Abstract
At the end of the cell cycle, the nascent cross wall is laid down within a transient membrane compartment referred to as the cell plate. Tethering factors, which act by capturing vesicles and holding them in the vicinity of their target membranes, are likely to play an important role in the first stages of cell plate assembly. Factors required for cell plate biogenesis, however, remain to be identified. In this study, we used a reverse genetic screen to isolate tethering factors required for cytokinesis in Arabidopsis (Arabidopsis thaliana). We focused on the TRAPPI and TRAPPII (for transport protein particle) tethering complexes, which are thought to be required for the flow of traffic through the Golgi and for trans-Golgi network function, as well as on the GARP complex, thought to be required for the tethering of endocytotic vesicles to the trans-Golgi network. We found weak cytokinesis defects in some TRAPPI mutants and strong cytokinesis defects in all the TRAPPII lines we surveyed. Indeed, four insertion lines at the TRAPPII locus AtTRS120 had canonical cytokinesis-defective seedling-lethal phenotypes, including cell wall stubs and incomplete cross walls. Confocal and electron microscopy showed that in trs120 mutants, vesicles accumulated at the equator of dividing cells yet failed to assemble into a cell plate. This shows that AtTRS120 is required for cell plate biogenesis. In contrast to the TRAPP complexes, we found no conclusive evidence for cytokinesis defects in seven GARP insertion lines. We discuss the implications of these findings for the origin and identity of cell plate membranes.
Cytokinesis is the division of the cytoplasm following nuclear division. This process differs considerably in plant and animal cells, due predominantly to the presence of the plant cell wall. In animal cells, a contractile ring pinches a dividing cell in two. In plants, by contrast, cross walls are deposited from the middle out within a transient membrane compartment, the cell plate, which is assembled during the anaphase-to-telophase transition. In addition to extensive membrane trafficking and cross wall deposition, plant cytokinesis requires the assembly of two plant-specific cytoskeletal structures, the preprophase band (PPB) and the phragmoplast (Assaad, 2001a). The PPB is a cortical ring of microtubules and actin filaments that marks the division site (Van Damme, 2009). The phragmoplast is a large array consisting of both microtubules and actin filaments, which delivers vesicles to the cell equator and guides the expanding cell plate to the division sites formerly marked by the PPB (Samuels et al., 1995; Seguí-Simarro et al., 2004; Van Damme, 2009).
A genetic dissection of plant cytokinesis has identified a relatively large proportion of trafficking genes (Jürgens, 2005). The cellular processes underlying vesicle traffic are largely conserved in all eukaryotic cells and can be broken down into six steps: budding, transport, tethering, docking, fusion, and recycling (Mellman and Warren, 2000; Assaad, 2001b; Assaad, 2009). With the exception of factors required for tethering, players potentially required for each of these six steps have been identified as being involved in plant cytokinesis. Budding of vesicles from donor membranes requires the action of ARF GTPases. These are encoded by a large family with 45 members in Arabidopsis (Arabidopsis thaliana), of which at least one (TITAN5) has been shown to play a role in mitosis (McElver et al., 2000). A number of motor proteins and microtubule-associated proteins (for review, see Jürgens, 2005) have been identified that could play a role in the transport of cell plate vesicles to the equator of a dividing cell. Membrane fusion events are mediated by the assembly of a four-helix bundle in which one helix is derived from an R (or v) SNARE, one from a Q (or t) SNARE, and two additional helices are provided by a protein referred to as SNAP25 (Mellman and Warren, 2000; Assaad, 2009). To date, two Q-SNAREs, KNOLLE and NPSN11, as well as a SNAP25 homolog (SNAP33) have been shown to play a role in cytokinesis (Lukowitz et al., 1996; Heese et al., 2001; Zheng et al., 2002), but no R-SNARE has been described. A Sec1/Munc18 protein, KEULE, as well as two Rab GTPases have been implicated in plant cytokinesis, and these could act to regulate fusion events at the cell plate in space and in time as well as to ensure the specificity and fidelity of membrane traffic (Assaad et al., 2001; Chow et al., 2008). Dynamin-related proteins are thought to constrict and shape the thin tubular membrane networks that characterize the initial stages of cell plate formation (Seguí-Simarro et al., 2004; Fujimoto et al., 2008). Finally, ESCRT (for endosomal sorting complexes required for transport) proteins involved in endosomal sorting and protein degradation have been shown to be required for cytokinesis (Spitzer et al., 2006, 2009).
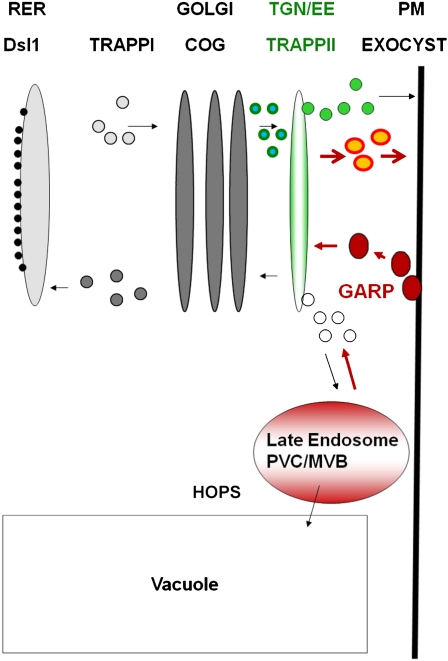
Notable for their absence from the above list are tethering factors. Tethering is a process that brings and holds vesicles in close proximity to their target membranes. There are two classes of tethering factors. The first class consists of long coiled-coil proteins, and the second class consists of large multisubunit complexes (Grosshans et al., 2006; Cai et al., 2007). Seven multisubunit tethering complexes have been conserved among all eukaryotes (Koumandou et al., 2007). Each of these tethering complexes resides on a specific cellular compartment and mediates a specific series of membrane fusion events in the secretory pathway. Thus, based on the yeast literature and as depicted in Figure 1, the TRAPPI (for transport protein particle) complex would mediate endoplasmic reticulum (ER)-to-Golgi, COG intra-Golgi, and TRAPPII exit from the Golgi as well as trans-Golgi network (TGN) functions. From the trans-Golgi, secretion to the plasma membrane would be mediated by the Exocyst complex, whereas vesicles destined for the prevacuolar compartment and the vacuole would be tethered by the HOPS (or VPS) complex. The tethering of early or late endosomes to the TGN would be mediated by the GARP complex. Finally, retrograde traffic from the Golgi to the ER would be mediated by the Dsl1 complex on the ER membrane (Koumandou et al., 2007). Studies on the GARP, HOPS, VPS, and Exocyst complexes in plants (see below) support this general model (Fig. 1).
Figure 1.
Conserved tethering complexes and the plant secretory pathway. Based on the yeast literature and as adapted from Koumandou et al. (2007), the TRAPPI complex would mediate ER-to-Golgi traffic and the COG complex would mediate intra-Golgi traffic; the TRAPPII complex would mediate exit from the Golgi as well as TGN functions. The TRAPPII complex is thought to reside on the TGN and to tether Golgi-derived as well as endocytotic vesicles in yeast (Cai et al., 2005). The GARP complex is also thought to reside on the TGN, where it tethers early (large, red) and late (small, white) endocytotic vesicles (Conibear et al., 2003; Quenneville et al., 2006). From the trans-Golgi, secretion to the plasma membrane would be mediated by the Exocyst complex, whereas vesicles destined for the prevacuolar compartment and the vacuole would be tethered by the HOPS (or VPS) complex. Finally, retrograde traffic from the Golgi to the ER would be mediated by the Dsl1 complex on the ER membrane. The TGN has been shown to function as an early endosome in plants (Dettmer et al., 2006; Lam et al., 2007). It thereby functions as a sorting station, being the first point of entry for vesicles coming from the plasma membrane (endocytotic vesicles; red) and delivering secretory vesicles (light green) and recycling endosomes (orange) to the plasma membrane as well as clathrin-coated vesicles to the vacuole. The cell plate is thought to be a specialized TGN compartment (Dettmer et al., 2006; Chow et al., 2008). As such, it could tether both Golgi-derived (small, blue-green) as well as endocytotic (red) vesicles. It follows that the TRAPPII complex may play a critical role in cytokinesis. We also predicted that if endocytosis contributes significant membranes to the cell plate, GARP mutants might be impaired in cytokinesis as well. MVB, Multivesicular body; PM, plasma membrane; RER, rough endoplasmic reticulum. See text for a more extensive description and for further references.
