Abstract
Some plants can avoid shaded conditions via rapid shoot elongation, thus growing into better lit areas in a canopy. Cell wall-modifying mechanisms promoting this elongation response, therefore, are important regulatory points during shade avoidance. Two major cell wall-modifying protein families are expansins and xyloglucan endotransglucosylase/hydrolases (XTHs). The role of these proteins during shade avoidance was studied in Arabidopsis (Arabidopsis thaliana). In response to two shade cues, low red to far-red light (implying neighbor proximity) and green shade (mimicking dense canopy conditions), Arabidopsis showed classic shade avoidance features: petiole elongation and leaf hyponasty. Measurement of the apoplastic proton flux in green shade-treated petioles revealed a rapid efflux of protons into the apoplast within minutes, unlike white light controls. This apoplastic acidification probably provides the acidic pH required for the optimal activity of cell wall-modifying proteins like expansins and XTHs. Acid-induced extension, expansin susceptibility, and extractable expansin activity were similar in petioles from white light- and shade-treated plants. XTH activity, however, was high in petioles exposed to shade treatments. Five XTH genes (XTH9, -15, -16, -17, and -19) were positively regulated by low red to far-red light conditions, while the latter four and XTH22 showed a significant up-regulation also in response to green shade. Consistently, knockout mutants for two of these XTH genes also had reduced or absent shade avoidance responses to these light signals. These results point toward the cell wall as a vital regulatory point during shade avoidance.
Crowding in natural plant communities or in crop fields leads to resource limitation and competition for the same. In order to survive in such a situation, plants need to be able to outgrow competing vegetation to get to the light. Rapid shoot elongation, coupled with an upward movement of the leaves, are two obvious morphological characteristics displayed by plants that are being shaded. Other features include reduced branching and, when the shading is persistent, an acceleration of flowering to produce seeds and thus ensure reproduction. Collectively, this suite of responses triggered by shade is referred to as the shade avoidance syndrome (SAS; Vandenbussche et al., 2005; Franklin, 2008). SAS is set into motion due to the modification of the spectral composition of light in a canopy. Light reflected from leaves gets enriched in far-red wavelengths due to the preferential absorption of red light. This reduction in red to far-red (R/FR) light is a very reliable signal of impending shade (Ballare et al., 1990). In closed canopies, light reflected from, as well as transmitted through, leaves has not just a low R/FR but also low blue fluence rates and a lower total light intensity. Perception of these light quality changes is possible in plants due to the presence of Pr and Pfr (Smith, 2000; Ishimaru et al., 2007), blue light receptors, the cryptochromes, and the phototropins (Christie and Briggs, 2001). Rapid shoot elongation during SAS involves, primarily, cellular expansion fueled by turgor-driven uptake of water, leading to increased pressure within cells. In order to allow further water uptake, the cell walls yield to this pressure by becoming more extensible (Cosgrove, 2000). Cell wall loosening is defined as a process where molecular modifications of the cell wall make a rigid, inextensible wall extensible (Cleland, 1971; Cosgrove, 1999). This results from the action of different proteins on the cell wall matrix, which weakens the cell wall, allowing it to yield to turgor pressure. Two cell wall-modifying protein families that are well characterized and implicated in cell expansion during growth and development are the expansins and the xyloglucan endotransglucosylase/hydrolases (XTHs; Cosgrove, 1999, 2005; Rose et al., 2002). Expansins were first identified as the mediators of acid-induced extension (AIE) in isolated cell walls. They are believed to act via disruption of the noncovalent interactions between cellulose and hemicelluloses (xyloglucan in most dicots) in the cell wall, thus allowing cell wall loosening (McQueen-Mason et al., 1992; Cosgrove, 2000).
Expansins are required for plant growth (Cho and Kende, 1997b; Vreeburg et al., 2005) and in developmental processes where modification of the cell wall is required, such as fruit softening (Brummell et al., 1999), abscission (Belfield et al., 2005), and plant-pathogen interactions (Cantu et al., 2008). Although manipulation of expansin gene expression has confirmed the role of these proteins as important players in the regulation of cell wall extensibility, in a few instances such studies have also revealed unexpected and counterintuitive effects (Caderas et al., 2000; Rochange et al., 2001) where the correlation between growth and expansin activity did not hold. XTHs are another family of wall-modifying proteins that exist as a large gene family in most plant species. XTHs also act on the xyloglucan-cellulose network in the cell wall, but, unlike expansins, they employ enzymatic mechanisms (hydrolysis and/or transglucosylation) to modify the cell wall (Nishitani and Tominaga, 1992; Rose et al., 2002). These proteins are also involved in plant growth and development, where wall modification is required (Campbell and Braam, 1999; Rose et al., 2002).
In the model plant Arabidopsis (Arabidopsis thaliana), SAS and the importance of the regulation of cell wall extensibility by wall-modifying proteins have been studied extensively, albeit separately. Both expansins and XTHs are present as large multigene families in Arabidopsis (Rose et al., 2002; Sampedro and Cosgrove, 2005) and have been implicated in Arabidopsis growth (Hyodo et al., 2003; Schopfer and Liszkay, 2006), in response to various environmental stresses such as drought and salt tolerance (Schopfer and Liszkay, 2006), and in response to pathogen/insect attack (Wieczorek et al., 2006; Divol et al., 2007). Rosette plants like Arabidopsis mostly respond to shade by moving their leaves (which normally have a prostrate orientation) upward, accompanied by an elongation of the petiole (Mullen et al., 2006; Djakovic-Petrovic et al., 2007). Numerous microarray studies have shown that this response involves massive and rapid changes in gene expression (Devlin et al., 2003; Salter et al., 2003; Sessa et al., 2005). In addition to transcription factors and hormone regulatory genes, a number of cell wall-related genes including XTHs and expansins are also regulated following treatment with shade signals (Ma et al., 2001; Sessa et al., 2005; Roig-Villanova et al., 2006). The aim of this study was to make use of this vast amount of available microarray data as well as other genomic resources in order to study the regulation of cell wall properties mediated by expansins and XTHs during shade avoidance responses in Arabidopsis. We used two types of light manipulations to mimic different stages of shading. A low R/FR indicates the presence of proximal neighbors and provides an early warning of impending canopy closure. Green shade replicates conditions that would occur in a dense canopy where there is already persistent shading and crowding from neighboring plants. Using these two distinct shade cues allowed any differences in the detection and response to these signals to be observed. Furthermore, it was of interest to see which particular members of the expansin or XTH multigene families might be involved and whether the regulation of these genes is light signal specific even though they ultimately bring about the same morphological response.
RESULTS
Arabidopsis Displays Typical Shade Avoidance Characteristics in Response to Shade Signals
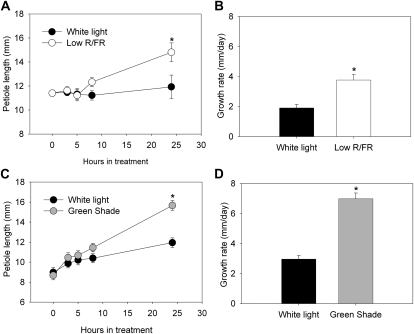
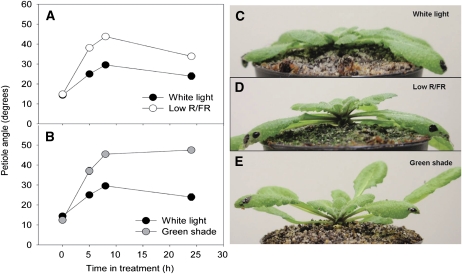
In response to low-R/FR (R/FR = 0.25) growth conditions, Arabidopsis Columbia (Col-0) plants showed a significant increase in the length of the petioles relative to control plants grown under white light (R/FR = 1.2) conditions after 24 h (Fig. 1A). The same trend was observed in plants grown under green shade conditions (combined reductions of blue, R/FR, and total light intensity), where a significant increase in the length of the petiole relative to control plants was observed after 24 h of treatment (Fig. 1C). The petiole growth rate of low-R/FR plants was almost double that of control plants, while the growth rate of green shade plants was more than twice that of controls (Fig. 1, B and D). Both these shade signals also induced an increase in leaf angle called hyponastic leaf movement. Under low R/FR, young leaves showed a significantly higher angle (to the horizontal axis) than their white light counterparts (Fig. 2A). At 5 h, this angle was already almost double that of white light controls and stayed higher until the 24-h measurement. The same was observed for green shade-treated plants, where the petiole angles were much higher at all the time points measured (except at 0 h; Fig. 2B). The increase in the angle of the petioles, however, was much higher in the green shade-treated plants compared with the low-R/FR-treated plants. Furthermore, while in low-R/FR-treated plants only the youngest leaves (also used for the measurements in Fig. 2) showed hyponasty, in green shaded plants, by 24 h almost all the leaves were hyponastic (Fig. 2, C–E). Also, in low R/FR, the petiole angles decreased somewhat at 24 h relative to the 8-h reading, while the green shade leaves retained their strongly hyponastic response until the 24-h time point (Fig. 2, A and B).
