been identified to have roles in those changes at VIN3 chromatin.
Here, we show that VIN3 is in a constitutively silenced state, which is mediated by the presence of a transposable element (TE)-derived sequence in its promoter region and by the components of repressive complexes, including PRC2 and LHP1. In addition, the full extent of VIN3 induction by vernalization requires components of activating complexes, including PAF1 and EFS. Thus, VIN3 expression is under the influence of chromatin level regulators. Furthermore, VIN3 chromatin is in a transiently bivalent state when VIN3 mRNA is induced, having both a repressive histone mark and an active histone mark at VIN3 chromatin. Our results show that VIN3 is under a constitutively repressed state, which is transiently relieved from repression only when sufficient cold is provided.
RESULTS
A Transposable Element Is Present in the VIN3 Promoter Region, and H3K9 Dimethylation Is Enriched at VIN3 Chromatin Prior to Vernalization
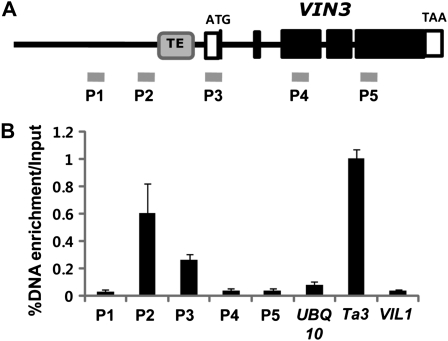
We surveyed possible regulatory sequences in 5′ upstream regions of VIN3. Interestingly, we located a MuDR-type transposable element-derived sequence at −185 to −765 from the transcription start site (Fig. 1A). The presence of a transposable element-derived sequence at VIN3 promoter region has been assigned as AT5TE83600 (www.arabidopsis.org). The sequence showed 55% and 53% nucleotide sequence identities with transposable elements, AT3G29634 and AT5G28495, respectively (Supplemental Fig. S1). The presence of a transposable element-derived sequence raised the possibility that the VIN3 upstream region contains a heterochromatin island by the insertion of a transposable element. It is not unprecedented that an insertion of a transposable element could create local heterochromatin. In fact, some allelic variants of FLC are due to insertions of transposable elements in its first intron (Michaels et al., 2003; Liu et al., 2004). In the case of FLC, two different classes of transposable elements create silenced FLC alleles due to extensive Histone H3 Lys 9 dimethylation (H3K9me2) that originated from the transposable element and spread throughout the entire FLC genomic region (Michaels et al., 2003; Liu et al., 2004). Similarly, we observed a localized enrichment of H3K9me2 around the transposable element into the VIN3 genomic region in a nonvernalized condition (Fig. 1B). This suggests that VIN3 chromatin is in a repressed state prior to the exposure to cold. Previous genomic studies to identify H3K9me2-enriched regions using genomic tiling arrays did not identify the VIN3 promoter region as an H3K9me2-enriched region (Rehrauer et al., 2010; Zhou et al., 2010). It is possible that the level of enrichment at VIN3 promoter is relatively lower than other regions and localized around the transposable element-derived sequence (Fig. 1B).
Figure 1.
H3K9me2 enrichment at VIN3 chromatin. A, Schematic diagram showing the gene structure of VIN3. The solid boxes indicate coding regions, and the open boxes indicate untranslated regions (UTRs). Gray box indicates the transposable element-derived sequence (TE). Solid bars (P1 to P5) indicate regions for primer pairs used for ChIP assays. B, ChIP-quantitative PCR to quantify the relative enrichment of H3K9me2. UBQ10, Ta3, and VIL1 regions were used to compare the levels of H3K9me2 enrichments. Primer sequence information used for ChIP assay is shown in Supplemental Table S1.