Among the plant tethering complexes, the HOPS/VPS, GARP, and Exocyst complexes have been the most extensively studied. In Arabidopsis, the HOPS/VPS complex resides on the vacuole and prevacuolar compartment and is required for vacuolar biogenesis (Rojo et al., 2001, 2003). The Exocyst is encoded by a number of single and expanded gene families in Arabidopsis and has been shown to play a role in pollen and root hair tip growth and in hypocotyl elongation (Cole et al., 2005; Synek et al., 2006; Hála et al., 2008; Chong et al., 2010). In addition, the Exocyst has recently been shown to be required in the stigma for the acceptance of compatible pollen in both Brassica and Arabidopsis (Samuel et al., 2009). Some Arabidopsis GARP mutants have also been implicated in pollen tip growth (Lobstein et al., 2004; Guermonprez et al., 2008) as well as in resistance to heat and osmotic stress (Lee et al., 2006). The TRAPPI and -II complexes, however, have not been studied in plants to date. The TRAPPI complex consists of seven subunits, and the TRAPPII complex consists of the TRAPPI complex and three additional subunits in yeast (Kim et al., 2006; Sacher et al., 2008). All seven TRAPPI and two TRAPPII subunits are conserved in Arabidopsis (Table I).
Table I. TRAPP and GARP subunits and their Arabidopsis homologs.
The amino acid conservation between the Arabidopsis proteins and their yeast and mammalian homologs is given in the last four columns. Alignment data were obtained from pairwise, global alignments of the protein sequences by EMBOSS (Rice et al., 2000 [Blossom62; gap open, 10.0; gap extend, 0.5]). Our phylogenetic analyses are consistent with the published data of Koumandou et al. (2007) and of Cox et al. (2007). % Sim., Percent similar; % Ident., percent identical. Dashes indicate that the yeast gene has no homolog in mammals or in Arabidopsis, or that the column header does not apply.
| Yeast Subunit | Mammalian Subunit | TRAPP Complex | Arabidopsis Homolog | GenBank Accession No. |
Saccharomyces cerevisiae |
Homo sapiens |
||
| % Sim. | % Ident. | % Sim. | % Ident. | |||||
| Bet5 | C1 | I and II | At1g51160 | 841539 | 39.5 | 23.1 | 53.9 | 37.1 |
| Trs20 | C2 | I and II | At2g20930 | 816627 | 49.2 | 31.6 | 69.3 | 49.3 |
| Bet3 | C3 | I and II | At5g54750 | 835565 | 67.0 | 42.8 | 70.4 | 50.0 |
| Trs23 | C4 | I and II | At5g02280 | 831680 | 38.5 | 24.3 | 46.8 | 33.2 |
| Trs31 | C5 | I and II | At5g58030 | 835915 | 37.0 | 26.3 | 65.0 | 45.7 |
| Trs33 | C6 | I and II | At3g05000 | 819661 | 32.2 | 20.3 | 58.2 | 42.4 |
| Trs85 | KIAA1012 | I and II | At5g16280 | 831488 | 17.8 | 9.3 | 40.2 | 23.7 |
| Trs65 | – | II | – | – | – | – | – | – |
| Trs120 | C9 | II | At5g11040 | 830971 | 33.2 | 18.6 | 36.2 | 20.0 |
| Trs130 | C10 | II | At5g54440 | 835532 | 34.6 | 17.3 | 31.6 | 19.1 |
| Vps52 | Vps52/SACM2L | – | At1g71270 | 843471 | 38.0 | 19.9 | 56.3 | 33.6 |
| Vps53 | Vps53 | – | At1g50500 | 841472 | 37.8 | 18.8 | 51.3 | 30.9 |
| Vps54 | Vps54 | – | At4g19490 | 827690 | 31.4 | 17.2 | 41.5 | 23.8 |
Several questions pertaining to the membrane dynamics of plant cytokinesis are still open. First, the origin of the cell plate is currently debated. Some reports suggest that Golgi-derived vesicles build the cell plate (Reichardt et al., 2007; Chow et al., 2008), others that extensive endocytosis contributes cell wall components as well as enzymes required for polysaccharide biosynthesis to emerging cross walls (Dhonukshe et al., 2006). The question that follows is whether the cell plate arises via the homotypic fusion of like vesicles or whether it arises via heterotypic fusion between different populations of vesicles. Second, little is known about the biogenesis of the cell plate. As tethering factors promote the first contact between donor and acceptor membranes and define specific membrane compartments and trafficking steps, identifying which tethering factors are required for cytokinesis could help answer some of the questions outlined above.
In this study, we used a reverse genetic approach to identify tethering factors required for cytokinesis in Arabidopsis. We focused on the TRAPPI, TRAPPII, and GARP tethering complexes because they mediate the flow of traffic through the Golgi apparatus and from the cell surface. Both the TRAPPII and GARP complexes have been shown to reside on the TGN in yeast (Conibear et al., 2003; Cai et al., 2005; Quenneville et al., 2006). Whereas the TRAPPII complex is required for both exocytosis and endocytosis, the GARP complex has only been implicated in endocytosis (Conibear et al., 2003; Cai et al., 2005; Quenneville et al., 2006). The TGN has been shown to function as an early endosome in plants (Dettmer et al., 2006; Lam et al., 2007; Wang et al., 2010). It thereby functions as a sorting station, being the first point of entry for vesicles coming from the plasma membrane and delivering secretory vesicles and recycling endosomes to the plasma membrane (Fig. 1) as well as clathrin-coated vesicles to the vacuole. The cell plate is thought to be a TGN-derived compartment (Dettmer et al., 2006; Chow et al., 2008). As such, it could tether both Golgi-derived and endocytotic vesicles (Fig. 1). If the cell plate is assembled from both Golgi-derived and endocytotic vesicles, we predicted that the TRAPP and GARP complexes might play equally important roles in its biogenesis.
RESULTS
Screening Insertion Mutants in Arabidopsis Tethering Factors for Cytokinesis Defects
All the insertion lines surveyed in this study are listed in Table II. Insertion lines were available for four TRAPPI subunits: Bet3, Bet5, Trs31, and Trs33. The TRAPPII complex consists of the TRAPPI complex and three additional subunits in yeast, of which two (Trs120 and Trs130) are encoded by unique genes in Arabidopsis. Four insertion lines were surveyed for Trs120. The GARP complex consists of four subunits in yeast and animal cells. Of these, only three, Vps52, Vps53, and Vps54, are encoded by unique genes in the Arabidopsis genome. Insertion lines were obtained for all three loci. T-DNA flanking sequences were amplified by PCR on genomic DNA isolated from individual soil-grown plants (for an example, see Supplemental Fig. S1). We were able to confirm the insertion sites by PCR analysis for all the lines listed in Table II.