Figure 1.
Petiole elongation in Arabidopsis in response to shade signals. Petiole length (A and C) and growth rates (B and D) of Arabidopsis (Col-0) plants in low R/FR and green shade as compared with plants grown under white light (spectral composition unaltered). Data are means ± se (n = 10). Asterisks indicate significant differences between light treatments (P < 0.05, Student’s t test).
Figure 2.
Petiole angles of Arabidopsis plants in response to shade signals. A and B, Petiole angles of Arabidopsis (Col-0) plants in low R/FR (white circles) and green shade (gray circles) compared with plants grown in white light (black circles; unaltered spectral composition). Data points represent means ± se (n = 10). Values were significantly different from each other at all time points measured (5, 8, and 24 h) except at 0 h (P < 0.05, Student’s t test). Error bars are smaller than the symbols depicted. C to E, Representative images showing petiole angles of Arabidopsis plants after 24 h in white light (C), low R/FR (D), and green shade (E). [See online article for color version of this figure.]
Shade Treatment Induces Rapid Apoplastic Acidification in Arabidopsis Petioles
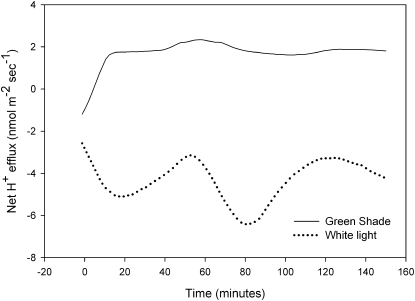
The microelectrode ion flux estimation technique was used in order to estimate the H+ fluxes in the apoplast of Arabidopsis petioles. Under white light conditions, there was a net negative efflux, which indicates an influx of protons from the apoplast into the cytoplasm. This influx was maintained, with some variation in the magnitude, throughout the period of measurement. In contrast, application of shade conditions using a green filter resulted in a rapid increase in the net proton flux into the apoplast (an efflux) that reached a maximum value in around 10 min (Fig. 3). Thereafter, although there was no increase in the flux, a steady efflux of protons was maintained for the remaining duration of the measurement (approximately 2.5 h).
Figure 3.
Apoplastic acidification in Arabidopsis petioles in response to green shade. Typical traces are shown for the net proton efflux at the abaxial side of an Arabidopsis (Col-0) petiole when illuminated with white light (dotted line) or green shade (solid line) conditions. Treatments started at 0 min. For each treatment, at least three runs were performed.
Expansin Activity Measured as AIE Is Not Regulated in Response to Shade Signals
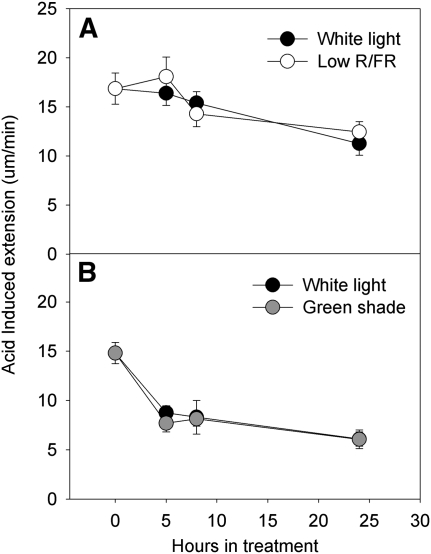
AIE in the petioles of low-R/FR-grown plants was not significantly different from AIE values measured in control plants (Fig. 4A). The same was observed for the petioles of plants grown in green shade (Fig. 4B). At all time points tested, there was no difference between the AIE of the petioles from plants in green shade and those in white light control growth conditions. However, for both light treatments, AIE values decreased after 24 h in treatment.
Figure 4.
AIE of Arabidopsis is not affected by shade signals. AIE is shown for petioles of Arabidopsis (Col-0) plants grown under low R/FR (white circles; A) and green shade (gray circles; B) compared with white light (black circles; spectral composition unaltered) controls. Data points represent means ± se (n = 8–10). Experiments were repeated twice with similar results.
Extractable Expansin Activity as Well as Susceptibility of Petioles to Expansins Are Similar in Control and Shade-Treated Plants
AIE values reflect the amount of extractable expansin activity in the petioles as well as the susceptibility of the cell walls in the petioles to an added expansin extract. Both these parameters were checked in plants grown in low-R/FR and green shade conditions. In response to the addition of equal amounts of a cucumber (Cucumis sativus) crude cell wall extract, the petioles from plants grown under both low R/FR and green shade showed similar extension rates as petioles from white light-grown control plants (Fig. 5). Thus, low-R/FR and green shade conditions did not make the petioles of the plant more sensitive to expansin action as compared with petioles from white light-grown control plants. The amount of extractable expansin present in petioles was checked via measurement of the extension produced in cucumber hypocotyls in response to the addition of expansin extracts from Arabidopsis petioles. Crude extracts from the petioles of plants grown in low R/FR and green shade produced comparable extension rates to that from petioles of plants grown in white light (Fig. 6).
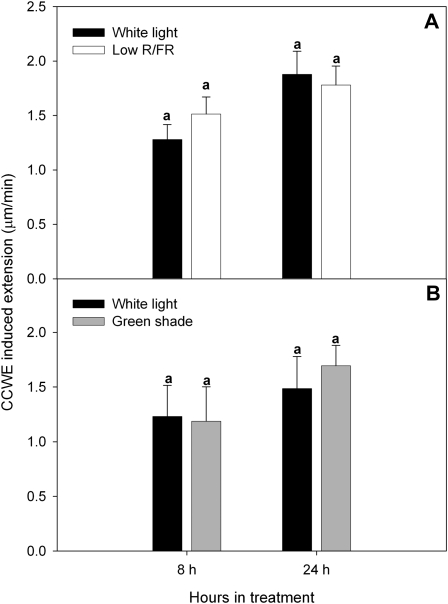
Figure 5.
Susceptibility of Arabidopsis petioles to a crude expansin extract. Petioles from Col-0 plants grown under low R/FR (A) and green shade (B) for 8 and 24 h were tested for their susceptibility to equal amounts of a crude cell wall expansin extract (CCWE) in an extensometer relative to white light control petioles. Data points represent means ± se (n = 5). Similar letters above the bars indicate nonsignificant differences (P < 0.05, Tukey’s b test).
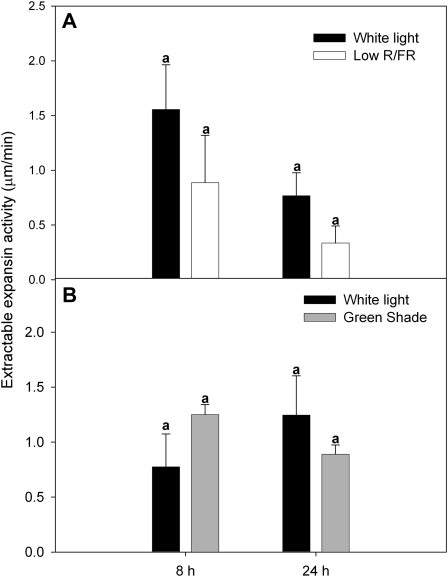
Figure 6.
