Electron microscopy and light microscopy both have been essential tools for investigating molecular distribution and cell structure. While electron microscopy is capable of much higher resolution compared to light microscopy, it is prone to artifacts introduced by sample preparation and it produces only static images, making the analysis of dynamic processes challenging. In addition, methods for specific molecular labeling and detection in electron microscopy have faced significant limitations. Light microscopy, on the other hand, excels at specific labeling, particularly with genetically encoded tags, and can be used in living cells where the dynamics of molecules and organelles can be visualized. However, light microscopy has been limited in comparison to electron microscopy in its ability to detect single proteins and protein complexes and to resolve structure and molecular distributions below the diffraction limit of light.
Recent technical developments address the shortcomings in both electron and light microscopy, while retaining their strengths. New strategies for introducing genetically encoded tags are bringing greater molecular specificity to electron microscopy (for review, see Giepmans, 2008), and advances in light detectors and imaging methods have permitted single molecule detection in vivo and have increased the resolving power of light microscopy below the diffraction limit (Fig. 1). Here, we discuss recent advances in light microscopy and how they can be applied to further our understanding of plant cell structure and function, with an emphasis on the lateral organization of the plasma membrane (PM).
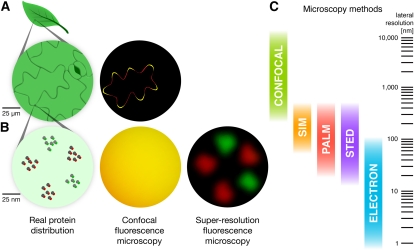
Figure 1.
Super-resolution fluorescence microscopy can reveal PM subcompartmentalization that is unresolvable by conventional techniques. A, Confocal microscopy is sufficient to visualize localization patterns at a scale of hundreds of nanometers. Two hypothetical PM proteins are labeled with different fluorescent tags in a pavement cell. One localizes to the tips of the cell lobes (green), much as observed for the small G-protein ROP2 (Fu et al., 2005), and the other is dispersed more uniformly (red). The overlap appears as yellow. B, At the nanoscale, these proteins also cluster into disparate microdomains. Super-resolution microscopy is necessary to resolve the two types of microdomains. All images are simulated. C, Comparison of the spatial scales at which different microscopy techniques are useful.
CELL MEMBRANES APPEAR TO BE HIGHLY ORGANIZED AND SUBCOMPARTMENTALIZED
The PM is the key site for molecular transport and signaling, the interface for communication with other cells, and an anchoring platform for cellular components. Although the molecular composition of the PM is well documented, surprisingly little is known about its organization. The fluid mosaic model, which represents the membrane as a random distribution of lipids and proteins undergoing free lateral diffusion (Singer and Nicolson, 1972), has been outdated by mounting evidence portraying a much more complicated architecture.
Fluorescence microscopy has revealed molecular sorting at the gross level, including PM polarization and segregation of proteins into domains or specialized structures such as caveolae and microvilli. In yeast (Saccharomyces cerevisiae), membrane proteins appear to be partitioned into at least two major domains (Malínská et al., 2003) that exhibit different rates of protein turnover for certain transporters (Grossmann et al., 2008). Furthermore, lateral diffusion of PM components seems to be partially restricted to 100- to 300-nm domains, as first shown for the transferrin receptor in animal cells (Sako and Kusumi, 1994). At yet smaller scales, on the order of tens of nanometers, the formation of protein-lipid clusters, termed microdomains, is thought to be important for the function of proteins that depend on interaction with specific lipids, the separation of biochemical and signaling pathways, and the cooperative interaction of some receptors (compare with e.g. Grossmann et al., 2008; Owen et al., 2010). For example, in animal cells, signaling events have been shown to trigger the clustering of receptors as shown by atomic force microscopy combined with fluorescence imaging (Ianoul et al., 2005).
