Plants and insects comprise the most species-rich lineages of multicellular eukaryotes. Most insects are herbivores, and generally have specific adaptations for feeding on a very limited group of evolutionarily or biochemically related host plants in their natural habitats. Coevolutionary interactions (reciprocal natural selection) between plants and insects are hypothesized to be the crucible for generating the majority of terrestrial organismic diversity. Therefore, understanding the mechanistic and evolutionary basis of plant-insect interactions provides insight into the processes that have led to the great diversification of metazoan life. Future research on the ecology of plant-insect interactions will be strongly influenced by recent advances in molecular biology, in particular the rapidly dropping price of DNA sequencing.
GENOMIC STUDIES INVOLVING BOTH SIDES OF A PLANT-INSECT INTERACTION
Whereas several plant genome projects have been completed, there are currently only two sequenced insect herbivores, the silkworm Bombyx mori (Xia et al., 2004) and the pea aphid Acyrthosiphon pisum (International Aphid Genomics Consortium, 2010). Honeybees (Apis mellifera), red flour beetles (Tribolium castaneum), and fruit flies (Drosophila melanogaster) are technically also herbivores with sequenced genomes, but this discussion is focused on herbivorous insects that feed from vegetative tissue and elicit specific plant defense responses. Numerous additional projects to sequence the genomes of insect herbivores and their host plants are in progress and will be completed in the near future.
With the imminent publication of the Medicago truncatula (barrel medic) genome (www.medicago.org/genome/) and the already completed A. pisum genome, this will be the first plant-insect model system (Fig. 1; Table I) where it is possible to conduct genetic and genomic studies using both partners in the interaction. Just as research on plant-pathogen interactions has benefited greatly from pairs of sequenced genomes, e.g. Arabidopsis (Arabidopsis thaliana) and Pseudomonas syringae, there will be synergistic effects in studying not only plant responses to insect herbivory, but also insect responses to plant defenses. Whereas there are well-developed tools for the genetic analysis of M. truncatula, such resources are more limited in the case of A. pisum (Table I). However, transcriptional profiling studies of nonmodel organisms, which were previously difficult to conduct, can be performed overnight with next-generation sequencing technologies. In addition to identifying candidate genes for ecologically interesting phenotypes, these approaches may also reveal sequence polymorphisms linking genotypes to phenotypes.
Figure 1.
A fully sequenced plant-herbivore-bacterial interaction. The genomes of M. truncatula, A. pisum, and its bacterial endosymbiont B. aphidicola have been sequenced.
Table I. Tools for genetic analysis of a plant-insect interaction.
Chr., Chromosomes.
| Organisms | Genome Size | Chr. | Available Tools for Genetic Analysis |
| Mb | |||
| M. truncatula | 500 | 8 | Database: medicago.org |
| cDNA sequences | |||
| T-DNA insertions | |||
| TILLING | |||
| Transformation | |||
| RNA interference | |||
| HapMap | |||
| Natural variants | |||
| A. pisum | 460 | 4 | Database: aphidbase.com |
| cDNA sequences | |||
| Expression silencing Natural variants | |||
| B. aphidicola | 0.64 | 1 | Database: BuchneraBase |
| Natural variants |
GENE DISCOVERY AND FUNCTIONAL ANALYSIS
Although many of the most important genes involved in plant-pathogen interactions have been identified through molecular mapping of both plant and pathogen mutations, such approaches have rarely been applied to studying plant-insect interactions. However, this situation will likely change through genome-enabled mutation mapping, which will involve not only the more genetically tractable host plants but also their insect herbivores. To date, most genetic mapping in herbivorous insects has been focused on insecticide resistance traits. New approaches such as bulked segregant analysis using next-generation sequencing technology (Schneeberger et al., 2009) will facilitate genetic mapping on the insect side of the interaction, making it possible to identify genes underlying host plant choice, detoxification of secondary metabolites, and other complex traits. Even in the absence of an assembled genome, it will be possible to identify molecular markers that are tightly linked to the phenotype of interest. Reverse genetics approaches for silencing the expression of specific genes are not yet available for most insect species. However, once such methods are developed, gene knockdown and knockout studies will benefit from the prior identification of target genes in known genomic sequence data.
Although transgenerational resistance effects in plant-insect interactions have been reported (Agrawal, 2001), the genetic and molecular mechanisms of such signaling interactions remain unknown. Since there is increasing evidence for mobile siRNA signals and inheritance of gene expression changes through DNA methylation, future research on plant-insect interactions must consider not only genetic effects, but also epigenetic regulation of plant defense pathways and likely also insect responses. To date, consistent epigenetic effects in insect defense have not been reported for any model plant species. Therefore, it will be necessary to either seek out such epigenetic effects in current model species or use next-generation sequencing approaches to investigate epigenetic defense regulation in less-well-studied species.