Extractable expansin activity in Arabidopsis petioles. The amount of extractable expansin activity in the petioles of Col-0 plants grown for 8 and 24 h in low R/FR (A) and green shade (B) relative to white light controls was measured as the extension produced upon addition of the crude extract to cucumber hypocotyls. Data points represent means ± se (n = 3). Similar letters above the bars indicate nonsignificant differences (P < 0.05, Tukey’s b test).
XTH Activity Is Up-Regulated in Response to Shade Signals
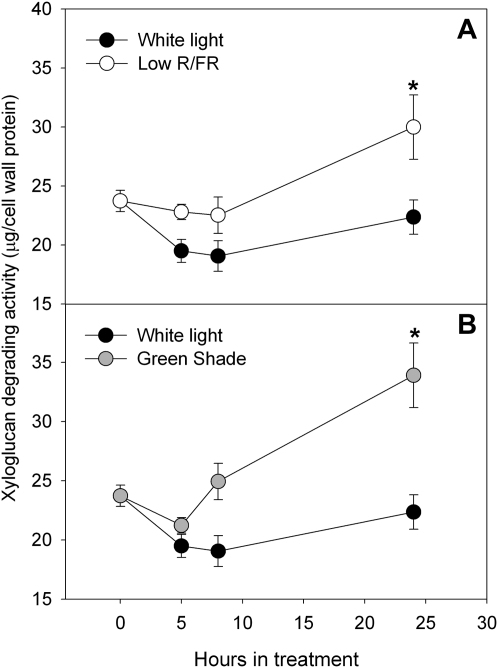
An in vitro assay was used to check for XTH activity in the petioles of Arabidopsis plants. XTH activity was measured as the amount of xyloglucan degraded by XTHs present in a crude enzyme extract made from petioles and was referred to as XDA. Enzyme extracts from the petioles of plants grown under low R/FR and green shade for 24 h had higher XDA values than corresponding white light controls (Fig. 7). In both treatments, the values were significantly higher than controls after 24 h of treatment. After 24 h in treatment, there were approximately 27% and 35% increases in the XTH activity in low-R/FR and green shade treatments, respectively, as compared with controls (Fig. 7).
Figure 7.
XTH activity in Arabidopsis petioles in response to shade signals. XTH activity was measured as XDA in the petioles of Arabidopsis (Col-0) plants grown in low-R/FR (white circles; A) and green shade (gray circles; B) conditions for 5, 8, and 24 h. Control refers to petioles from plants grown in white light (black circles) conditions. Data points represent means ± se (n = 4). Asterisks indicate significant differences (P < 0.05, Student’s t test). Experiments were repeated twice with similar results.
XTHs Are Differentially Regulated in Response to Shade Signals
The Genevestigator Web site (https://www.genevestigator.com/; Hruz et al., 2008) was mined for light quality control of XTH gene expression data. These light quality manipulations involved changes in the R/FR in a white light background as well as exposures to monochromatic red, far-red, or blue light intensities. These data were used for the selection of XTH genes that showed a 2-fold or more change in expression in response to light quality manipulations, resulting in an initial list of 20 XTH genes (Table I; Supplemental Fig. S1). Data for organ-specific expression profiles of XTH genes were also obtained from the Genevestigator Web site (Supplemental Fig. S2). The initial list of 20 XTH genes was further filtered for genes that were highly expressed in the petioles (Table I). We also used available XTH pro::GUS lines to identify genes that showed GUS staining in shaded petioles (data not shown; Supplemental Table S1). The final list of candidate genes thus included genes that were found to be light quality regulated and that were either highly expressed in the petioles or showed positive GUS staining in the petioles, or both. This list was composed of the genes XTH6, -9, -15, -17, -19, and -22. In addition, two other XTHs were included in this final list: Because of the high sequence homology between XTH15 and XTH16 (Rose et al., 2002), the latter was added to the final list of candidate XTH genes. XTH17 was included since it shares promoter sequences with XTH19 (Vissenberg et al., 2005). The expression pattern of these eight XTH genes was studied using real-time quantitative reverse transcription (RT)-PCR.
Table I. XTH genes showing a minimum 2-fold change in expression based on Genevestigator data.
A selection of XTH genes based on expression data compiled from relevant light quality manipulation microarray experiments extracted from the Genevestigator Web site is shown.
| Gene Name | Arabidopsis Genome Initiative Identifier | High Petiole Expressiona | GUS Staining in Petioleb |
| AtXTH2 | At4g13090 | NA | |
| AtXTH3 | At3g25050 | NA | |
| AtXTH5 | At5g13870 | − | |
| AtXTH6 | At5g65730 | + | NA |
| AtXTH8 | At1g11545 | + | − |
| AtXTH10 | At2g14620 | NA | |
| AtXTH11 | At3g48580 | NA | |
| AtXTH12 | At5g57530 | − | |
| AtXTH13 | At5g57540 | NA | |
| AtXTH14 | At4g25820 | − | |
| AtXTH15 | At4g14130 | + | |
| AtXTH19 | At4g30290 | + | |
| AtXTH22 | At5g57560 | + | NA |
| AtXTH23 | At4g25810 | + | − |
| AtXTH24 | At4g30270 | + | − |
| AtXTH25 | At5g57550 | − | |
| AtXTH29 | At4g18990 | − | |
| AtXTH31 | At3g44990 | NA | |
| AtXTH32 | At2g36870 | + | − |
| AtXTH33 | At1g10550 | − |
Based on the data in Supplemental Figure S2, a gene is designated as highly expressed (+) in petioles or not.
Based on GUS staining in shaded petioles relative to white light controls: +, staining observed in shaded petioles of XTHpro::GUS reporter line plants; −, no staining; NA, no XTHpro::GUS lines available.
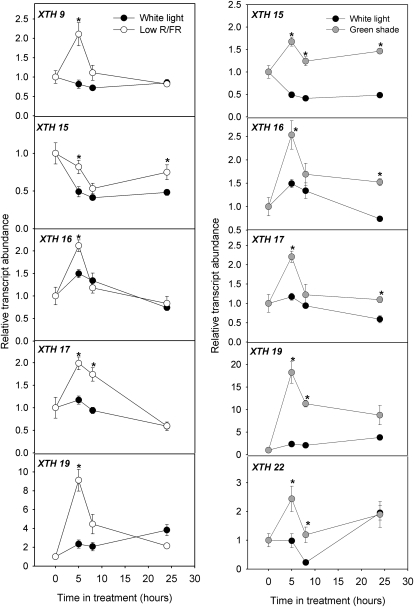
In response to low R/FR, five genes, XTH9, -15, -16, -17 and -19, were up-regulated. For all genes, significant increases in transcript abundance were observed at 5 h. This up-regulation was maintained until the last time point measured only for XTH15 (Fig. 8). In response to green shade, the expression of five XTH genes, XTH15, -16, -17, -19, and -22, was up-regulated relative to white light controls (Fig. 8). XTH9 showed no differential regulation in response to green shade (data not shown). XTH15 and -19 transcripts were significantly higher compared with controls at all time points measured. For all the genes, the highest fold up-regulation was observed at the 5-h time point (Fig. 8). XTH6 showed no differences in expression between controls and shade-treated plants (data not shown).
Figure 8.
Differential regulation of XTHs in response to low R/FR and green shade. XTH expression profiles are shown for the petioles of Arabidopsis (Col-0) plants in response to low R/FR (white circles) and green shade (gray circles) as compared with white light controls (black circles). Values are expressed relative to the transcript abundance at 0 h, which was set as 1. Data points represent means ± se (n = 4). Asterisks beside data points indicate statistically significant differences (P < 0.05, Student’s t test) for the low-R/FR and green shade treatments relative to white light controls at corresponding time points.