In plants, our knowledge of PM organization is yet more limited, but evidence for PM microdomain formation is growing (Zappel and Panstruga, 2008). For example, the hexose transporter HUP1 of Chlorella kessleri shows clustering in both the Chlorella PM, as detected by antibody staining, and in the PM of yeast when heterologously expressed as a GFP fusion (Grossmann et al., 2006). The K+ channel KAT1 appears to be distributed in specific fine-scale patterns in different cell types when tagged by GFP: It exhibits punctate localization in tobacco (Nicotiana tabacum) epidermal cells (Sutter et al., 2006) and a striated pattern in Vicia faba guard cells (Homann et al., 2007). Fluorescently labeled cellulose synthase was observed as motile punctae at the PM, moving along linear tracks coincident with cortical microtubules (Paredez et al., 2006). Two recently reported examples are members of the peripheral PM protein families of remorins and flotillins, which play important roles in nodulation of legumes and show striking punctate membrane localization (Raffaele et al., 2009; Haney and Long, 2010).
LIPID RAFTS AND MICRODOMAINS
The concepts of microdomains and lipid rafts have been closely linked. The term lipid raft was introduced to describe microdomains of tightly packed lipids and proteins, enriched in sterols and sphingolipids, hypothesized to be dynamic structures of approximately 10 to 200 nm in diameter (Simons and Ikonen, 1997; Pike, 2006). Lateral partitioning of proteins into lipid rafts is likely spurred by specific interaction with membrane lipids (Anderson and Jacobson, 2002). Lipid rafts are thought to exhibit reduced susceptibility to detergent solubilization—a property that was extensively used to identify raft constituents. Detergent-resistant membranes have been isolated from several eukaryotic systems, including plants (Mongrand et al., 2004; Borner et al., 2005). However, such results have been criticized to not reliably represent in vivo situations (Munro, 2003). As a result, the attention that lipid rafts had once attracted waned over recent years, while a movement away from detergent extraction toward visualization has been undertaken (Lingwood and Simons, 2010). Based on biochemical evidence, some plant membrane-associated proteins have been suggested to localize and be stabilized in microdomains defined by lipid rafts, prominent among them the PIN proteins (Titapiwatanakun et al., 2009), but clear resolution of these domains has been elusive. New imaging technologies will likely galvanize a renaissance of this field, revealing lateral sorting patterns that were previously hidden from view.
STUDYING MICRODOMAINS IN VIVO IS LIMITED BY OPTICAL RESOLUTION
Optical resolution is limited by the diffraction of light, presenting a challenge for single protein complexes and raft-like microdomains, hypothesized to be 10 to 200 nm in diameter. Even with perfect optics, light can only be focused to a fuzzy spot, called a point spread function (PSF). If two fluorophores are in close proximity, their PSFs in the image plane may overlap to such an extent that it is impossible to distinguish the two spots. Diffraction sets the lower resolution limit of light microscopy at approximately 200 nm.
In recent years, several ingenious fluorescence microscopy techniques have broken the diffraction resolution barrier; methods now known as super-resolution imaging (Huang et al., 2009). Accomplishing this feat requires modification of the microscope’s excitation/emission pathway, computer-assisted image analysis, or both. Some of the most promising technologies include stimulated emission depletion (STED) microscopy, structured illumination microscopy (SIM), and the family of stochastic methods exemplified by photoactivated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM). Each has its advantages and drawbacks (Combs, 2010).
STED microscopy uses two lasers to produce a sharpened excitation spot, which is scanned across the specimen to build an image (Hell and Wichmann, 1994). The intensity of the depletion laser controls the effective spot size and ultimately the spatial resolution. The resolving power of this technique is, in theory, limitless; however, very high laser intensities are difficult to generate and potentially damaging to biological tissue, imposing practical constraints on achievable resolution. With a resolution obtained below 50 nm, STED nanoscopy helped to visualize the existence of nano-sized lipid domains for the first time in living cells (Eggeling et al., 2009). Even higher resolution below 20 nm had been achieved for biological samples (Donnert et al., 2006).
In SIM, a series of excitation light patterns are applied to the specimen. The resulting emission patterns are diffraction limited, yet contain information that can be decrypted to construct a super-resolution image. Resolution below 50 nm has been demonstrated (Gustafsson, 2005).
PALM and STORM share the same underlying principle (Betzig et al., 2006; Hess et al., 2006; Rust et al., 2006). Specimens are labeled with photoswitchable probes whose fluorescence can be turned on or off using particular wavelengths of light. A small subset of fluorophores is activated, imaged, and then deactivated. The PSF generated by each fluorophore is fitted with a Gaussian distribution to estimate the molecule’s coordinates. The imaging cycle is repeated multiple times to build a complete image. This methodology requires efficient detection of single fluorophores, a feat greatly aided by highly quantum efficient electron-multiplying CCD cameras. The precision of PSF fitting, which determines image resolution, improves as the number of detected photons increases. Resolutions of <20 nm have been obtained in biological samples (Betzig et al., 2006; Rust et al., 2006). Super-resolution techniques were initially limited to only two dimensions with monochrome fluorophores in fixed tissue; however, three-dimensional, multicolor, and live-cell imaging are now possible (Huang et al., 2009).