NEW GENOME-ENABLED MODEL SYSTEMS
Research on plant-insect interactions is currently limited by the lack of a genetically tractable herbivore that feeds from a well-studied model plant species. To address this, Scaptomyza flava, which is nested phylogenetically among the >12 sequenced Drosophila species, is being developed as a genetic model (Whiteman et al., 2010). Based on flow cytometry, the genome sizes of S. flava females and males are estimated as 293 and 287 Mb, respectively (Whiteman et al., 2010), which is smaller than that of Drosophila virilis, the largest Drosophila genome sequenced to date. Scaptomyza has five chromosomes and, across the lineage, the metaphase configuration of the chromosomes is one V shaped, three rods, and one dot (O’Grady et al., 2003).
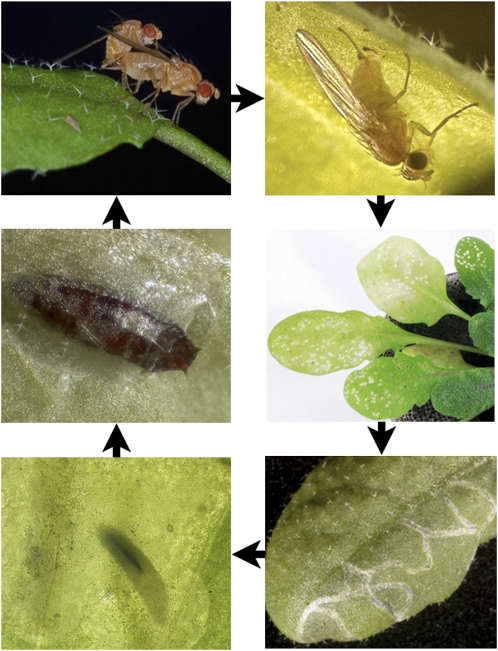
Unlike most Drosophila, S. flava is a true leaf-feeding herbivore that mines the leaves of Arabidopsis in the wild (Fig. 2) and is susceptible to jasmonate-mediated plant defenses. Research methods developed for the fruit fly, together with the sequenced transcriptome and likely imminent genomic sequence of S. flava, will bring research on plant-insect interactions to a new level by allowing genetic analyses on both sides of the interaction. In combination with the extensive Arabidopsis genetic tools, S. flava represents a promising system to explore host plant resistance, herbivore counter responses, and host selection. Although much of what is known about arthropod xenobiotic metabolism and even host plant choice has come from the genus Drosophila, none of the species studied to date are true herbivores and instead have larvae that feed primarily on microbes living in rotting plant tissue.
Figure 2.
S. flava can complete its entire larval life cycle on Arabidopsis. From top left, clockwise: a pair of S. flava adults mating on an Arabidopsis leaf, a female creating a feeding puncture with her dentate ovipositor, eggs laid within feeding punctures on the underside of a leaf, serpentine mine of a first instar larva in a leaf, a blotch mine of a second or third instar larva, a puparium under the epidermis of a leaf. Egg to adult development time is roughly 21 d at 22°C.
With the availability of genome sequence data, other insect species that are easily cultured and have well-studied model species as host plants will also become more amenable to genetic analysis. For instance, the diamondback moth (Plutella xylostella) feeds readily from Arabidopsis, has a generation time of less than 3 weeks, and has already been studied extensively due to its role as an agricultural pest and its propensity to develop insecticide resistance. Genome sequencing and genetic mapping, combined with implementation of gene silencing approaches that are also being developed for other Lepidoptera, will facilitate the functional analysis of diamondback moth genes mediating host plant selection and insect responses to plant defenses.
EVOLUTIONARY ECOLOGY
Current and future plant genome projects will make it progressively easier to move research on the molecular biology of plant-insect interactions from the laboratory to natural ecosystems. General conclusions in the field of plant-insect interactions often have been developed based on the analysis of one or only a few isolates of a given species. For instance, Arabidopsis thrives in a wide variety of habitats, but it is unlikely that all insect defenses are regulated in the same manner as they are in the extensively studied Columbia-0 accession. An ongoing project to sequence a large collection Arabidopsis accessions (www.1001genomes.org) will make it possible to address this deficit at the molecular level. Importantly, many additional plant genome projects are taking place within phylogenetic proximity to well-studied model species, providing new insight into the evolution of plant defense pathways. Comparative analyses will determine the extent to which plant defense pathways are truly conserved and how microevolutionary forces have shaped both plant defense against insect herbivores and herbivore responses to these plant defenses.