VIN3 Is Derepressed in lhp1 and clf;swn Double Mutants
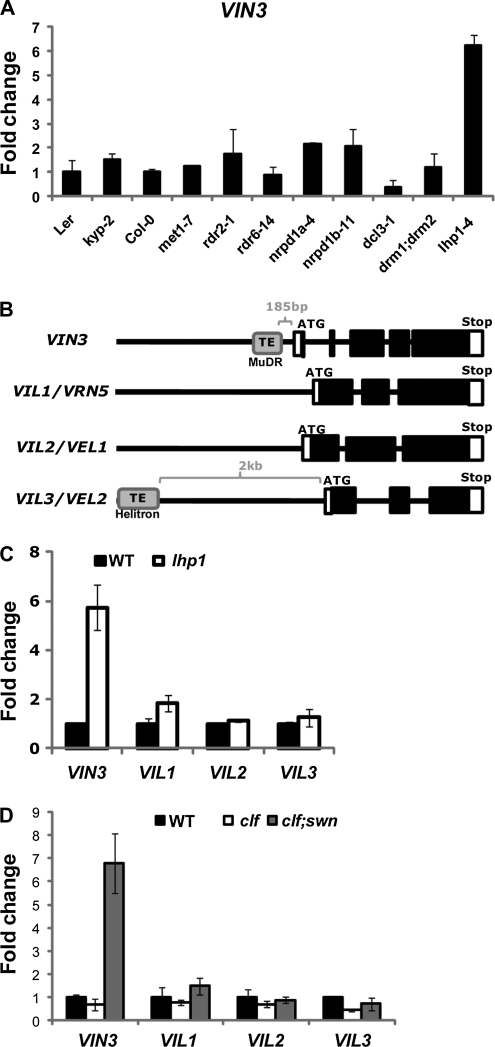
The presence of a transposon-derived sequence at VIN3 promoter region prompted us to test a series of mutants that are involved in gene silencing for their effects on the VIN3 expression. We tested a series of Arabidopsis mutants involved in (1) DNA methylation, met1-7 and drm1;drm2; (2) RNA interference production, rdr2-1, rdr6-14, dcl3-1, nrpd1a-4, and nrpd1b-11; and (3) Histone methylation, kyp-2 and lhp1-4. Among tested mutants, we observed a significantly elevated level of VIN3 mRNA expression prior to the cold exposure in lhp1 mutants (Fig. 2A).
Figure 2.
Involvement of LHP1 and PRC2 in the repression of VIN3 gene in a nonvernalized condition. A, Real-time reverse transcription (RT)-PCR analysis on VIN3 expression between wild types (Col-0 and Ler) and a series of mutants. B, Schematic diagram showing the gene structures of VIN3 family genes (VIN3, VIL1/VRN5, VIL2/VEL1, and VIL3/VEL2). The solid boxes indicate the coding regions, and the open boxes indicate UTRs. C, Real-time RT-PCR analysis on VIN3 family genes between the wild type (WT; FRI-Col) and lhp1-4 mutants in NV condition. D, Real-time RT-PCR analysis on VIN3 expression in the wild type (Col), clf-29, and clf-29/swn-2 mutants in NV condition. C and D, Relative fold change was determined by normalization with the levels of PP2A gene as previously reported (Czechowski et al., 2005). Primer sequence information used for real-time RT-PCR analysis is shown in Supplemental Table S1.
VIN3 exists as a small gene family including VIL1/VRN5, VIL2/VEL1, and VIL3/VEL2. Some members of the gene family also show vernalization-mediated regulation at the transcriptional level (Sung et al., 2006b; Greb et al., 2007). A survey of promoter regions of other members of the family revealed no other members harbor a transposable element in their promoter regions, with an exception of VIL3, which has a Helitron-related transposable element 2 kb upstream of the transcription start site (Fig. 2B). Furthermore, only VIN3 exhibits derepressed expression without the vernalization treatment in lhp1 mutants (Fig. 2C). Taken together, the regulation by the insertion of a transposable element and by LHP1 is specific to VIN3 among the gene family.
LHP1 shows a functional overlap with PRC2 in Arabidopsis (Hennig and Derkacheva, 2009). CURLY LEAF (CLF) and SWINGER (SWN) are two Arabidopsis homologs of E(z), which is a core component of PRC2. To address the functional overlap of PRC2 with LHP1 in the regulation of VIN3, we obtained clf;swn double mutants. Indeed, clf;swn double mutants do have an elevated level of VIN3 mRNA before cold exposure similar to lhp1 (Fig. 2D). clf single mutant alone does not cause the derepression of VIN3, suggesting the functional redundancy between CLF and SWN. Similar to the derepression of VIN3 in lhp1 mutants, mutations in clf;swn only affect VIN3 and not other gene family members (Fig. 2D).