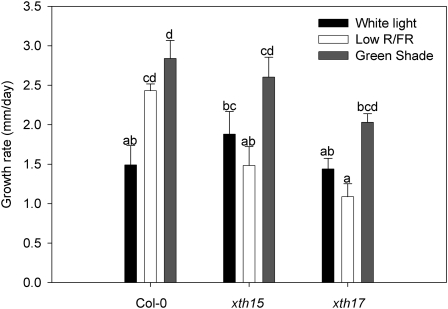
XTH Mutants Have Reduced Petiole Growth Rates under Shade Conditions
Two knockout mutant lines for the XTH genes XTH15 and XTH17 showed reduced petiole growth rates in response to both low R/FR and green shade (Fig. 9). This reduction was more pronounced for a plant growing under low-R/FR conditions, where the normally enhanced petiole growth rates seen in wild-type plants was completely abolished. In green shade, although there was an increase in growth rates relative to white light controls, this increase appears reduced compared with the trend observed for wild-type plants.
Figure 9.
Petiole growth rates of wild-type Col-0, xth15, and xth17 in response to low R/FR and green shade. Data points represent means ± se (n = 5–7). Similar letters above the bars indicate nonsignificant differences (P < 0.05, Tukey’s b test).
DISCUSSION
The control of cell wall extensibility is an important regulatory point during fast elongation responses. The primary goal of this study was to investigate this regulation in Arabidopsis during shade avoidance, when timely and rapid elongation of the petiole is crucial when competing for limited resources in a dense canopy. We investigated the possible role of two candidate wall-modifying proteins, expansins and XTHs, in regulating cell wall extensibility and hence growth responses to two shade signals, low R/FR (an early warning signal of impending shade) and green shade (mimicking a dense canopy).
Shade Avoidance and Arabidopsis
Rosette plants like Arabidopsis are more prone to shading because they form leaves that are close to the ground. Perception of shade in such plants usually results in a repositioning of the leaves, which helps to avoid shading imposed by neighboring plants. Similar morphological responses were observed when Arabidopsis was exposed to low-R/FR and green shade conditions (Figs. 1 and 2). Petiole growth rates were significantly higher in Arabidopsis plants exposed to low R/FR and green shade as compared with control plants grown in white light. The magnitude of the response, however, was greater in green shade than in low R/FR (Fig. 1). Similar trends were observed when the angle of the petiole was measured. Although both shade signals triggered a hyponastic response, angles were much higher under green shade than in low R/FR (Fig. 2). These responses are consistent with the implication of the individual signals. Low R/FR implies a threat of shading and elicited a milder response than green shade, which mimicked an already present shading stress and therefore would require a stronger and persistent response. In Arabidopsis, both petiole growth and hyponasty require, primarily, cellular expansion, hyponasty being the result of differential growth in the petiole (Millenaar et al., 2005). This rapid cellular expansion is the result of turgor-driven water uptake and concomitant modification of the cell wall by proteins such as expansins and XTHs, allowing extension of the cell wall.
Cellular Expansion and Cell Wall Loosening during SAS
Among wall-modifying agents, expansins and XTHs have received particular attention, since their activity and gene expression profiles have most often matched growth trends during plant development and responses to biotic and abiotic stresses (Cosgrove et al., 2002; Rose et al., 2002). However, to our knowledge, this study is the first to characterize expansin and XTH activity during shade avoidance in Arabidopsis. Expansins are considered the primary mediators of cell wall loosening, and expansin gene expression and protein activity are usually found to be highly correlative to growth changes (Cosgrove, 2000; Cosgrove et al., 2002). The experiments performed here, however, suggested otherwise. There were no differences in the AIE values (reflecting in planta expansin activity) between low-R/FR- or green shade-treated petioles relative to their white light controls (Fig. 4). AIE values reflect the amount of extractable expansins in a tissue as well as the sensitivity of that tissue to expansins. However, these two factors were also not different between shade-treated and white light controls (Figs. 5 and 6). Expansin activity, therefore, did not show a clear correlation with petiole elongation during shade avoidance in Arabidopsis. However, it could be argued that in low-R/FR- and green shade-treated petioles, only a certain part of the petiole undergoes rapid elongation and hence would also have a correspondingly higher AIE value. Such differential cell wall extensibility in different parts of the same plant organ has been observed for rice (Oryza sativa; Cho and Kende, 1997a). However, experiments on Arabidopsis plants in our shade treatment setup suggest that petiole growth under shaded conditions occurs uniformly along the length of the petiole (data not shown). Furthermore, this is not the first instance where expansin activity has not correlated with growth responses. Similar findings have been cited in studies with tomato (Solanum lycopersicum) and Festuca pratensis, where growth trends did not coincide with expansin gene expression or protein levels (Caderas et al., 2000; Reidy et al., 2001; Rochange et al., 2001).
Since expansin activity did not seem to be regulated during shade avoidance, we turned our attention to XTHs as a possible means of regulation of cell wall properties. XTH activity was up-regulated in response to both low R/FR and green shade relative to white light controls (Fig. 7). The XTH gene family in Arabidopsis consists of 33 members with distinct organ- and developmental stage-specific expression (Rose et al., 2002; Becnel et al., 2006; Kurasawa et al., 2006). In order to identify which of these candidates were required during shade avoidance, we used a combination of available XTH::GUS reporter lines and published microarray data. This allowed us to narrow our search for potential candidates. These were further analyzed for the regulation of gene expression using real-time quantitative RT-PCR. In response to low R/FR, five genes, XTH9, -15, -16, -17, and -19, were found to be positively regulated, while green shade induced the up-regulation of XTH15, -16, -17, -19, and -22 transcripts (Fig. 8). Increase in XTH transcript abundance occurred already at the first time point measured (5 h), while XTH activity showed a significant increase only after 24 h of shade treatment. High-resolution growth measurements on Arabidopsis petioles exposed to low R/FR have demonstrated an enhancement of petiole growth relative to white light controls already within 2 h of the treatment (Djakovic-Petrovic et al., 2007). Discrepancies between XTH activity and growth profiles could be due to the following reasons. (1) Higher enzyme activities might be associated with particular fast-growing regions or even particular cell types of the petiole, which could get diluted when whole petioles were used for activity measurements. In fact, among all the XTH genes, XTH15 and XTH17 are found to be relatively enriched in epidermal cells compared with other cell types (Mustroph et al., 2009; J. Bailey-Serres, personal communication). This also lends more credence to the involvement of these genes in cellular expansion, since the epidermis has been implicated in driving expansive growth (Savaldi-Goldstein et al., 2007). (2) XTHs can also be involved in wall strengthening (Antosiewicz et al., 1997). It is possible that some XTH genes are also down-regulated during shade avoidance, leading to a total lower XTH activity in shade-treated petioles. (3) Rapid growth rates as seen within 2 h probably involve mechanisms in addition to XTH gene expression regulation. Cell wall dynamics are affected by the action of numerous cell wall proteins, the production of reactive oxygen species, which can also affect the cell wall structure (Schopfer and Liszkay, 2006), and a balance between the rates of wall loosening and wall synthesis.
The functionality of XTHs in the context of shade avoidance responses was confirmed in a screen of XTH T-DNA insertion lines exposed to shade conditions. Specifically, the mutant lines xth15 and xth17 showed decreased petiole elongation responses, particularly to low R/FR but also to green shade. Both these genes show strong up-regulation in plants growing under low-R/FR and green shade conditions (Fig. 9). This confirms the functional role of XTHs in the restructuring of the cell wall required for the cellular expansion process that fuels rapid petiole elongation during shade avoidance.