Table II. Insertion lines used in this study.
n = refers to the total number of individuals analyzed. Asterisks indicate that homozygous mutant individuals were recovered.
| Locus | Polymorphism | Position in Gene | Accession No. | Allele | Cytokinesis Defectivea | Transmissionb |
| TRAPPI | ||||||
| Bet3 | GABI_318C08 | Fourth intron | N430464 | bet3-1 | 0% (n = 200) | 7.6% (n = 66) |
| Bet5 | SALK_099482 | Second intron | N599482 | bet5-1 | 1% (n = 192) | 15.2% (n = 79) |
| SAIL_634_G07 | Second intron | N827313 | bet5-2 | 1% (n = 98) | 67.2%* (n = 64) | |
| Trs31 | FLAG_488E06 | Third intron | EWSTV26T3 | trs31-1 | 1% (n = 211) | 13.6% (n = 59) |
| Trs33 | SALK_109244 | Third exon | N609244 | trs33-1 | 5.7% (n = 614) | 55.6% (n = 36) |
| SALK_109724 | Third intron | N609724 | trs33-2 | 2.9% (n = 104) | 15.9% (n = 69) | |
| TRAPPII | ||||||
| Trs120 | SALK_125246 | First intron | N625246 | trs120-1 | 6.3% (n = 221) | 11.8% (n = 34)c |
| SALK_021904 | First intron | N521904 | trs120-2 | 8.4% (n = 455) | 7.7% (n = 78) | |
| SALK_111574 | Seventh exon | N611574 | trs120-3 | 11% (n = 349) | 13.1% (n = 107) | |
| SAIL_1285_D07 | Seventh intron | N879232 | trs120-4 | 11% (n = 617) | 23.8% (n = 105) | |
| GARP | ||||||
| Vps52 | SALK_055433 | 16th exon | N555433 | vps52-2d | 0% (n = 169) | 50% (n = 56) |
| Vps53 | SAIL_87_D06 | Fifth intron | N870946 | vps53-2 | 0% (n = 540) | 27.5% (n = 69) |
| GABI_400C01 | Seventh exon | N344145 | vps53-3 | 0% (n = 620) | 82% (n = 62) | |
| SAIL_117_D11 | 13th intron | N871277 | vps53-4 | 0% (n = 221) | 50.8% (n = 59) | |
| GABI_463D10 | 23rd exon | N344181 | vps53-5 | 0% (n = 600) | 87.8%* (n = 49) | |
| Vps54 | SALK_036485 | First exon | N536485 | vps54-1d | 0% (n = 196) | 25.5% (n = 55) |
| SALK_062261 | Eighth exon | N562261 | vps54-2d | 0% (n = 340) | 39.1% (n = 69) | |
Cytokinesis-defective mutant seedlings on the plate in the progeny of a selfed hemizygous line (%). In the absence of defects in transmission of the mutant alleles, 25% would be expected.
Hemizygous or homozygous segregants in the progeny of a hemizygous line grown on soil (%). In the absence of defects in transmission of the T-DNA insertion, and in case of lethality, 67% would be expected. All numbers are based on PCR analysis of genomic DNA isolated from soil-grown individuals (Supplemental Fig. S1), except as noted in subsequent footnotes.
For trs120-1, we scored cytokinesis-defective mutants in the progeny of individual lines.
These alleles have been described by Guermonprez et al. (2008).
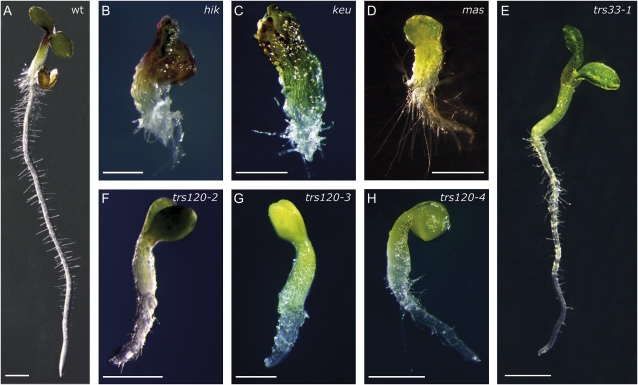
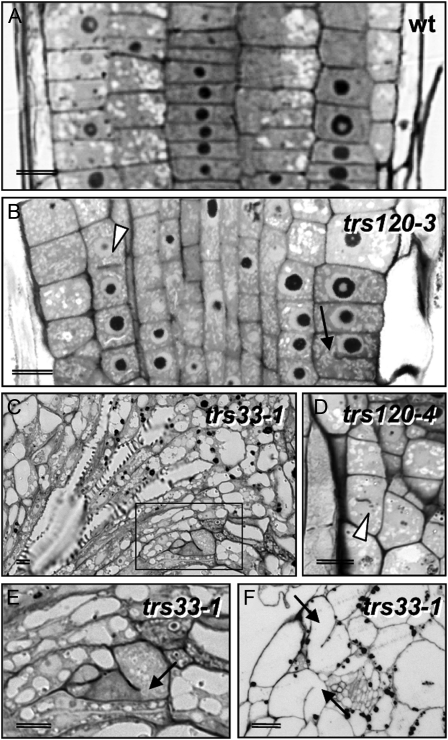
We plated all the lines and surveyed seedlings for cytokinesis-defective phenotypes. The strongest cytokinesis-defective mutants described to date in the plant literature are typically seedling lethal (Söllner et al., 2002, and refs. therein). Therefore, we specifically screened for the canonical seedling-lethal cytokinesis-defective phenotype, which includes a reduced growth of the shoot and root meristems, a rough surface layer with enlarged cells, and a somewhat bloated general appearance (Söllner et al., 2002). We found no evidence of cytokinesis defects in the progeny of seven GARP insertion lines, even though transmission rates were reduced in five of these lines (Table II; reduced transmission has also been reported for GARP mutants by Guermonprez et al. [2008]). In TRAPPI lines, we occasionally observed an attenuated version of the canonical cytokinesis-defective phenotype (Fig. 2E; Table II). The TRAPPII lines segregated 6% to 11% cytokinesis-defective seedlings (Fig. 2, F–H; Table II), whereby seedling morphogenesis was not as severely disrupted in TRAPPII mutants as it was for hinkel and keule (Fig. 2, compare B and C with F–H), which have some of the more extreme phenotypes (Assaad et al., 1996; Söllner et al., 2002; Strompen et al., 2002). Histological sections confirmed that TRAPPI and -II mutant seedlings were cytokinesis defective, as inferred from the presence of cell wall stubs and floating walls in the mutants (Fig. 3). As the canonical cytokinesis-defective mutant phenotype occurred more reproducibly and reliably in TRAPPII than in TRAPPI lines, we focused on the TRAPPII complex. An insertion mutant in the TRAPPII gene Trs130 was found to be allelic to club-1 (Söllner et al., 2002); therefore, we named it trs130/club-2. club mutants, which were identified as being cytokinesis defective in a forward genetic screen, are described elsewhere (Jaber et al., 2010). In this study, we characterize the Arabidopsis Trs120 ortholog (AtTRS120).
Figure 2.
Seedling phenotypes of TRAPPI and TRAPPII mutants and their comparison with canonical cytokinesis-defective mutants. Five-day-old seedlings were grown on Murashige and Skoog Suc plates. A, The wild type. B, hinkel. C, keule. D, massue (mas-5). E, trs33-1. F, trs120-2. G, trs120-3. H, trs120-4. Note the reduced root and apical meristem growth and bloated appearance of the mutants. Bars = 1 mm.
Figure 3.
Toluidene blue-stained histological sections of trs120 and trs33 seedlings. A, The wild type. B, trs120-3. C, E, and F, trs33-1. D, trs120-4. E corresponds to the boxed area in C. Black arrows (B, E, and F) point to selected cell wall stubs, and the arrowheads in B and D point to “floating” walls.
Characterization of AtTRS120 Insertion Lines
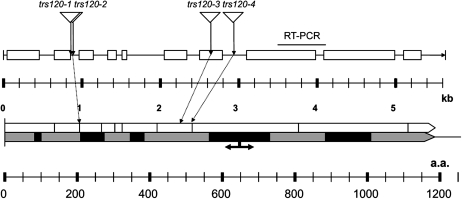
All four insertion lines in AtTRS120 were verified by sequencing (Table III; Fig. 4) and shown to be transcriptionally null by reverse transcription (RT)-PCR (Fig. 4; Supplemental Fig. S2). The AtTRS120 gene spans 5,636 bp and encodes a protein of 1,186 amino acids (Fig. 4). The Trs120p signature spans the entire length of the protein, with five well-conserved domains (designated by black boxes in Fig. 4) ranging from 20 to 165 amino acids in length and characterized by high-quality sequence similarities and by the presence of highly conserved amino acids in the alignment (Cox et al., 2007). In budding yeast, Trs120 is an essential gene, and truncations upstream of the functional break point (designated by an arrow in Fig. 4) are lethal, while truncations downstream of the arrow yield temperature-sensitive phenotypes (Cox et al., 2007). All four insertions in AtTRS120, depicted in Figure 4, are upstream of the yeast break point, consistent with their seedling-lethal phenotype.