LHP1 and CLF Directly Associate with VIN3 Chromatin But Do Not Dissociate from VIN3 Chromatin When VIN3 Is Induced by Vernalization
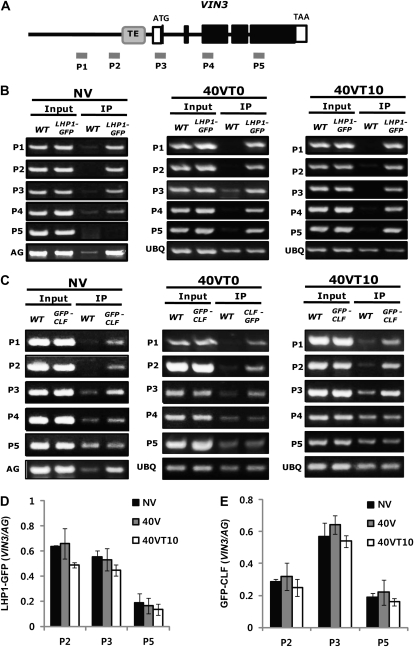
Because lesions in LHP1 or CLF/SWN result in the derepression of VIN3 in a nonvernalized condition, we address whether LHP1 and CLF/SWN directly regulate VIN3 expression through physical association with VIN3 chromatin using chromatin immunoprecipitation (ChIP) assays. Using GFP-tagged LHP1 and GFP-tagged CLF transgenic lines, both of which fully rescue lhp1 and clf mutants, respectively (Schubert et al., 2006; Sung et al., 2006a), we observed strong enrichments of LHP1 and CLF at VIN3 chromatin by ChIP (Fig. 3, B and C). The associations of LHP1 and CLF with VIN3 chromatin appear to be robust throughout the course of vernalization (Fig. 3, B and C). To confirm constitutive associations of LHP1 and CLF with VIN3 chromatin, we compared the relative enrichments of LHP1 and CLF at VIN3 chromatin during the course of vernalization using ChIP followed by real-time quantitative PCR (Fig. 3, D and E). We did not detect significant difference in the enrichments of LHP1 and CLF at VIN3 chromatin even when VIN3 is highly expressed during the cold exposure (Fig. 3, D and E). This shows that LHP1 and CLF are constitutively present at VIN3 chromatin regardless of the expression state of VIN3.
Figure 3.
ChIP assay using LHP1:GFP and GFP:CLF transgenic plants. A, Schematic diagram showing the gene structure of VIN3. Solid boxes indicate the coding regions, and open boxes indicate UTR. Gray box indicates the transposable element-derived sequence (TE). Solid bars (P1 to P5) indicate regions for primer pairs used for ChIP assays. B, ChIP assay using LHP1:GFP transgenic lines. C, ChIP assay using GFP:CLF transgenic line. AGAMOUS (AG) and UBQ10 were used as controls. As an input control, the preimmunoprecipitated DNA after sonication was used (B and C). D, ChIP assay using LHP1:GFP transgenic lines followed by real-time quantitative PCR. E, ChIP assay using GFP:CLF transgenic line followed by real-time quantitative PCR. For quantifications, the relative enrichments compared to input were calculated, and relative enrichments are shown compared to those of the control, AG (D and E). IP, Immunoprecipitation; NV, nonvernalized; 40V and 40VT0, 40 d of vernalization; 40VT10, 40 d of vernalization followed by 10 d of normal growth temperature; WT, wild type.
VIN3 Is Induced Faster by Cold in lhp1 Mutants
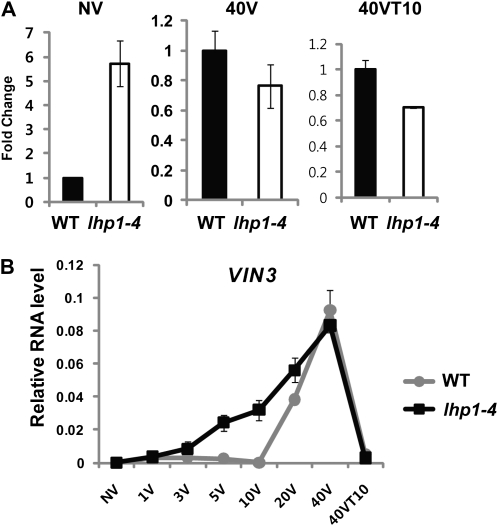
Constitutive association of LHP1 with VIN3 chromatin during the course of vernalization suggests that cold-induced expression of VIN3 must overcomes the repressive activity of LHP1. Although VIN3 is derepressed in lhp1 mutants prior to the cold exposure, the vernalization treatment still significantly up-regulates VIN3 to levels comparable to those of the wild type (Fig. 4A). In addition, derepressed levels of VIN3 in lhp1 or clf;swn double mutants do not fully recapitulate the levels of vernalization-induced VIN3 expression (Figs. 2, C and D, and 4A). Thus, the level of VIN3 induction by vernalization cannot be achieved solely by the removal of LHP1 or CLF/SWN.