Apoplastic Acidification
Another theory that could account for the rapid elongation growth in response to shade, while reconciling the discrepancy between the timing of the growth rate increase and the protein activity profiles, is the acid growth theory. This theory describes the rapid elongation of plant organs in response to acidic conditions and states that the extrusion of protons into the apoplast allows immediate wall modification via the activation of wall-modifying proteins already present in the apoplast (Hager, 2003). Within minutes of a green shade treatment, Arabidopsis petioles showed a net increase in proton efflux as compared with white light-treated petioles, where this efflux was absent (Fig. 3). This rapid apoplastic acidification probably sets the optimal milieu for the activity of wall-modifying proteins such as expansins (Cosgrove, 2000), XTHs (Fry, 1998), and yieldins (Okamoto-Nakazato et al., 2000), all of which require acidic conditions for activity. Low pH is also known to enhance the production of hydroxyl radicals, which have been demonstrated to induce cell wall extension (Fry, 1998). In such a scenario, even with equal amounts of wall-loosening proteins in the cell walls of white light- and shade-treated plants, an apoplastic acidification in the latter situation would result in elongation growth only in response to shade. Such apoplastic acidification has also been reported in submergence-induced petiole elongation (Vreeburg et al., 2005).
Synergism of Cell Wall Loosening
We recently published a study implicating expansins in the shade-induced elongation growth in two ecotypes of the Caryophyllaceae species Stellaria longipes (Sasidharan et al., 2008). It was expected that the basic wall-modifying mechanism underlying elongation responses to identical cues (low R/FR and green shade) would be similar. Therefore, it was surprising to observe that, unlike S. longipes, in Arabidopsis XTHs show a much stronger correlation with shade-induced elongation growth. Even though the cell wall contains numerous enzyme activities, expansins have been considered primary wall-loosening agents mostly due to their ability to bring about extension of isolated wall specimens (Cosgrove, 2005). Most studies investigating elongation growth in response to environmental cues have supported this notion (Wu et al., 1996; Cho and Kende, 1997a; Vriezen et al., 2000; Cosgrove et al., 2002; Sasidharan et al., 2008). However, there have also been a few studies where expansin activity and/or gene expression have not matched growth trends (Caderas et al., 2000; Rochange et al., 2001). In addition, XTHs have recently been demonstrated to be able to bring about wall-loosening activity in vitro (Van Sandt et al., 2007). Therefore, XTHs, although sometimes considered secondary wall-loosening agents, do have the ability to be primary candidates. This study also supports the role of XTHs as primary wall-modifying agents during shade avoidance in Arabidopsis. However, it is likely not to be the sole mechanism. Cell wall properties are very often the result of coordinate activities of cell wall assembly, disassembly, polymer synthesis, and secretion. Most large-scale analyses of cell walls have revealed a number of cell wall-related proteins to be regulated during different developmental stages and in response to environmental stresses. These proteins act on specific parts of the cell wall and have characteristic effects on it. In all likelihood, wall loosening requires a synergism between these different proteins. In Arabidopsis as well, the primary action of XTHs is probably enhanced or supplemented by other wall-modifying agents such as expansins, endoglucanases, and free radicals.
During expansion, regulation occurs not just at the level of wall modification but also for the delivery of wall components to the cell wall (Cosgrove, 2005). Furthermore, cell wall composition differs not just between species but also between different developmental stages and in response to different environmental stresses, as has been demonstrated for Arabidopsis roots (Freshour et al., 1996). Thus, the composition of a particular cell wall at a particular time, and in a particular species and organ, would probably dictate the kind of enzymatic activities required. This suggestion is supported by large-scale transcriptomic and proteomic studies that have revealed how not just different protein families, but also specific members within these protein families, are coordinately expressed at different developmental stages (Yokoyama et al., 2004; Imoto et al., 2005; Irshad et al., 2008). In addition, comparisons have been drawn with cellulose-digesting microbes that utilize multiprotein complexes to coordinate multiple protein activities while acting on their target. It is speculated that such a complex might exist in plants and would allow multiple proteins to be present together at the right location in the cell wall, thereby increasing the efficiency of wall modification (Rose and Bennett, 1999). Finally, the activities of both expansins and XTHs can be affected by conditions in the cell wall, such as substrate availability, accessibility to the target, the presence of activators/inhibitors, and also the pH of the apoplast. There was a rapid efflux of protons into the apoplastic space in Arabidopsis petioles when exposed to shaded conditions. Thus, the amount of acidification can also determine which proteins are active and to what extent. Therefore, coordination with other pathways that determine the optimal milieu for enzyme activities would also be essential.
CONCLUSION
In conclusion, these results demonstrate that Arabidopsis can clearly distinguish between different shade signals that imply either neighbor proximity (low R/FR) or shade (green shade). In response to each of these signals, Arabidopsis plants responded with characteristic morphological responses that differed in magnitude and that corresponded with an increase in XTH activity. Different XTH genes were expressed in response to each shade cue, and this increase in gene expression is probably responsible for the observed increase in XTH activity. Furthermore, the reduced ability of xth mutants to enhance petiole elongation when shaded points toward a functional role for XTHs in regulating cell wall properties that are essential for elongation responses during shade avoidance. Although expansin activity appeared not to be regulated during responses to shade, they may play a role in regulating wall extensibility through a collaborative or synergistic action with other wall-modifying mechanisms, including XTHs. Rapid apoplastic acidification was initiated upon green shade conditions and probably provides the optimum environment for wall-modifying processes to occur.
MATERIALS AND METHODS
Plant Growth
Both wild-type and transgenic Arabidopsis (Arabidopsis thaliana) plants used were in the Col-0 wild-type background. Seeds were sown on a moist filter paper placed in sealed petri dishes. After a 4-d stratification period in the dark at 4°C, the petri dishes were placed in a growth chamber for another 4 d in order to allow seed germination. Seedlings were then transferred to 70-mL pots filled with a soil and perlite (1:2, v/v) mixture containing 0.14 mg of MgOCaO (17%; Vitasol) and 0.14 mg of slow-release fertilizer (Osmocote “plus mini”; Scotts Europe). Prior to seedling transfer, the pots containing soil were allowed to soak up 20 mL of nutrient solution containing 2.6 mm KNO3, 2.0 mm Ca(NO3)2, 0.6 mm KH2PO4, 0.9 mm MgSO4, 6.6 mm MnSO4, 2.8 mm ZnSO4, 0.5 mm CuSO4, 66 mm H3BO3, 0.8 mm Na2MoO4, and 134 mm Fe-EDTA, pH 5.8. All chemicals were analytical reagent grade and obtained from Merck. The pots were placed on irrigation mats that were automatically watered every day in a short-day (9-h photoperiod, 200 μmol m−2 s−1 photosynthetically active radiation [PAR]) growth chamber for 4 weeks before being used for experiments.
Light Treatments
Four-week-old plants were used for experiments. Light quality manipulations took place in a white light background (Philips Master HPI-T Plus 400 W and Philips Plus Line Pro 150 W). The R/FR was reduced from 1.2 to 0.25 by supplemental far-red light (730-nm light-emitting diode; Shinko Electronics [http://www.shinkohelecs.com]). PAR for this light treatment was maintained at 140 μmol m−2 s−1. Green shading mimicking light conditions in a dense canopy was achieved using two layers of Lee 122 Fern Green, which reduced the PAR to 65 μmol m−2 s−1, the R/FR to 0.19, and the blue light photon fluence rate to 2 μmol m−2 s−1. Wherever mentioned, white light control refers to data from plants grown in light conditions with an unaltered spectral composition and PAR of 140 μmol m−2 s−1. All light treatments were started at approximately 10 am each time the experiments were performed.
Measurement of Plant Growth
Care was taken to choose similar-sized plants. The same leaf in each pot was marked with ink, and the corresponding petiole was measured using a digital caliper at relevant time points at the start and for the duration of specific treatments. For each treatment, petiole lengths were measured from at least 10 individual plants. Measurements were made for three independent trials.