Table III. Four AtTRS120 alleles.
| Allele | Polymorphism | Position in Gene | Position on the Open Reading Framea | Plate Phenotype |
| trs120-1 | SALK_125246 | First intron | 517 | Canonical cytokinesis defective |
| trs120-2 | SALK_021904 | First intron | 542–562 | Canonical cytokinesis defective |
| trs120-3 | SALK_111574 | Seventh exon | 2,626 | Canonical cytokinesis defective |
| trs120-4 | SAIL_1285_DO7 | Seventh intron | 2,832 | Canonical cytokinesis defective |
Nucleotide position on the full-length genomic sequence, including introns, starting from the start (ATG) codon. The polymorphisms are depicted graphically in Figure 4.
Figure 4.
Four insertion lines in AtTRS120, a putative TRAPPII tethering factor. The top line represents the AtTRS120 gene. A single open reading frame is depicted by a line, and the exons are drawn as boxes. Four alleles, depicted with triangles, show a cytokinesis-defective phenotype (Table III). The scale is for nucleotides in kb. A line above the gene designates the interval spanned by RT-PCR primers, with which no transcripts were detected downstream of the four insertions. The bottom line represents the AtTRS120 protein. The Trs120p signature (designated by a gray arrow) spans the entire length of the protein, with five well-conserved domains (designated by black rectangles) characterized by the presence of highly conserved amino acids in the alignment (Cox et al., 2007). The scale is for amino acid (a.a.) residues. The black arrow represents a functional break point in yeast, with mutations upstream of the break point being lethal and mutations downstream being temperature sensitive (Cox et al., 2007). Note that all four insertions are upstream of the functional break point.
To assess T-DNA transmission in the insertion lines, DNA was isolated from viable plants grown on soil, and the presence or absence of the T-DNA was monitored via PCR. For a seedling-lethal mutation, one would expect 67% hemizygous and 33% wild-type plants in the progeny of a selfed hemizygous line. For the four AtTRS120 lines, which segregated the highest frequency of cytokinesis-defective seedlings (Table II), we observed only between 7.7% and 23.8% hemizygous segregants among the progeny of selfed hemizygous lines (Table II). Thus, these insertion lines showed reduced transmission of the mutant allele. To test whether the T-DNA segregated with the phenotype, we selfed hemizygous plants and analyzed their progeny. In 290 segregants we looked at (for three alleles: trs120-2, trs120-3, and trs120-4), lines harboring a T-DNA insertion (as shown in Supplemental Fig. S1) segregated cytokinesis-defective mutants, but wild-type segregants did not. Thus, the data (Table II) show an absolute segregation of the T-DNA insertion with the cytokinesis-defective phenotype. In a total of 324 plants, we did not recover a single individual that was homozygous for the T-DNA insertion, indicating that null mutations in AtTRS120 are lethal.
AtTRS120 Is Required for Cell Plate Assembly
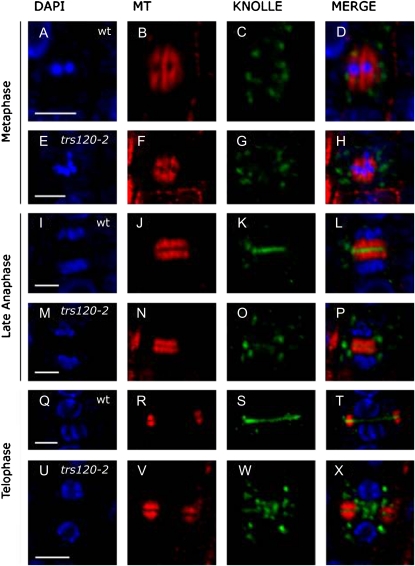
To determine the role of AtTRS120 in cytokinesis, we used the KNOLLE polyclonal rabbit antibody (Lauber et al., 1997) to label cell plate vesicles and membranes. In addition, a 4′,6-diamino-2-phenylindole (DAPI) stain of the nucleus and a microtubule monoclonal antibody stain were used to monitor cell cycle progression. Confocal analysis of a total of 139 wild-type and 296 trs120 dividing cells showed that trs120 mutants are impaired in cell plate assembly. The comparison between the wild type and mutants is depicted in Figure 5 and described in detail for each cell cycle stage below. Although we looked at root tips of three alleles, trs120-2, trs120-3, and trs120-4, and ascertained that they shared the same phenotypes, for the sake of consistency we depict only the trs120-4 allele in Figure 5. A quantitative analysis was carried out for two alleles, trs120-3 and trs120-4 (Table IV).
Figure 5.
Cell plate assembly during anaphase in wild-type and trs120-4 root tips. Columns are as follows: DAPI/nucleus (blue, first column); microtubules (red, second column); KNOLLE protein (green, third column); and merge (fourth column). The cell cycle stage is indicated on the left, and the genotype (wild-type [wt], A–D, I–L, and Q–T; trs120-4, E–H, M–P, and U–X) is indicated in the first column. In all cases, representative cells are shown (see Table IV). Note that cell plate formation, as judged with the KNOLLE antibody, is complete by late anaphase in the wild type (K). In trs120 mutants, by contrast, the cell plate has failed to assemble throughout cytokinesis (late anaphase [O] and telophase [W]). See text for a detailed description. Bars = 5 μm.
Table IV. Confocal analysis.
The extent of cell plate assembly correlates negatively with vesicle abundance. Some of the more pronounced differences between the mutant and the wild type are highlighted in boldface.
| Phase | Cell Plate Assembly |
Vesicle Abundance |
Total Cells | ||||||
| Cell Plate Absent | Patchily Assembled | Intermediate or Thin | Complete | None | Few | Intermediate | Abundant | ||
| % | % | ||||||||
| Late anaphase | |||||||||
| Wild type | 6.1 | 2.0 | 4.1 | 87.8 | 30.6 | 20.4 | 38.8 | 10.2 | n = 49 |
| trs120-3 | 33.3 | 28.6 | 26.2 | 11.9 | 4.8 | 21.4 | 59.5 | 14.3 | n = 42 |
| trs120-4 | 25.0 | 50.0 | 12.5 | 12.5 | 0.0 | 12.5 | 54.2 | 33.3 | n = 24 |
| Telophase | |||||||||
| Wild type | 0.0 | 1.4 | 7.0 | 91.6 | 50.7 | 22.5 | 25.4 | 1.4 | n = 71 |
| trs120-3 | 37.2 | 45.1 | 11.8 | 5.9 | 2.0 | 7.8 | 58.8 | 31.4 | n = 51 |
| trs120-4 | 50.9 | 29.1 | 5.5 | 14.5 | 3.6 | 5.5 | 50.9 | 40.0 | n = 55 |
Metaphase
At this stage, chromosomes are aligned along the equatorial plane of the cell (Fig. 5, A and E) and are surrounded by the spindle apparatus (Fig. 5, B and F). In both wild-type and trs120 mutant cells, KNOLLE-positive vesicles were distributed throughout the cell (Fig. 5, C and G). We did not notice any difference in the abundance or appearance of vesicles between the mutant and wild type at this stage.
Late Anaphase
At the end of anaphase, once the daughter chromosomes have been brought to opposite poles of the cell (Fig. 5, I and M), the spindle apparatus disintegrates and phragmoplast microtubules can be seen to form two bands at the cell equator (Fig. 5, J and N). As of this stage, the wild type and mutant differ. Indeed, 87.8% of wild-type cells but only 11.9% to 12.5% of mutant cells had a fully assembled cell plate (Fig. 5K) at this stage. In the mutant cells, the cell plate was entirely missing, abnormally thin, or patchily assembled (Table IV). The failure to assemble a cell plate correlated with the presence of loose vesicles that could be seen at the equator of trs120 cells (Fig. 5O; Table IV).