Figure 4.
Real-time RT-PCR analysis on VIN3 expression between the wild type (FRI-Col) and lhp1-4 (in FRI-Col) mutants during the course of vernalization. A, Nonvernalized (NV); 40V, 40 d of vernalization; 40VT10, 40 d of vernalization followed by 10 d of normal growth temperature. WT, Wild type. B, Shorter periods of cold exposure. 1V, 1 d of cold; 3V, 3 d of cold; 5V, 5 d of cold; 10V, 10 d of cold; 20V, 20 d of cold. Relative fold change was determined by normalization with the levels of PP2A gene as previously reported (Czechowski et al., 2005). Primer sequence information used for real-time RT-PCR analysis is shown in Supplemental Table S1.
To address the contribution of LHP1 in the VIN3 induction kinetics, we examined the level of VIN3 mRNA expression during the course of vernalization, including shorter periods of cold treatment in lhp1 mutants (Fig. 4B). Interestingly, VIN3 is induced by shorter period of cold exposure in the absence of LHP1 (Fig. 4B). With 5 to 10 d of cold exposure, VIN3 is significantly induced in lhp1 mutants, while no significant induction of VIN3 is observed in the wild type (Fig. 4B). This suggests that the slow kinetics of VIN3 induction in the wild type is at least in part due to the repression by LHP1.
Histone Modification Landscapes Change at VIN3 Chromatin during the Course of Vernalization
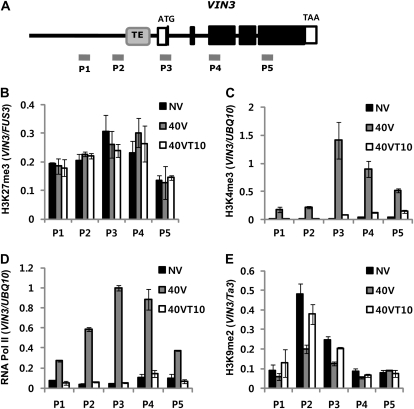
The presence of a transposable element at the promoter region of VIN3 and the involvement of LHP1 and CLF/SWN in the repression of VIN3 led us to examine chromatin modifications at VIN3 chromatin during the course of vernalization (Fig. 5). Histone H3 Lys-27 trimethylation (H3K27me3) is often correlated with gene repression and is mediated by the PRC2 complex. Interestingly, we did not observe any significant change in the level of H3K27me3 at VIN3 chromatin during the course of vernalization (Fig. 5B). This result is consistent with the constitutive association of CLF, which is an H3K27 methyltransferase, with VIN3 chromatin. On the other hand, an active mark, H3K4me3, is enriched when VIN3 is highly expressed during cold exposure (Fig. 5C). Consistent with the active transcription of VIN3, RNA polymerase II (Pol II) enrichment at VIN3 chromatin increases during the cold exposure (Fig. 5D). Thus, during the cold exposure, VIN3 chromatin contains both a repressive mark and an active mark, creating a bivalent state. We observed that H3K9me2 is enriched around VIN3 coding regions that originated from a transposable element prior to the cold exposure (Fig. 1B). To evaluate the dynamics of H3K9me2 enrichment at VIN3 by vernalization, we examined the relative enrichments of H3K9me2 during the course of vernalization. Interestingly, the level of enrichment of H3K9me2 is transiently reduced when VIN3 is highly expressed during cold exposure (Fig. 5E), suggesting that the induction of VIN3 by vernalization overcomes the effect of a transposable element at its promoter region.