Measurement of Leaf Angle
The leaves to be measured were marked with ink before the start of the treatments. Leaves obstructing the leaves to be measured were cut in order to be able to make angle measurements. Angle measurements were made by photographing the plants at relevant time points and then measuring the angle X between the two marked leaves using the angle measurement tool of the image software ImageJ (Abramoff et al., 2004). This angle X was then used to estimate the angle of the marked leaf (Y) from the soil surface using the formula Y = (180 – X)/2. At least 10 plants were used per treatment per time point. The experiments were repeated twice.
Measurement of Apoplastic Proton Fluxes
The microelectrode ion flux estimation (Newman, 2001) technique was used to measure net H+ fluxes in petioles from 28-d-old Arabidopsis plants. Microelectrodes were pulled from borosilicate glass capillaries (GC150-10; Harvard Apparatus) and silanized with tributylchlorosilane (Fluka). Before use, the microelectrodes were backfilled with 15 mm NaCl plus 40 mm KH2PO4 and the tip was filled with Hydrogen Ionophore II (Cocktail A; Fluka). After calibration of the microelectrode (pH 5.1–7.8), a young Arabidopsis leaf was detached from the plant, its petiole was gently abraded using Carborundum, and the leaf was then mounted in a 3-mL cuvette filled with 1 mm KCl. The cuvette was then placed in front of an inverted microscope (Nikon), and the H+ selective microelectrode was mounted vertically in a holder connected to a three-way piezo-controlled micromanipulator (Luigs and Neumann) driven by a computer-controlled motor (M061-CE08; Superior Electric). The electrode was then brought into position next to the abaxial surface of the petiole at a distance of 20 μm. During measurements, the distance between the microelectrode and the petiole surface changed from 20 to 60 μm with a frequency of 0.1 Hz. For controls, measurements were performed with the leaf illuminated using two fiber-optic cables (PAR of 180 μm m−2 s−1). For shade treatments, two layers of green filter (Lee 122 Fern Green) were placed across the two fiber-optic cables. The solution used to submerge the leaf was bubbled overnight with CO2-free air, and during the experiment CO2-free air was blown over the surface of the cuvette to minimize the effect of changing CO2 concentrations on the pH of the experimental solution. Chemical activity of H+ was recorded at two distances, and these data were then used to generate H+ flux values according to Newman (2001).
Measurement of AIE
Leaves with petioles that were at least 6 mm in length were marked before the start of the light treatments. This was the minimal starting length required in order to be able to place the petiole in the extensometer setup. Petioles were harvested at relevant time points and immediately frozen in liquid nitrogen. Thawed, abraded, and pressed petioles were clamped in the extensometer cuvette under a constant weight of 20 g. The petioles were first bathed in 160 μL of a 50 mm HEPES (pH 6.8) buffer for 30 min, after which the buffer was replaced with a 50 mm sodium acetate (pH 4.5) buffer for another 30 min. AIE was measured as the difference in the slopes of lines fitted through 10-min intervals before and after the observed bending point obtained upon the change in pH of the buffer. For each treatment, at least six to eight petioles from individual plants were measured. Experiments were repeated at least two times.
Extraction of Cell Wall Proteins and Measurement of Extractable Expansin Protein Activity
Crude cell wall extracts were made as described by Rochange et al. (2001) and modified to extract in 1.5-mL tubes. Briefly, 30 Arabidopsis petioles (two petioles per plant) were harvested and pooled per biological replicate, per treatment, per time point. The petioles were immediately frozen and then ground in a homogenization buffer. The cell walls were retained via centrifugation (21,000g, 1 min). The cell wall pellet was then washed several times with a wash buffer. This was followed by the addition of the extraction buffer containing 1 m NaCl. The cell walls were then allowed to incubate in this extraction buffer, after which the supernatant was recovered via centrifugation. The proteins were precipitated via the addition of ammonium sulfate and then recovered via centrifugation. Protein pellets were suspended in 50 mm sodium acetate buffer, pH 4.5, and the amount of protein was estimated using the method of Bradford (1976).
Susceptibility of Arabidopsis Petioles to Crude Cell Wall Extracts from Cucumber Hypocotyls
Arabidopsis petioles were harvested from plants grown under different light treatments (control, low R/FR, and green shade) and then frozen immediately and stored at −80°C. Frozen, thawed petioles were then boiled for 5 min to eliminate any endogenous expansin activity. These petioles were then pressed for 2 min and mounted in the extensometer setup under a constant pulling weight of 20 g. Extension was first measured for 30 min in an acidic (50 mm sodium acetate, pH 4.5) buffer, after which the buffer was replaced with a crude cell wall cucumber (Cucumis sativus) extract in the same acidic buffer. All petioles received an equal amount of the protein extract. Susceptibility of the petioles to this crude protein extract was measured as the difference in the slopes of the lines produced before and after the addition of the protein extract.
Measurement of XDA
Petioles from Arabidopsis Col-0 plants grown under different light treatments were harvested at different (0, 5, 8, and 24 h) time points. Harvested material was immediately frozen in liquid nitrogen and stored at −80°C until it was ready to be used. Enzyme extracts were prepared as described by Soga et al. (1999). Briefly, frozen petioles were homogenized in ice-cold sodium phosphate buffer (10 mm, pH 7). The homogenate was centrifuged, and the supernatant was discarded. The remaining cell wall pellet was washed twice with sodium phosphate buffer (10 mm, pH 7), after which the wall pellet was resuspended in sodium phosphate buffer (10 mm, pH 6) containing 1 m NaCl. The walls were then allowed to extract in this buffer for 24 h at 4°C before centrifugation and removal of the supernatant. This supernatant was then used as a crude enzyme extract to measure XDA as described by Sulova et al. (1995). The reaction mixture contained 15 μL of the enzyme extract, 0.4 mg mL−1 xyloglucan (Megazyme International), and 0.2 mg mL−1 xyloglucan oligosaccharides (XGOs; Megazyme International) in 0.2 mL of 0.1 m sodium phosphate buffer, pH 6. XGOs act as additional glycosyl acceptors for the transglucosylation reaction and thereby stimulate the depolymerization of xyloglucan by the enzyme. Following incubation of the reaction mixture for 1 h at 37°C, the reaction was terminated via the addition of 0.1 mL of 1 n HCl. The remaining xyloglucan was then detected by the iodine staining method, followed by spectrophotometric measurement of the resulting xyloglucan-iodine complex as described by Sasidharan et al. (2008). The XDA measured includes the transglucosylating activity and hydrolytic activity of XTHs as well as the hydrolytic activity of nonspecific endoglucanases. However, parallel assays run without XGOs (wherein the enzyme would function only as a hydrolase) indicated that the measured values had negligible hydrolytic activity at the termination of the assay. The values measured, therefore, are indicative of XET activity and are expressed as percentage XDA per microgram of cell wall protein. Protein estimation was performed using the Bradford (1976) assay using a commercially available Bradford Reagent (Bio-Rad). Twenty petioles from different plants were pooled together to obtain a crude enzyme extract, and this constituted one biological replicate. Each experiment used at least three biological replicates. Experiments were repeated twice.
Selection of Candidate XTH Genes
In order to identify which of the 33 Arabidopsis XTH genes are regulated during shade-induced petiole elongation, we mined the Genevestigator Web site (https://www.genevestigator.com/; Hruz et al., 2008) for a comprehensive analysis of XTH expression regulation. Expression data for the effect of light quality (R/FR, red, far-red, and blue light wavelengths) manipulations were compiled. The raw data from the Web site were used to generate a heat map using the TM4 MeV software (download available from http://www.tm4.org/; Saeed et al., 2006). The Genevestigator Web site was also used to extract XTH organ-specific expression data, which were plotted using the same software (TM4 MeV). We also screened available XTHpro::GUS lines (Vissenberg et al., 2005; Becnel et al., 2006; Liu et al., 2007) for candidate genes regulated upon shade treatment. The Genevestigator and XTHpro::GUS reporter data were combined to get the final list of XTH candidate genes. The regulation of these XTH genes was studied using real-time RT-PCR.