Telophase
Telophase cells were identified based on a DAPI stain showing an ovoid nucleus in which the DNA is decondensed and the nucleoli are visible (Fig. 5, Q and U), together with the presence of a phragmoplast in which the microtubules had depolymerized from the middle and repolymerized at the leading edges (Fig. 5, R and V). A total of 91.6% of wild-type telophase cells (Fig. 5S) but only 5.9% to 14.6% of trs120 mutant cells had formed a complete cell plate by this stage (Table IV). A cloud of loose vesicles could be seen at the equator in more than 90% of the mutant cells (Fig. 5W), but we scored vesicles as being abundant in only 1.4% of wild-type cells (Table IV).
Conclusion
In wild-type cells, cell plate formation was complete by late anaphase. In contrast, in trs120 mutants, cell plate assembly was impaired and untethered or unfused vesicles were abundant at the cell equator throughout cytokinesis.
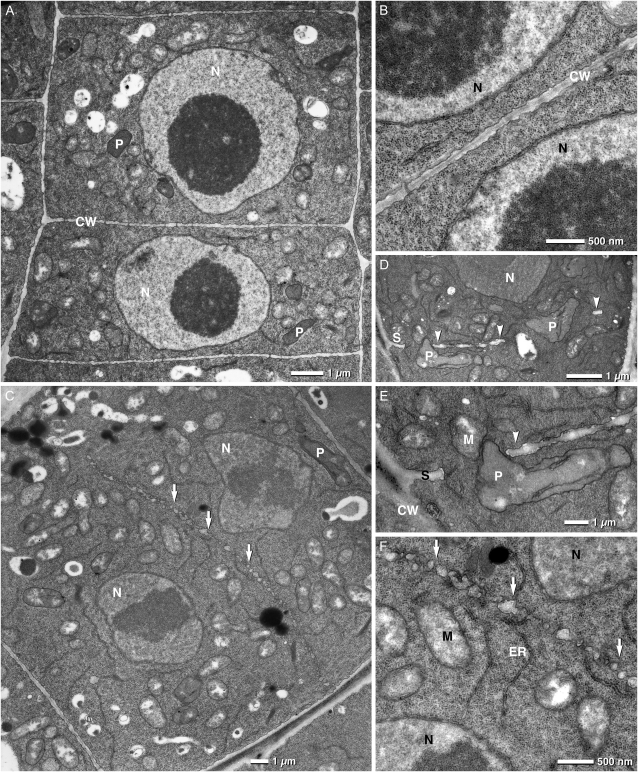
Electron microscopy on root tips confirmed the impaired cell plate assembly we observed with confocal analysis. Figure 6C, depicts a trs120-4 cell in which large vesicles (arrows in C and closeup in F) are aligned along the equator of a cell but have failed to form a contiguous cell plate. The nuclei in this cell, however, have decondensed chromosomes and a nuclear envelope (Fig. 6F), which indicates that the cell is well past the anaphase stage and therefore should have a more mature cell plate or a cross wall, as shown in the wild-type cells depicted in Figure 6, A and B. Similarly, Figure 6, D and E, show a patchily assembled incomplete cell plate in a trs120-3 mutant, resulting in a short stub at one end of the cell and two small walls that, in the focal plane of this sample, appear to be floating (arrowheads). Collectively, the confocal and electron micrographs point to a requirement of AtTRS120 for cell plate biogenesis.
Figure 6.
Incomplete cell plate assembly in trs120 mutants. Electron micrographs of 5-d-old root tips are shown. A and B, The wild type. Note the regular shape of the cells and the complete cross wall. C and F, trs120-4. D and E, trs120-3. F is an enlargement of C, and E is an enlargement of D. CW, Cell wall; M, mitochondria; N, nucleus; P, proplastid; S, cell wall stub; V, vacuole. In C and F, white arrows point to vesicles or compartments that have failed to assemble into a contiguous cell plate or cross wall. In fact, the cell plate appears to be in an initial assembly stage characteristic of anaphase. The nuclei, however, are at telophase or interphase, as judged by the presence of decondensed chromosomes and a nuclear envelope (G). Thus, the nuclear cycle is complete but the cell plate has failed to assemble. This is in contrast to the wild type shown in B. D and E show an incomplete, patchy cell plate or cross wall. Arrowheads point to walls that appear to be floating, at least in the focal plane of the micrograph. Note that in E, a protoplastid (P) can be seen inside a large gap in the cross wall.
DISCUSSION
TRAPPII Mutants Have a Strong Cytokinesis-Defective Phenotype
In this study, we specifically screened insertion lines for strong cytokinesis defects. The strongest cytokinesis-defective mutants described to date in the plant literature are typically seedling lethal (Söllner et al., 2002). Embryo lethality has been reported only in rare cases, most notably in a double mutant between keule and knolle (Waizenegger et al., 2000). Weak cytokinesis-defective mutants are viable but small and compromised, with seedling phenotypes that are uninformative in the absence of extensive histological analyses (Heese et al., 2001). Canonical seedling-lethal cytokinesis defects were most frequently observed in TRAPPII mutants. This correlated with an impaired biogenesis of the cell plate, a transient compartment thought to be derived from the TGN.
In yeast, the TRAPPII complex has been shown to reside on the TGN, where it mediates both exocytosis and endocytosis (Cai et al., 2005). The KNOLLE syntaxin, which is cell cycle regulated at the transcriptional level, remains the best marker for the cell plate. KNOLLE-positive vesicles appear at the onset of the cell cycle, and KNOLLE antibodies label the cell plate throughout cytokinesis (Lauber et al., 1997). The cell plate is also labeled by Rab GTPases Rab-A2 and Rab-A3, which colocalize with KNOLLE throughout mitosis (Chow et al., 2008). Rab-A2/A3 reside on an early endosome (EE)/TGN compartment, implicated in both endocytosis and exocytosis (Chow et al., 2008). The Rab-A2/A3 compartment only partially overlaps with the TGN marker VHA-a1, which is excluded from the cell plate (Dettmer et al., 2006; Chow et al., 2008). The partial colocalization of the VHA-a1 and Rab-A2/3 markers and their difference at the cell plate reflect the existence of subdomains or subcompartments within the TGN.
As the major sorting station within the plant cell (Dettmer et al., 2006; Lam et al., 2007), the TGN contributes membranes in the form of secretory or clathrin-coated vesicles to the plasma membrane, to the late endosome/prevacuolar compartment (PVC), and (in retrograde traffic) to the Golgi. Yet neither the PVC nor the Golgi colocalizes with Rab-A2/A3 gene fusions, which also predominantly label punctate mobile structures (Chow et al., 2008). The observation that Rab-A2/A3 labels the cell plate throughout cytokinesis suggests that the cell plate has a Rab-A2/A3 EE/TGN identity. Chow et al. (2008) propose that the Rab-A2/A3 TGN/EE compartment is the precursor to the cell plate and that the cell plate may even arise substantially by KNOLLE-mediated homotypic fusion of Rab-A2/A3-like membranes. Our observations are entirely compatible with this view. We show that KNOLLE-positive compartments fail to assemble into a cell plate in the absence of a tethering factor thought, by orthology, to reside on the TGN.
Tethering factors comprise an important addition to existing markers, such as SNAREs and Rab GTPases, that have been shown to colocalize with other markers thought to best define a given cellular compartment (Robinson et al., 2008; Spitzer and Otegui, 2008; Richter et al., 2009). This is because tethers define the target membrane as well as the nature of the vesicles that fuse with this target membrane. As such, they not only define membrane compartments but also specific trafficking steps. We postulate that TRAPPII mutants have strong cytokinesis defects because they impact a crucial early phase in cytokinesis, namely cell plate biogenesis. Tethering factors have been implicated in the biogenesis of other compartments, such as the vacuole. Indeed, a very striking, embryo-lethal phenotype has been reported for the HOPS or VPS tethering complex. Electron micrographs of Arabidopsis VPS mutants show a severe defect in cell elongation due to the absence of a vacuole, and the mutants were accordingly named vacuoless1 (Rojo et al., 2001). The HOPS complex is thought to tether prevacuolar compartments to each other, as the initial step in vacuolar biogenesis (Rojo et al., 2003). Similarly, and based on our confocal and electron microscopy, we postulate that the TRAPPII complex tethers Golgi-derived and possibly endocytotic vesicles at the cell equator as an initial step in cell plate biogenesis. While there are 26 different Rab-A’s in Arabidopsis (Vernoud et al., 2003), the TRAPPII AtTRS120 and AtTRS130 proteins are encoded by single genes. These would thus be expected to fulfill all TRAPPII-mediated TGN functions. In this context, cell plate biogenesis might be considered to be a specialized TGN function in mitotic cells.