Figure 5.
ChIP assays using anti-H3K27me3, H3K9me2, Pol II, and H3K4me3 antibody during the course of vernalization. A, Schematic diagram showing the gene structure of VIN3. Solid boxes indicate the coding regions, and open boxes indicate UTRs. Gray box indicates the transposable element-derived sequence (TE). Solid bars (P1 to P5) indicate regions for primer pairs used for ChIP assays. B, ChIP assays followed by real-time quantitative PCR using the anti-H3K27me3 antibody during the course of vernalization. For quantifications, the relative enrichments compared to input were calculated, and relative enrichments are shown compared to those of the control, FUS3. A FUS3 region was used as a positive control as previously reported (Makarevich et al., 2006). NV, Nonvernalized. C, ChIP assay followed by real-time quantitative PCR using the anti-H3K4me3 antibody during the course of vernalization. For quantifications, the relative enrichments compared to input were calculated, and relative enrichments are shown compared to those of the control, UBQ10. D, ChIP assay followed by real-time quantitative PCR using the anti-Pol II antibody (8WG16) during the course of vernalization. The Pol II antibody (8WG16) detects both phosphorylated and nonphosphorylated forms of RNA Pol II. For quantifications, the relative enrichments compared to input were calculated, and relative enrichments are shown compared to those of the control, UBQ10. E, ChIP assay followed by real-time quantitative PCR using the anti-H3K9me2 antibody during the course of vernalization. For quantifications, the relative enrichments compared to input were calculated, and relative enrichments are shown compared to those of the control, Ta3. Primer sequence information used for ChIP assay is shown in Supplemental Table S1. NV, Nonvernalized; 40V, 40 d of vernalization; 40VT10, 40 d of vernalization followed by 10 d of normal growth temperature.
Activating Components Are Involved in the Induction of VIN3 by Vernalization
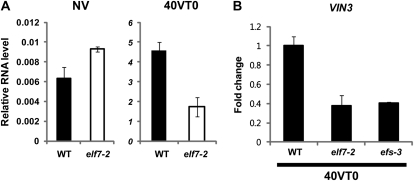
It is interesting to note that common repressive components (i.e. LHP1 and CLF/SWN) are involved in the repressions of both VIN3 and its target, FLC. Furthermore, the contributions of PRC2 components, CLF and SWN, to the repressed state of VIN3 suggest that other FLC regulators may also be involved in the regulation of VIN3. Particularly, increased enrichment of H3K4me3 at VIN3 chromatin during cold exposure indicates that H3K4me3-mediated activating complexes are also involved in the regulation of VIN3. To test the involvement of such activating components, we examined the VIN3 expression pattern in elf7 mutants. ELF7 encodes a homolog of PAF1, which commonly associates with Trithorax-mediated gene activations. Interestingly, the induced levels of VIN3 by vernalization are significantly lower in elf7 mutants than in the wild type (Fig. 6A). Similar reduction in the induced level of VIN3 is observed in efs mutants (Fig. 6B), another mutation in activating components for FLC. However, it should be noted that VIN3 is still induced by 40 d of vernalizing cold treatment both in elf7 and efs mutants (Fig. 6). Thus, activating components that are required for the FLC expression are also necessary for the full extent of VIN3 induction by vernalization, in which VIN3 expression must overcome the repressive effects of LHP1 and the PRC2 complex.
Figure 6.
ELF7 and EFS are required for the full induction of VIN3 by vernalization. A, Real-time RT-PCR analysis on VIN3 expression in the wild type (WT; Col-0) and elf7-2 mutants in nonvernalized (NV) and 40 d of vernalization (40VT0) conditions. B, Real-time RT-PCR analysis on VIN3 expression in the wild type, elf7-2, and efs-3 mutants during vernalization. Relative fold change was determined by normalization with the levels of PP2A as previously reported (Czechowski et al., 2005).
CONCLUSION
We identified multiple factors that contribute to the repressed state of VIN3 prior to cold exposure. We observed derepression of VIN3 in the absence of components of PRC2, CLF and SWN, and LHP1. In addition, a transposable element at the VIN3 promoter region appears to contribute the repressed state of VIN3 prior to cold.