Real-Time RT-PCR
Petioles from 4-week-old Arabidopsis Col-0 plants grown under different light treatments (low R/FR, green shade, and control light conditions) were harvested at different time points (5, 8, and 24 h) during the treatments. Two petioles per plant were harvested, and at least 10 petioles were pooled to form one biological replicate. The same two petioles were harvested from each plant. Harvested material was immediately frozen and stored at −80°C. All the data shown are means of four biological replicates. Total RNA from these samples was extracted using the RNeasy plant mini kit (Qiagen). RT of total RNA using random hexamers was performed as described above. Real-time RT-PCR was performed using Arabidopsis ubiquitin (AtUBQ10) as an internal standard in a 20-μL reaction that contained 11 μL of SYBR Green Supermix (Bio-Rad; catalog no. 170-8882), 30 ng of cDNA (0.1 ng for 18S rRNA), and gene-specific primers (Table I). A Bio-Rad MyiQ single-color real-time PCR detection system was used. The following program was used for all the genes tested: 3 min at 95°C, followed by 40 cycles of 30 s at 95°C, 30 s at gene-specific annealing temperature, and 60 s at 72°C. The annealing temperature was optimized for each primer pair to result in specific amplification of the transcript of interest while avoiding the formation of primer dimers. Primer sequences and annealing temperatures are given in Table II. In addition, for every primer combination used, efficiency and melting curves were obtained. PCR products were also resolved on 1% agarose gels in order to confirm single products of the expected size. The cycle threshold (Ct) value for each gene was normalized relative to the Ct value of AtUBQ10. Relative transcript levels were calculated using the comparative Ct method (Livak and Schmittgen, 2001) and expressed relative to the average value at day 0, which was set as 1.
Table II. Sequences (5′–3′) of primer combinations and annealing temperatures for the genes studied using real-time RT-PCR.
| Arabidopsis Genome Initiative Identifier | Gene Name | Forward Primer | Reverse Primer | Annealing Temperature |
| °C | ||||
| At4g03210 | AtXTH9 | TACCATGAATACAACACTGCGTTTACT | TACCATGAATACAACACTGCGTTTACT | 62 |
| At4g14130 | AtXTH15 | CGGCACCGTCACTGCTTAC | GAAACTCAAAGTCTATCTCGTCATGTG | 62 |
| At3g23730 | AtXTH16 | CCGGTAACTCCGCTGGAA | TCTCGTCGTGTGTTGGTCCTT | 62 |
| At1g65310 | AtXTH17 | ATGGGCTAATGGAAAATCATCTTGTT | TACTTTGCACACCTTTCATTCTTGTC | 62 |
| At4g30290 | AtXTH19 | TGCAGCTAAATGATTGATTCTTTGAT | CCATTGAGTTACAAAGACAACGTCA | 62 |
| At5g57560 | AtXTH22 | CTAAAGAGTGCTTAGCTGCATAGAGAGA | CAAATCAATAAAATTCACGTGATCTACAA | 62 |
| At4g05320 | AtUBQ10 | GGCCTTGTATAATCCCTGATGAATAAG | AAAGAGATAACAGGAACGGAAACATAGT | 62 |
Mutant Screen
T-DNA insertion lines for several XTH genes were obtained from the Salk Institute. xth17 (SALK_008429) has been previously characterized (Osato et al., 2006). xth15 (SALK_039464) was subjected to the following analyses after three backcrosses to the wild type. The genotype of the T-DNA insertion allele was determined by PCR of genomic DNA using primer sets of an XTH15-specific forward primer (5′-CTGGAAGCTTAATCACTATGTAGGAGGATGCG-3′), a T-DNA left border-specific primer (5′-GCGTGGACCGCTTGCTGCAACT-3′), and an XTH15-specific reverse primer (5′-CCAGGAATGCTTTATTGATCTTGAC-3′). Mutant lines were exposed to low R/FR and green shade conditions, which were as described before. There were no size differences between wild-type Col-0 and mutant plants. Petiole measurements were made at 0 and 24 h after the start of the light treatments. Measurements were made as described before. Experiments were repeated twice.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genevestigator Arabidopsis XTH gene expression in response to light quality manipulations.
Supplemental Figure S2. Genevestigator XTH organ-specific expression.
Supplemental Table S1. XTH genes screened for shade-induced differential gene expression using XTHpro::GUS lines.
Supplementary Material
Acknowledgments
We thank Prof. Janet Braam (Rice University) for supplying XTH::GUS lines that were used in screening for candidate XTH genes.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. (2004) Image processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Antosiewicz DM, Purugganan MM, Polisensky DH, Braam J. (1997) Cellular localization of Arabidopsis xyloglucan endotransglycosylase-related proteins during development and after wind stimulation. Plant Physiol 115: 1319–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballare CL, Scopel AL, Sanchez RA. (1990) Far-red radiation reflected from adjacent leaves: an early signal of competition in plant canopies. Science 247: 329–332 [DOI] [PubMed] [Google Scholar]
- Becnel J, Natarajan M, Kipp A, Braam J. (2006) Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol Biol 61: 451–467 [DOI] [PubMed] [Google Scholar]
- Belfield EJ, Ruperti B, Roberts JA, McQueen-Mason S. (2005) Changes in expansin activity and gene expression during ethylene-promoted leaflet abscission in Sambucus nigra. J Exp Bot 56: 817–823 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH, Dunsmuir P. (1999) Differential expression of expansin gene family members during growth and ripening of tomato fruit. Plant Mol Biol 39: 161–169 [DOI] [PubMed] [Google Scholar]
- Caderas D, Muster M, Vogler H, Mandel T, Rose JK, McQueen-Mason S, Kuhlemeier C. (2000) Limited correlation between expansin gene expression and elongation growth rate. Plant Physiol 123: 1399–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P, Braam J. (1999) Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci 4: 361–366 [DOI] [PubMed] [Google Scholar]
- Cantu D, Vicente AR, Greve LC, Dewey FM, Bennett AB, Labavitch JM, Powell ALT. (2008) The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. Proc Natl Acad Sci USA 105: 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Kende H. (1997a) Expression of expansin genes in rice is correlated with growth. Plant Physiol (Suppl) 114: 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Kende H. (1997b) Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell 9: 1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Briggs WR. (2001) Blue light sensing in higher plants. J Biol Chem 276: 11457–11460 [DOI] [PubMed] [Google Scholar]
- Cleland R. (1971) Cell wall extension. Annu Rev Plant Physiol 22: 197–222 [Google Scholar]
- Cosgrove DJ. (1999) Enzymes and other agents that enhance cell wall extensibility. Annu Rev Plant Physiol 50: 391–417 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2000) Expansive growth of plant cell walls. Plant Physiol Biochem 38: 109–124 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Li LC, Cho HT, Hoffmann-Benning S, Moore RC, Blecker D. (2002) The growing world of expansins. Plant Cell Physiol 43: 1436–1444 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA. (2003) A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol 133: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divol F, Vilaine F, Thibivilliers S, Kusiak C, Sauge MH, Dinant S. (2007) Involvement of the xyloglucan endotransglycosylase/hydrolases encoded by celery XTH1 and Arabidopsis XTH33 in the phloem response to aphids. Plant Cell Environ 30: 187–201 [DOI] [PubMed] [Google Scholar]
- Djakovic-Petrovic T, de Wit M, Voesenek LACJ, Pierik R. (2007) DELLA protein function in growth responses to canopy signals. Plant J 51: 117–126 [DOI] [PubMed] [Google Scholar]
- Franklin KA. (2008) Shade avoidance. New Phytol 179: 930–944 [DOI] [PubMed] [Google Scholar]