The Role of Endosomal Trafficking in Plant Cytokinesis
Cross wall deposition requires extensive recycling, reorganization, and remodeling, presumably via distinct and specialized forms of endosomal trafficking. KNOLLE marks the juvenile phase of cross wall deposition, as it appears at the onset of mitosis and disappears once cytokinesis is complete (Lauber et al., 1997; Thiele et al., 2009). As a membrane compartment, the cell plate is initially distinct from, yet after cytokinesis contiguous with, the plasma membrane. Similarly, cross walls are considerably remodeled shortly after they are deposited, with callose being the dominant luminal component of the cell plate yet absent from intact cross walls (Samuels et al., 1995; Thiele et al., 2009). Thus, cell plate or cross wall maturation requires the active removal of certain juvenile traits such as KNOLLE or callose. Proteins targeted for degradation are sorted into luminal vesicles of multivesicular bodies by the ESCRT machinery (Winter and Hauser, 2006; Otegui and Spitzer, 2008). Consistently, ESCRT mutants of Arabidopsis have been reported to have cytokinesis defects (Spitzer et al., 2006, 2009).
The role of GARP proteins is distinct from the role of ESCRT proteins, in that GARP proteins are required not for the degradation but for the recycling of proteins back to the TGN. One potential role for GARP proteins in this context would be to recycle membranes and cell plate materials from the middle of the cell plate to its leading edges as the cell plate expands and joins the parental walls. In a recent study, endocytosis has been suggested to be required for the spatial restriction of the KNOLLE syntaxin to the cell plate (Boutté et al., 2009). Indeed, an impairment of endocytosis via drugs or mutation resulted in the lateral diffusion of KNOLLE after the cell plate had fused with the parental plasma membrane (Boutté et al., 2009). If the role of GARP proteins is restricted to limiting lateral diffusion at late stages of cytokinesis, then one might predict a weaker, more subtle phenotype than the seedling lethality we screened for. It is to be noted, however, that the TRAPPII complex could in principle fulfill all potential endocytotic functions at the cell plate, such that our inability to observe cytokinesis defects in GARP lines does not necessarily exclude a role for endocytosis in cell plate biogenesis.
However, there are three lines of evidence that contest the model, put forth by Dhonukshe et al. (2006), that endocytosis contributes substantially to the cell plate. First, the observation that RabF2b, RabF2a, and GNOM, which are markers for the late endosome (PVC) in plants, were found to colocalize with KNOLLE at the cell plate (Dhonukshe et al., 2006) has been contested (Chow et al., 2008). Second, a pharmacological inhibition of endocytosis did not impact cytokinesis (Reichardt et al., 2007). Third, mutations in retromer proteins that impact endosomal recycling (Niemes et al., 2010) have not been reported to have an effect on cytokinesis (Jaillais et al., 2007; Kleine-Vehn et al., 2008; Yamazaki et al., 2008).
The GARP and TRAPPII Tethering Complexes and the TGN
Both the GARP and TRAPPII tethering complexes have been shown to reside on the TGN in yeast, but their distinct functions at the TGN in space and in time remain to be elucidated. The TGN is emerging as the predominant sorting station in the plant secretory pathway and as a hub in the flow of information to and from the cell surface. Electron tomography has shown that the TGN cannot be perceived as a single compartment but as a continuum of early, late, and mature compartments that release their vesicles by means of cisternal fragmentation (Staehelin and Kang, 2008). TGN cisternae can be seen to generate budding secretory vesicles targeted to the cell surface as well as clathrin-coated vesicles targeted to the vacuole. Interestingly, the ratio of secretory to clathrin-coated vesicles can vary from 5:1 to 1:3 within a single cell (Staehelin and Kang, 2008). This is an example of the plasticity of TGN compartments and of their ability to rapidly adapt to changes in the nature of the cargo molecules present. While electron tomography and live imaging (Nebenführ and Staehelin, 2001; Yang et al., 2005; Moreau et al., 2007; Staehelin and Kang, 2008) have greatly contributed to our understanding of the plant secretory pathway, little is known at a molecular level about how the TGN is subcompartmentalized to fulfill its diverse functions.
The cell plate is widely accepted to be a TGN-derived compartment (Dettmer et al., 2006; Chow et al., 2008) whose assembly can readily be monitored in space and in time. In this study, we show that the TRAPPII AtTRS120 locus is required for cell plate biogenesis. By contrast, we found no evidence for an involvement of the GARP tethering complexes in cytokinesis in our analysis, but we are not able to exclude this possibility. There are two possible interpretations for these findings. The first is that endocytosis is not required for cell plate biogenesis. A second interpretation is that the TRAPPII complex tethers both Golgi-derived and endocytotic vesicles at the cell plate. Regardless of which interpretation turns out to be correct, an interesting point is that the GARP and TRAPPII complexes differ with respect to their role in the biogenesis of a TGN-derived compartment. This may open up new avenues for research into the different facets of the TGN, a plastic, complex compartment of pivotal importance for the regulation of differential growth in plants.
MATERIALS AND METHODS
Lines
Due to the lethality of the majority of Arabidopsis (Arabidopsis thaliana) TRAPP and GARP mutants, and more specifically of all trs120 mutants, lines were propagated as hemizygotes. Insertion mutants were selected via the www.arabidopsis.org (Swarbreck et al., 2008) or www.signal.salk.edu (Alonso et al., 2003) Web sites and were obtained from the Nottingham Arabidopsis Stock Centre or the Arabidopsis Biological Resource Center (http://Arabidopsis.info; Scholl et al., 2000) as well as from the Institut National de la Recherche Agronomique (http//www-ijpb.versailles.inra.fr; Samson et al., 2002) and GABI (http://www.gabi-kat.de; Rosso et al., 2003). The insertion lines are in Columbia, and this ecotype was subsequently used as a wild-type control. The hinkel allele used was G235 (F.F. Assaad, unpublished data), the keule allele was MM125 (Assaad et al., 2001), and the massue allele was mas-5 (Thiele et al., 2009). Plants were grown in the greenhouse throughout the year under controlled temperature conditions and with supplemental light or under controlled growth chamber conditions (16-h/8-h photoperiod at 250 μmol m−2 s−1). For an analysis of embryo lethality in seed pods, we used healthy plants grown in the greenhouse during the fall or in controlled growth chambers. Seedlings were surface sterilized, stratified at 4°C for 2 d, and plated on Murashige and Skoog medium supplemented with 1% Suc and B5 vitamins (Sigma; http://www.sigmaaldrich.com) as described previously (Assaad et al., 1996). The root tips of 5-d-old plate-grown seedlings were used for light, confocal, and electron microscopy.
Characterization of Insertion Lines
DNA was isolated from rosette leaves or inflorescences of soil-grown plants. Cetyl-trimethyl-ammonium bromide minipreps were prepared as described by Assaad et al. (2001), and RT-PCR was carried out as described in Supplemental Materials and Methods S1. A full description of the PCR and RT-PCR primers we designed for the analysis of the insertion lines can be found in Supplemental Materials and Methods S1 or in Supplemental Table S1.
Antibody Stains and Confocal Microscopy
Antibody stains were carried out on root tips as described (Völker et al., 2001). The KNOLLE rabbit polyclonal antiserum was generated as described (Lauber et al., 1997) and used at a 1:2,000 dilution. Monoclonal anti-tubulin antibody (Sigma) was used at a dilution of 1:2,500. Goat anti-rabbit monoclonal antibody was coupled to Alexa-m488 (Molecular Probes; http://www.invitrogen.com), and goat anti-mouse secondary antibody was coupled to Cy3 (Dianova; http://www.dianova.de). Nuclei were stained with 1 mg mL−1 DAPI (Sigma). Slides were analyzed with a Fluoview 1000 confocal laser scanning microscope (Olympus; http://www.olympus.de). A 40× water-immersion 0.9 numerical aperture objective (Olympus) was used, and scanning was carried out with 4- to 6-fold electronic magnification. Images were acquired and processed with the Fluoview 1000 acquisition software (Olympus) and subsequently processed with Adobe Photoshop. Images were assembled with Adobe Illustrator (http://www.abode.com).