Interestingly, there is no significant change in the level of H3K27me3, a repressive chromatin mark, on VIN3 chromatin, even when VIN3 is highly expressed (Fig. 4B). This suggests that the induction of VIN3 can overcome the effect of H3K27me3, a representative repressive histone mark. On the other hand, an active histone mark, H3K4me3, is highly enriched at the 5′ end of VIN3 chromatin during the cold exposure (Fig. 4C). Thus, when VIN3 is induced during the cold exposure, VIN3 chromatin appears to become a bivalent domain (Bernstein et al., 2006), containing both a repressive mark, H3K27me3, and an active mark, H3K4me3. The bivalent domains are proposed to be characteristic of genes that are silent but poised to be activated or vice versa and thus provide flexibility of gene expression in undifferentiated cells (Azuara et al., 2006; Bernstein et al., 2006). Here, we observed a bivalent domain that is transiently activated by an environmental cue, winter cold.
PRC2 and LHP1 are constitutively associated with VIN3 chromatin regardless of the expression state of VIN3. However, the lack of repressive components (CLF/SWN or LHP1) cannot recapitulate the induced level of VIN3 expression without vernalization. It is possible that the induction of VIN3 by cold is due to the combined contributions of both derepression of transposons and the removal of LHP1-PRC2 from VIN3 chromatin by cold. However, examination of the LHP1 enrichment at VIN3 chromatin indicates that LHP1 enrichment at VIN3 chromatin is static throughout the course of vernalization (Fig. 3, B and D). Accordingly, a similar finding was observed for the CLF enrichment at VIN3 chromatin (Fig. 3, C and E). This is consistent with the lack of changes at the level of enrichment of H3K27me3 at VIN3 chromatin during the course of vernalization (Fig. 5B).
The derepressed VIN3 in lhp1 mutants does show ectopic induction by cold. This suggests that the slow kinetic of VIN3 induction by vernalization is due to in part by the LHP1-mediated repression. Furthermore, activating chromatin remodeling complexes are also necessary for the full extent of VIN3 induction by vernalization (Fig. 6). Thus, during the earlier cold exposure, activating complexes (i.e. PAF1 and EFS) need to overcome the repressive effect by LHP1 and PRC2. This may explain, at least in part, the slow kinetics of the VIN3 induction by cold. However, it is worth noting that the full extent of VIN3 induction in lhp1 mutants still requires more than 20 d of cold. In addition, the rerepression of VIN3 after the exposure to vernalizing cold still occurs in the absence of LHP1 (Fig. 4B).
Activating complex components (i.e. PAF1 and EFS) are required for the full extent of VIN3 induction by vernalization (Fig. 6). PAF1 often functionally associates with Trithorax-like components for the FLC activation (Tamada et al., 2009). Furthermore, the enrichment of H3K4me3 at VIN3 chromatin is transiently increased when VIN3 is induced. Thus, it is likely that Trithorax-like proteins are also involved in the induction of VIN3 (Fig. 7). Although activating complex components are necessary for the full extent of VIN3 induction by vernalization, the VIN3 induction by vernalization still occurs in the absence of either PAF1 or EFS. Thus, it is probable that there are unidentified cold-induced activators and/or cold-repressed repressors involved in the induction of VIN3 by vernalization, which could be enhanced by activating complex components (Fig. 7).
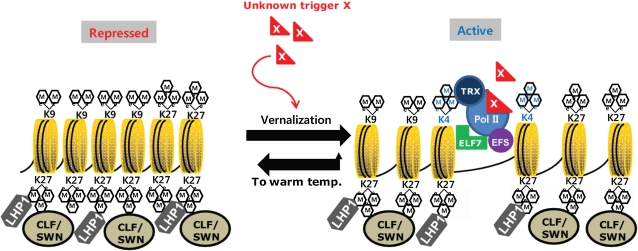
Figure 7.