- Freshour G, Clay RP, Fuller MS, Albersheim P, Darvill AG, Hahn MG. (1996) Developmental and tissue-specific structural alterations of the cell-wall polysaccharides of Arabidopsis thaliana roots. Plant Physiol 110: 1413–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC. (1998) Oxidative scission of plant cell wall polysaccharides by ascorbate-induced hydroxyl radicals. Biochem J 332: 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager A. (2003) Role of the plasma membrane H+-ATPase in auxin-induced elongation growth: historical and new aspects. J Plant Res 116: 483–505 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyodo H, Yamakawa S, Takeda Y, Tsuduki M, Yokota A, Nishitani K, Kohchi T. (2003) Active gene expression of a xyloglucan endotransglucosylase/hydrolase gene, XTH9, in inflorescence apices is related to cell elongation in Arabidopsis thaliana. Plant Mol Biol 52: 473–482 [DOI] [PubMed] [Google Scholar]
- Imoto K, Yokoyama R, Nishitani K. (2005) Comprehensive approach to genes involved in cell wall modifications in Arabidopsis thaliana. Plant Mol Biol 58: 177–192 [DOI] [PubMed] [Google Scholar]
- Irshad M, Canut H, Borderies G, Pont-Lezica R, Jamet E. (2008) A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: confirmed actors and newcomers. BMC Plant Biol 8: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru M, Smith DL, Gross KC, Kobayashi S. (2007) Expression of three expansin genes during development and maturation of Kyoho grape berries. J Plant Physiol 164: 1675–1682 [DOI] [PubMed] [Google Scholar]
- Kurasawa K, Matsui A, Saitou K, Yokoyama R, Nishitani K. (2006) A comprehensive analysis of all members of the group III subfamily of xyloglucan endotransglucosylase/hydrolase (XTH) family of Arabidopsis. Plant Cell Physiol 47: S72 [Google Scholar]
- Liu YB, Lu SM, Zhang JF, Liu S, Lu YT. (2007) A xyloglucan endotransglucosylase/hydrolase involved in growth of primary root and alters the deposition of cellulose in Arabidopsis. Planta 226: 1547–1560 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW. (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. (1992) Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4: 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millenaar FF, Cox MCH, de Jong van Berkel YEM, Welschen RAM, Pierik R, Voesenek LACJ, Peeters AJM. (2005) Ethylene-induced differential growth of petioles in Arabidopsis: analyzing natural variation, response kinetics, and regulation. Plant Physiol 137: 998–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JL, Weinig C, Hangarter RP. (2006) Shade avoidance and the regulation of leaf inclination in Arabidopsis. Plant Cell Environ 29: 1099–1106 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman IA. (2001) Ion transport in roots: measurement of fluxes using ion-selective microelectrodes to characterize transporter function. Plant Cell Environ 24: 1–14 [DOI] [PubMed] [Google Scholar]
- Nishitani K, Tominaga R. (1992) Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem 267: 21058–21064 [PubMed] [Google Scholar]
- Okamoto-Nakazato A, Takahashi K, Kido N, Owaribe K, Katou K. (2000) Molecular cloning of yieldins regulating the yield threshold of cowpea cell walls: cDNA cloning and characterization of recombinant yieldin. Plant Cell Environ 23: 155–164 [Google Scholar]
- Osato Y, Yokoyama R, Nishitani K. (2006) A principal role for AtXTH18 in Arabidopsis thaliana root growth: a functional analysis using RNAi plants. J Plant Res 119: 153–162 [DOI] [PubMed] [Google Scholar]
- Reidy B, McQueen-Mason S, Nosberger J, Fleming A. (2001) Differential expression of alpha- and beta-expansin genes in the elongating leaf of Festuca pratensis. Plant Mol Biol 46: 491–504 [DOI] [PubMed] [Google Scholar]
- Rochange SF, Wenzel CL, McQueen-Mason SJ. (2001) Impaired growth in transgenic plants over-expressing an expansin isoform. Plant Mol Biol 46: 581–589 [DOI] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou J, Sorin C, Devlin PF, Martinez-Garcia JF. (2006) Identification of primary target genes of phytochrome signaling: early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol 141: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Bennett AB. (1999) Cooperative disassembly of the cellulose-xyloglucan network of plant cell walls: parallels between cell expansion and fruit ripening. Trends Plant Sci 4: 176–183 [DOI] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K. (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43: 1421–1435 [DOI] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li JW, Thiagarajan M, White JA, Quackenbush J. (2006) TM4 microarray software suite. Kimmel AR, Oliver B, , DNA Microarrays, Part B: Databases and Statistics, Vol 411. Elsevier Academic Press, San Diego, pp 134–193 [DOI] [PubMed] [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC. (2003) Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426: 680–683 [DOI] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ. (2005) The expansin superfamily. Genome Biol 6: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R, Chinnappa CC, Voesenek LACJ, Pierik R. (2008) The regulation of cell wall extensibility during shade avoidance: a study using two contrasting ecotypes of Stellaria longipes. Plant Physiol 148: 1557–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Peto C, Chory J. (2007) The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202 [DOI] [PubMed] [Google Scholar]
- Schopfer P, Liszkay A. (2006) Plasma membrane-generated reactive oxygen intermediates and their role in cell growth of plants. Biofactors 28: 73–81 [DOI] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I. (2005) A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev 19: 2811–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. (2000) Phytochromes and light signal perception by plants: an emerging synthesis. Nature 407: 585–591 [DOI] [PubMed] [Google Scholar]
- Soga K, Wakabayashi K, Hoson T, Kamisaka S. (1999) Hypergravity increases the molecular mass of xyloglucans by decreasing xyloglucan-degrading activity in azuki bean epicotyls. Plant Cell Physiol 40: 581–585 [DOI] [PubMed] [Google Scholar]
- Sulova Z, Lednicka M, Farkas V. (1995) A colorimetric assay for xyloglucan-endotransglycosylase from germinating seeds. Anal Biochem 229: 80–85 [DOI] [PubMed] [Google Scholar]
- Vandenbussche F, Pierik R, Millenaar FF, Voesenek LACJ, Van der Straeten D. (2005) Reaching out of the shade. Curr Opin Plant Biol 8: 462–468 [DOI] [PubMed] [Google Scholar]
- Van Sandt VST, Suslov D, Verbelen JP, Vissenberg K. (2007) Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann Bot (Lond) 100: 1467–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Oyama M, Osato Y, Yokoyama R, Verbelen JP, Nishitani K. (2005) Differential expression of AtXTH17, AtXTH18, AtXTH19 and AtXTH20 genes in Arabidopsis roots: physiological roles in specification in cell wall construction. Plant Cell Physiol 46: 192–200 [DOI] [PubMed] [Google Scholar]
- Vreeburg RAM, Benschop JJ, Peeters AJM, Colmer TD, Ammerlaan AHM, Staal M, Elzenga TM, Staals RHJ, Darley CP, McQueen-Mason SJ, et al. (2005) Ethylene regulates fast apoplastic acidification and expansin A transcription during submergence-induced petiole elongation in Rumex palustris. Plant J 43: 597–610 [DOI] [PubMed] [Google Scholar]
- Vriezen WH, De Graaf B, Mariani C, Voesenek LACJ. (2000) Submergence induces expansin gene expression in flooding-tolerant Rumex palustris and not in flooding-intolerant R. acetosa. Planta 210: 956–963 [DOI] [PubMed] [Google Scholar]
- Wieczorek K, Golecki B, Gerdes G, Heinen P, Szakasits D, Durachko DM, Cosgrove DJ, Kreil DP, Puzio PS, Bohlmann H, et al. (2006) Expansins are involved in the formation of nematode-induced syncytia in roots of Arabidopsis thaliana. Plant J 48: 98–112 [DOI] [PubMed] [Google Scholar]
- Wu Y, Sharp RE, Durachko DM, Cosgrove DJ. (1996) Growth maintenance of the maize primary root at low water potentials involves increases in cell-wall extension properties, expansin activity, and wall susceptibility to expansins. Plant Physiol 111: 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Rose JKC, Nishitani K. (2004) A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice: classification and expression analysis. Plant Physiol 134: 1088–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.