Light and Electron Microscopy
Seedlings for light and electron microscopy were fixed and prepared as described by Schumann et al. (2007), and infiltration was as described by Assaad et al. (1996). Five-day-old seedlings were used for light and electron micrographs. The light microscope was an Olympus BX61 microscope equipped with an F-view II digital CCD camera (Olympus). Transmission electron micrographs were taken with an EM 912 electron microscope (Zeiss; http://www.zeiss.com) equipped with an integrated OMEGA energy filter operated at 80 kV in the zero loss mode. For scanning electron microscopy of fresh material, samples were placed onto stubs and examined immediately in low vacuum with a Zeiss (LEO) VP 438 scanning electron microscope operated at 15 kV. Electron micrographs were digitally recorded from the back-scattered electron signal.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Table I.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. PCR analysis of the trs120-3 and trs120-4 T-DNA insertion.
Supplemental Figure S2. RT-PCR analysis of the four trs120 alleles.
Supplemental Table S1. PCR primers for an analysis of polymorphisms.
Supplemental Materials and Methods S1. PCR conditions.
Supplementary Material
Acknowledgments
We are especially grateful to Erwin Grill for his encouragement and useful suggestions. For transmission electron micrography analyses, Silvia Dobler and Connie Niemann are gratefully acknowledged for excellent technical assistance. Caroline Klaus, Lisa Held, and Michael Schmidt tended to our plants. Thanks to Theis Stüven and Peter Zehetmayer, to numerous members of the Botany and Genetics Departments, and to the Schneitz and Schwechheimer laboratories for their input. We are grateful to The Arabidopsis Information Resource (http://arabidopsis.org), to the Nottingham Arabidopsis Stock Centre (http://arabidopsis.org), to the Salk/Stanford/PGEC Consortium (http://signal.salk.edu), to GABI-KAT (www.gabi-kat.de), and to the Institut National de la Recherche Agronomique, Versailles (http://www-ijpb.versailles.inra.fr), for bioinformatic resources and T-DNA lines. We also thank anonymous reviewers and Maureen McCann for their contributions to improving the manuscript.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Assaad FF. (2001a) Plant cytokinesis: exploring the links. Plant Physiol 126: 509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad FF. (2001b) Of weeds and men: what genomes teach us about plant cell biology. Curr Opin Plant Biol 4: 478–487 [DOI] [PubMed] [Google Scholar]
- Assaad FF. (2009) The membrane dynamics of root hair morphogenesis. Emons AMC, Ketelar T, , Root Hairs. Springer Verlag, Heidelberg, pp 65–84 [Google Scholar]
- Assaad FF, Huet Y, Mayer U, Jürgens G. (2001) The cytokinesis gene KEULE encodes a Sec1 protein which binds the syntaxin KNOLLE. J Cell Biol 152: 531–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad FF, Mayer U, Wanner G, Jürgens G. (1996) The KEULE gene is involved in cytokinesis in Arabidopsis. Mol Gen Genet 253: 267–277 [DOI] [PubMed] [Google Scholar]
- Boutté Y, Frescatada-Rosa M, Men S, Chow CM, Ebine K, Gustavsson A, Johansson L, Ueda T, Moore I, Jürgens G, et al. (2009) Endocytosis restricts Arabidopsis KNOLLE syntaxin to the cell division plane during late cytokinesis. EMBO J 29: 546–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Reinisch K, Ferro-Novick S. (2007) Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell 12: 671–682 [DOI] [PubMed] [Google Scholar]
- Cai H, Zhang Y, Pypaert M, Walker L, Ferro-Novick S. (2005) Mutants in trs120 disrupt traffic from the early endosome to the late Golgi. J Cell Biol 171: 823–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong YT, Gidda SK, Sanford C, Parkinson J, Mullen RT, Goring DR. (2010) Characterization of the Arabidopsis thaliana exocyst complex gene families by phylogenetic, expression profiling, and subcellular localization studies. New Phytol 185: 401–419 [DOI] [PubMed] [Google Scholar]
- Chow CM, Neto H, Foucart C, Moore I. (2008) Rab-A2 and Rab-A3 GTPases define a trans-Golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20: 101–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RA, Synek L, Zarsky V, Fowler JE. (2005) SEC8, a subunit of the putative Arabidopsis exocyst complex, facilitates pollen germination and competitive pollen tube growth. Plant Physiol 138: 2005–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Cleck JN, Stevens TH. (2003) Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol Biol Cell 14: 1610–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R, Chen SH, Yoo E, Segev N. (2007) Conservation of the TRAPPII-specific subunits of a Ypt/Rab exchanger complex. BMC Evol Biol 7: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K. (2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P, Baluska F, Schlicht M, Hlavacka A, Samaj J, Friml J, Gadella TW., Jr (2006) Endocytosis of cell surface material mediates cell plate formation during plant cytokinesis. Dev Cell 10: 137–150 [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Arimura S, Nakazono M, Tsutsumi N. (2008) Arabidopsis dynamin-related protein DRP2B is co-localized with DRP1A on the leading edge of the forming cell plate. Plant Cell Rep 27: 1581–1586 [DOI] [PubMed] [Google Scholar]
- Grosshans BL, Ortiz D, Novick P. (2006) Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci USA 103: 11821–11827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermonprez H, Smertenko A, Crosnier MT, Durandet M, Vrielynck N, Guerche P, Hussey PJ, Satiat-Jeunemaitre B, Bonhomme S. (2008) The POK/AtVPS52 protein localizes to several distinct post-Golgi compartments in sporophytic and gametophytic cells. J Exp Bot 59: 3087–3098 [DOI] [PubMed] [Google Scholar]
- Hála M, Cole R, Synek L, Drdová E, Pecenková T, Nordheim A, Lamkemeyer T, Madlung J, Hochholdinger F, Fowler JE, et al. (2008) An exocyst complex functions in plant cell growth in Arabidopsis and tobacco. Plant Cell 20: 1330–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese M, Gansel X, Sticher L, Wick P, Grebe M, Granier F, Jürgens G. (2001) Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33 and its role in plant cytokinesis. J Cell Biol 155: 239–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaber E, Thiele K, Kindzierski V, Loderer C, Rybak K, Jürgens G, Mayer U, Söllner R, Wanner G, Assaad FF. (2010) A putative TRAPPII tethering factor is required for cell plate assembly during cytokinesis in Arabidopsis. New Phytol 187: 751–763 [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Santambrogio M, Rozier F, Fobis-Loisy I, Miège C, Gaude T. (2007) The retromer protein VPS29 links cell polarity and organ initiation in plants. Cell 130: 1057–1070 [DOI] [PubMed] [Google Scholar]
- Jürgens G. (2005) Plant cytokinesis: fission by fusion. Trends Cell Biol 15: 277–283 [DOI] [PubMed] [Google Scholar]
- Kim YG, Raunser S, Munger C, Wagner J, Song YL, Cygler M, Walz T, Oh BH, Sacher M. (2006) The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell 127: 817–830 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Leitner J, Zwiewka M, Sauer M, Abas L, Luschnig C, Friml J. (2008) Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc Natl Acad Sci USA 105: 17812–17817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumandou VL, Dacks JB, Coulson RM, Field MC. (2007) Control systems for membrane fusion in the ancestral eukaryote: evolution of tethering complexes and SM proteins. BMC Evol Biol 7: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SK, Siu CL, Hillmer S, Jang S, An G, Robinson DG, Jiang L. (2007) Rice SCAMP1 defines clathrin-coated, trans-Golgi-located tubular-vesicular structures as an early endosome in tobacco BY-2 cells. Plant Cell 19: 296–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jürgens G. (1997) The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol 139: 1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CF, Pu HY, Wang LC, Sayler RJ, Yeh CH, Wu SJ. (2006) Mutation in a homolog of yeast Vps53p accounts for the heat and osmotic hypersensitive phenotypes in Arabidopsis hit1-1 mutant. Planta 224: 330–338 [DOI] [PubMed] [Google Scholar]