Schematic model on the regulation at VIN3 chromatin by vernalization. In an NV condition, LHP1 and CLF-containing PRC2 complexes are necessary for the repressed state of VIN3. H3K9me2 and H3K27me3 are enriched at VIN3 chromatin prior to vernalization. When plants are exposed to a prolonged cold (i.e. winter), unknown upstream components (X) induce VIN3, although LHP1 and PRC2 complexes are still enriched at VIN3 chromatin. The induction of VIN3 causes the decrease in enrichments of H3K9me2 mark and the increase in enrichments of H3K4me3 mark at the transcription start site of VIN3. The increased enrichments of H3K4me3 are in part due to the activity of activating complexes (i.e. PAF1, EFS, and Trithorax-like proteins). Once plants return to warm growth temperature, H3K4me3 mark decreases and H3K9me2 mark increases again at VIN3 chromatin in the absence of an unknown trigger (X). [See online article for color version of this figure.]
In conclusion, PRC2-LHP1 and the transposable element affects the repressed state of VIN3 prior to cold and the effects of PRC2-LHP1 and that of the transposable element need to be overcome by unknown factors (X) along with the activity of activating chromatin remodeling complexes (Fig. 7).
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) Columbia-0 (Col-0) and Landsberg erecta (Ler) were used as the wild type in this study. clf-29 (Salk_021003), lhp1-4 (referred also as tfl2-2), clf-29;swn-2, rdr2-1, rdr6-14, nrpd1a-4, nrpd1b-11, drm1;drm2, met1-7 (salk_076522), kyp-2 (Ler background), elf7-2, and efs-3 were used in this study. Standard growth conditions were 22°C under illumination with white fluorescent light. The photoperiodic cycle was 16 h light/8 h dark (long day) or 8 h light/16 h dark (short day). For vernalization treatments, seeds were surface sterilized, placed on agar-solidified germination medium, and grown for 7 d at 22°C growth chamber and transferred to 4°C for 40 d under short-day photoperiods.
RNA Extraction and Quantitative Real-Time PCR Analysis
Seven-day-old seedling plants were harvested for RNA extraction. Total RNA was extracted using the Qiagen Plant RNeasy mini kit. Before reverse transcription, total RNA was treated for 30 min at 37°C with RNase-free DNase I (Invitrogen) to eliminate contaminated genomic DNA, and then total RNA (2 to 3 μg) was used for cDNA synthesis using M-MLV reverse transcriptase (Promega). Quantitative real-time PCR reaction was done using SYBR green dye reaction mixture (Applied Biosystems) according to the manufacturer’s instructions. Real-time PCR reaction was performed on a 7900HT Fast Real-Time PCR system (Applied Biosystems). PCR primers used for quantitative real-time PCR analysis are listed in Supplemental Table S1. The relative expression level of each gene was normalized using PP2A as described previously (Czechowski et al., 2005).
ChIP Assay
ChIP experiments were performed as previously reported (Johnson et al., 2002) with some modifications. Seven-day-old seedling plants were used for ChIP analysis. To elute DNA from immunoprecipitated complexes, the QIAquick PCR purification kit (Qiagen) was used. Antibodies recognizing H3K9me2 (Abcam catalog number ab1220), H3K27me3 (Abcam catalog number ab6002), H3K4me3 (Abcam catalog number ab8580), and Pol II (Abcam catalog number ab817) were purchased and used for ChIP assay with 10 μg per each reaction. All ChIP assays were done at least three times from at least two biological replicates and produced similar results. The quantitative real-time PCR reaction was done using SYBR green dye reaction mixture (Applied Biosystems) according to the manufacturer’s instructions. Real-time PCR reaction was performed on a 7900HT Fast Real-Time PCR system. For primer sequences used in ChIP assay, refer to Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. TE sequence alignment.
Supplemental Table S1. Primer sequence information.
Supplementary Material
Acknowledgments
We thank Jessica Sisavath, Evelyn Joo, Shahrin Tina, and Jorge Barajas for technical assistance.