Understanding interactions between the genes and genomes of free-living plants, insects, and other organisms is emerging as an important research theme. For example, the ecologically foundational North American tree species, Populus angustifolia, a congener of the fully sequenced Populus trichocarpa (Tuskan et al., 2006), is an up-and-coming model in the area of community genetics/genomics. This field aims to understand how genes in one species, or individual, influence those in another, and ultimately, how these interactions shape populations dynamics and trajectories of all interacting species (Whitham et al., 2008). The availability of newly genome-enabled tree species is likely to encourage the development of model herbivores that can be studied on these host plants.
When studying plant-insect interactions, it is important to consider that in nature these systems almost always consist of more than two interacting organisms. Three-way (and beyond) interactions between plants, microbes, and arthropods play an integral role in the evolution of terrestrial biodiversity. Microbes form symbiotic and mutualistic interactions with insects, and provide nutritional as well as protective benefits to their hosts. For instance, all phloem-feeding insects rely on microbial endosymbionts, e.g. Buchnera aphidicola in aphids (Fig. 1; Table I), for the biosynthesis of essential amino acids. As the genomes of additional species (both insects and bacteria) become sequenced, they will add an important comparative context in which to explore similarities and differences in parallel coevolutionary interactions. Microbes that live symbiotically with invertebrates and facilitate the herbivorous way of life are also providing new insight into herbivore manipulation of host plant signaling pathways (Kaiser et al., 2010).
Mutualism between plants and microbes is another important research area into which the genomics revolution will provide unprecedented insight. Such interactions include those involving symbiotic endophytic and mycorrhyzial fungi, their host plants, and insect herbivores. For instance, fine-scale studies of grass-fungal endophyte systems have shown that the presence and genotype of fungal endophytes influences resistance against herbivores, and has community-wide, cascading effects (Clay et al., 2005). The genome of the grass endophyte Epichloë festucae, which produces antiherbivore toxins, is being sequenced. The genetic model grass Brachypodium distachyon has congers that include species that are naturally infected with Epichloë species (e.g. Brachypodium sylvaticum; Brem and Leuchtmann, 2001), suggesting that this will soon be a genome-enabled system.
As research on these and numerous other ecologically important systems enters the genomics era, this will undoubtedly lead to new insights into the evolution of complex interactions between plants and insect herbivores, which dominate the terrestrial biodiversity of the planet. Given the staggering crop losses that result from insect herbivory and the environmental problems associated with insecticide use, genome-enabled research on the natural mechanisms that plants use to defend themselves against insects will undoubtedly have not only ecological, but also agricultural relevance.
References
- Agrawal AA. (2001) Transgenerational consequences of plant responses to herbivory: an adaptive maternal effect? Am Nat 157: 555–569 [DOI] [PubMed] [Google Scholar]
- Brem D, Leuchtmann A. (2001) Epichloë grass endophytes increase herbivore resistance in the woodland grass Brachypodium sylvaticum. Oecologia 126: 522–530 [DOI] [PubMed] [Google Scholar]
- Clay K, Holah J, Rudgers JA. (2005) Herbivores cause a rapid increase in hereditary symbiosis and alter plant community composition. Proc Natl Acad Sci USA 102: 12465–12470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Aphid Genomics Consortium (2010) Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol 8: e1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser W, Huguet E, Casas J, Commin C, Giron D. (2010) Plant green-island phenotype induced by leaf-miners is mediated by bacterial symbionts. Proc Biol Sci 277: 2311–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Grady P, Bonacum J, DeSalle R, Do Val F. (2003) The placement of Enigscaptomyza, Grimshawomyia, and Titanochaeta, three clades of endemic Hawaiian Drosophilidae (Diptera). Zootaxa 159: 1–16 [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jorgensen JE, Weigel D, Andersen SU. (2009) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Whiteman NK, Groen SC, Chevasco D, Bear A, Beckwith N, Gregory TR, Denoux C, Mamarella N, Ausubel FM, Pierce NE. (2010) Mining the plant-herbivore interface with a leafmining Drosophila of Arabidopsis. Mol Ecol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham TG, Difazio SP, Schweitzer JA, Shuster SM, Allan GJ, Bailey JK, Woolbright SA. (2008) Extending genomics to natural communities and ecosystems. Science 320: 492–495 [DOI] [PubMed] [Google Scholar]
- Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, Zhao P, Zha X, Cheng T, Chai C, et al. (2004) A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 306: 1937–1940 [DOI] [PubMed] [Google Scholar]