- Lobstein E, Guyon A, Férault M, Twell D, Pelletier G, Bonhomme S. (2004) The putative Arabidopsis homolog of yeast vps52p is required for pollen tube elongation, localizes to Golgi, and might be involved in vesicle trafficking. Plant Physiol 135: 1480–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jürgens G. (1996) Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84: 61–71 [DOI] [PubMed] [Google Scholar]
- McElver J, Patton D, Rumbaugh M, Liu C, Yang LJ, Meinke D. (2000) The TITAN5 gene of Arabidopsis encodes a protein related to the ADP ribosylation factor family of GTP binding proteins. Plant Cell 12: 1379–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I, Warren G. (2000) The road taken: past and future foundations of membrane traffic. Cell 100: 99–112 [DOI] [PubMed] [Google Scholar]
- Moreau P, Brandizzi F, Hanton S, Chatre L, Melser S, Hawes C, Satiat-Jeunemaitre B. (2007) The plant ER-Golgi interface: a highly structured and dynamic membrane complex. J Exp Bot 58: 49–64 [DOI] [PubMed] [Google Scholar]
- Nebenführ A, Staehelin LA. (2001) Mobile factories: Golgi dynamics in plant cells. Trends Plant Sci 6: 160–167 [DOI] [PubMed] [Google Scholar]
- Niemes S, Langhans M, Viotti C, Scheuring D, San Wan Yan M, Jiang L, Hillmer S, Robinson DG, Pimpl P. (2010) Retromer recycles vacuolar sorting receptors from the trans-Golgi network. Plant J 61: 107–121 [DOI] [PubMed] [Google Scholar]
- Otegui MS, Spitzer C. (2008) Endosomal functions in plants. Traffic 9: 1589–1598 [DOI] [PubMed] [Google Scholar]
- Quenneville NR, Chao TY, McCaffery JM, Conibear E. (2006) Domains within the GARP subunit Vps54 confer separate functions in complex assembly and early endosome recognition. Mol Biol Cell 17: 1859–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt I, Stierhof YD, Mayer U, Richter S, Schwarz H, Schumacher K, Jürgens G. (2007) Plant cytokinesis requires de novo secretory trafficking but not endocytosis. Curr Biol 17: 2047–2053 [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Richter S, Voss U, Jürgens G. (2009) Post-Golgi traffic in plants. Traffic 10: 819–828 [DOI] [PubMed] [Google Scholar]
- Robinson DG, Jiang L, Schumacher K. (2008) The endosomal system of plants: charting new and familiar territories. Plant Physiol 147: 1482–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E, Gillmor CS, Kovaleva V, Somerville CR, Raikhel NV. (2001) VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev Cell 1: 303–310 [DOI] [PubMed] [Google Scholar]
- Rojo E, Zouhar J, Kovaleva V, Hong S, Raikhel NV. (2003) The AtC-VPS protein complex is localized to the tonoplast and the prevacuolar compartment in Arabidopsis. Mol Biol Cell 14: 361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Sacher M, Kim YG, Lavie A, Oh BH, Segev N. (2008) The TRAPP complex: insights into its architecture and function. Traffic 12: 2032–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A. (2002) FLAGdb/FST: a database for mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res 30: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Chong YT, Haasen KE, Aldea-Brydges MG, Stone SL, Goring DR. (2009) Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell 21: 2655–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels AL, Giddings TH, Jr, Staehelin LA. (1995) Cytokinesis in tobacco BY-2 and root tip cells: a new model of cell plate formation in higher plants. J Cell Biol 130: 1345–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH. (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann U, Prestele J, O’Geen H, Brueggeman R, Wanner G, Gietl C. (2007) Requirement of the C3HC4 zinc RING finger of the Arabidopsis PEX10 for photorespiration and leaf peroxisome contact with chloroplasts. Proc Natl Acad Sci USA 104: 1069–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguí-Simarro JM, Austin JR, II, White EA, Staehelin LA. (2004) Electron tomographic analysis of somatic cell plate formation in meristematic cells of Arabidopsis preserved by high-pressure freezing. Plant Cell 16: 836–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner R, Glasser G, Wanner G, Somerville CR, Jürgens G, Assaad FF. (2002) Cytokinesis-defective mutants of Arabidopsis. Plant Physiol 129: 678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C, Otegui MS. (2008) Endosomal functions in plants. Traffic 9: 1589–1598 [DOI] [PubMed] [Google Scholar]
- Spitzer C, Reyes FC, Buono R, Sliwinski MK, Haas TJ, Otegui MS. (2009) The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell 21: 749–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C, Schellmann S, Sabovljevic A, Shahriari M, Keshavaiah C, Bechtold N, Herzog M, Müller S, Hanisch FG, Hülskamp M. (2006) The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development 133: 4679–4689 [DOI] [PubMed] [Google Scholar]
- Staehelin LA, Kang BH. (2008) Nanoscale architecture of endoplasmic reticulum export sites and of Golgi membranes as determined by electron tomography. Plant Physiol 147: 1454–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strompen G, El Kasmi F, Richter S, Lukowitz W, Assaad FF, Jürgens G, Mayer U. (2002) The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr Biol 12: 153–158 [DOI] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36: D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synek L, Schlager N, Eliás M, Quentin M, Hauser MT, Zárský V. (2006) AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development. Plant J 48: 54–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele K, Wanner G, Kindzierski V, Jürgens G, Mayer U, Pachl F, Assaad FF. (2009) The timely deposition of callose is essential for cytokinesis in Arabidopsis. Plant J 58: 13–26 [DOI] [PubMed] [Google Scholar]
- Van Damme D. (2009) Division plane determination during plant somatic cytokinesis. Curr Opin Plant Biol 12: 745–751 [DOI] [PubMed] [Google Scholar]
- Vernoud V, Horton AC, Yang Z, Nielsen E. (2003) Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol 131: 1191–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völker A, Stierhof YD, Jürgens G. (2001) Cell cycle-independent expression of the Arabidopsis cytokinesis-specific syntaxin KNOLLE results in mistargeting to the plasma membrane and is not sufficient for cytokinesis. J Cell Sci 114: 3001–3012 [DOI] [PubMed] [Google Scholar]
- Waizenegger I, Lukowitz W, Assaad FF, Schwarz H, Jürgens G, Mayer U. (2000) The Arabidopsis KNOLLE and KEULE genes interact to promote fusion of cytokinetic vesicles during cell plate formation. Curr Biol 10: 1371–1374 [DOI] [PubMed] [Google Scholar]
- Wang H, Tse YC, Law AH, Sun SS, Sun YB, Xu ZF, Hillmer S, Robinson DG, Jiang L. (2010) Vacuolar sorting receptors (VSRs) and secretory carrier membrane proteins (SCAMPs) are essential for pollen tube growth. Plant J 61: 826–838 [DOI] [PubMed] [Google Scholar]
- Winter V, Hauser MT. (2006) Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci 11: 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M, Shimada T, Takahashi H, Tamura K, Kondo M, Nishimura M, Hara-Nishimura I. (2008) Arabidopsis VPS35, a retromer component, is required for vacuolar protein sorting and involved in plant growth and leaf senescence. Plant Cell Physiol 49: 142–156 [DOI] [PubMed] [Google Scholar]
- Yang YD, Elamawi R, Bubeck J, Pepperkok R, Ritzenthaler C, Robinson DG. (2005) Dynamics of COPII vesicles and the Golgi apparatus in cultured Nicotiana tabacum BY-2 cells provides evidence for transient association of Golgi stacks with endoplasmic reticulum exit sites. Plant Cell 17: 1513–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Bednarek SY, Sanderfoot AA, Alonso J, Ecker JR, Raikhel NV. (2002) NPSN11 is a cell plate-associated SNARE protein that interacts with the syntaxin KNOLLE. Plant Physiol 129: 530–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.