References
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. (2006) Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bond DM, Wilson IW, Dennis ES, Pogson BJ, Jean Finnegan E. (2009) VERNALIZATION INSENSITIVE 3 (VIN3) is required for the response of Arabidopsis thaliana seedlings exposed to low oxygen conditions. Plant J 59: 576–587 [DOI] [PubMed] [Google Scholar]
- Choi K, Park C, Lee J, Oh M, Noh B, Lee I. (2007) Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134: 1931–1941 [DOI] [PubMed] [Google Scholar]
- Clarke JH, Dean C. (1994) Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Mol Gen Genet 242: 81–89 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AM, Greb T, Dean C. (2008) A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci USA 105: 16831–16836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis ES, Peacock WJ. (2007) Epigenetic regulation of flowering. Curr Opin Plant Biol 10: 520–527 [DOI] [PubMed] [Google Scholar]
- Finnegan JE, Kovac KA, Jaligot E, Sheldon CC, James Peacock W, Dennis ES. (2005) The downregulation of FLOWERING LOCUS C (FLC) expression in plants with low levels of DNA methylation and by vernalization occurs by distinct mechanisms. Plant J 44: 420–432 [DOI] [PubMed] [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C. (2003) Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol 132: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T, Mylne JS, Crevillen P, Geraldo N, An H, Gendall AR, Dean C. (2007) The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr Biol 17: 73–78 [DOI] [PubMed] [Google Scholar]
- He Y, Amasino RM. (2005) Role of chromatin modification in flowering-time control. Trends Plant Sci 10: 30–35 [DOI] [PubMed] [Google Scholar]
- He Y, Doyle MR, Amasino RM. (2004) PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev 18: 2774–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES. (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46: 183–192 [DOI] [PubMed] [Google Scholar]
- Hennig L, Derkacheva M. (2009) Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet 25: 414–423 [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G. (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21: 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Johnson L, Cao X, Jacobsen S. (2002) Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol 12: 1360–1367 [DOI] [PubMed] [Google Scholar]
- Kim SY, He Y, Jacob Y, Noh YS, Michaels S, Amasino R. (2005) Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17: 3301–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I. (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Amasino RM. (1995) Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol 108: 157–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Bleecker A, Amasino R. (1993) Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol Gen Genet 237: 171–176 [DOI] [PubMed] [Google Scholar]
- Liu J, He Y, Amasino R, Chen X. (2004) siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes Dev 18: 2873–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O, Goodrich J, Grossniklaus U, Kohler C. (2006) Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep 7: 947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, He Y, Scortecci KC, Amasino RM. (2003) Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci USA 100: 10102–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh YS, Amasino RM. (2003) PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15: 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh YS, Bizzell CM, Noh B, Schomburg FM, Amasino RM. (2004) EARLY FLOWERING 5 acts as a floral repressor in Arabidopsis. Plant J 38: 664–672 [DOI] [PubMed] [Google Scholar]
- Oh S, Zhang H, Ludwig P, van Nocker S. (2004) A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 16: 2940–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F. (2005) Flowering: A time for integration. Int J Dev Biol 49: 585–593 [DOI] [PubMed] [Google Scholar]
- Reeves PH, Murtas G, Dash S, Coupland G. (2002) early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development 129: 5349–5361 [DOI] [PubMed] [Google Scholar]
- Rehrauer H, Aquino C, Gruissem W, Henz SR, Hilson P, Laubinger S, Naouar N, Patrignani A, Rombauts S, Shu H, et al. (2010) AGRONOMICS1: A new resource for Arabidopsis transcriptome profiling. Plant Physiol 152: 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Yilmaz M, Le O, Hou G, Sadder M, Al-Abdallat A, Xia Y, Lu G, Ladunga I, et al. (2008) The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell 20: 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J. (2006) Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Krober S, Amasino RA, Coupland G. (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES. (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Amasino RM. (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164 [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM. (2005) Remembering winter: Toward a molecular understanding of vernalization. Annu Rev Plant Biol 56: 491–508 [DOI] [PubMed] [Google Scholar]
- Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM. (2006a) Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet 38: 706–710 [DOI] [PubMed] [Google Scholar]
- Sung S, Schmitz RJ, Amasino RM. (2006b) A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev 20: 3244–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada Y, Yun JY, Woo SC, Amasino RM. (2009) ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell 21: 3257–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. (2001) So what’s new in the field of plant cold acclimation? Lots! Plant Physiol 125: 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA. (2006) The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA 103: 14631–14636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, van Nocker S. (2002) The VERNALIZATION INDEPENDENCE 4 gene encodes a novel regulator of FLOWERING LOCUS C. Plant J 31: 663–673 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Yu Y, Meyer D, Wu C, Shen WH. (2005) Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol 7: 1256–1260 [DOI] [PubMed] [Google Scholar]
- Zhou J, Wang X, He K, Charron JB, Elling AA, Deng XW. (2010) Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol Biol 72: 585–595 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.