OPPORTUNITIES FOR SUPER-RESOLUTION MICROSCOPY IN PLANT CELL BIOLOGY
Three-dimensional SIM super-resolution microcopy was recently applied to plant cells to visualize the fine structure of plasmodesmata (Fitzgibbon et al., 2010). This technique permitted visualization of features that had only been previously resolved by electron microscopy, such as the apertures of individual plasmodesmal pores, and enabled the tracing in three dimensions of fine endoplasmic reticulum-derived tubules (sieve element reticulum) through these pores and between adjacent sieve element cells. To our knowledge, there are no published reports for the application of STED and PALM/STORM in plants to date. The super-resolution methods are usually paired with total internal reflection microscopy (TIRF-M), which restricts imaging to an approximately 100-nm slice adjacent to the cover glass. Confinement of excitation to this thin plane reduces background signal to enhance detection of single fluorophores. Cell walls are typically approximately 200 nm thick, lifting the PM away from the glass and out of range for TIRF imaging. However, workarounds may be available, such as electron-multiplying CCD camera-based spinning disc confocal microscopy and variable-angle epifluorescence microscopy (pseudo-TIRF; Konopka and Bednarek, 2008; Staiger et al., 2009). Access to the PM and peripheral cytosol might also be obtained with true TIRF-M by digesting the cell wall, but this manipulation would necessarily limit applications.
Live-cell imaging adds the dimension of time to data acquisition, enabling the study of molecular dynamics. However, tradeoffs between spatial resolution, temporal resolution, field of view, brightness, and number of frames must be weighed. STED is limited by photodamage (with high-depletion laser intensities) and long scanning times, the latter of which can be mitigated by a smaller field of view. A recent study using STED achieved a remarkable spatiotemporal resolution of 10 to 20 nm and <1 ms but was limited to an observation area of 0.05 μm2 (Sahl et al., 2010). Both SIM and PALM require multiple illumination cycles, resulting in lengthy acquisition times. Recent advances have pushed the resolution to 100 nm/91 ms (64 μm2 area) for SIM (Kner et al., 2009) and approximately 60 nm/25 s (>1,000 μm2 area) for PALM (Shroff et al., 2008). In addition, four-dimensional super-resolution techniques, such as the double-helix PSF method (Pavani et al., 2009), are promising but have yet to be optimized for biological specimen. Future improvements in equipment design and fluorophore engineering (e.g. greater photostability and brightness) will help to overcome unfavorable compromises researchers currently face with subdiffraction imaging.
Several outstanding questions in plant biology can only be addressed by live-cell super-resolution imaging. For instance, using confocal microscopy, cellulose synthase complexes have been observed to move bidirectionally along cortical microtubules (Paredez et al., 2006). It is unclear how these large complexes, confined within the plane of the PM, are able to move in opposite directions along the same tracks without mutual interference. One possible explanation is that complexes sort into two populations with respect to their direction of movement, one on either side of the microtubule bundle. Live-cell subdiffraction imaging may help solve these questions, as the two motile populations would be separated by less than 200 nm (microtubule diameter approximately 25 nm).
As super-resolution technologies mature, they will likely become the gold standard for colocalization studies as well as a valuable complement to techniques that detect heterotypic protein interaction. Positive colocalization by conventional confocal microscopy simply indicates that the two fluorophores are in close proximity—within the range of optical resolution—but does not allow us to discern whether two proteins physically interact (Lalonde et al., 2008). As resolution improves to molecular scales, colocalization becomes a more reliable indicator of possible protein-protein interaction in vivo. With its widespread utility and rising accessibility due to the recent introduction of commercial systems, super-resolution microscopy is poised to become an invaluable tool in plant cell biology.
References
- Anderson RGW, Jacobson K. (2002) A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science 296: 1821–1825 [DOI] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313: 1642–1645 [DOI] [PubMed] [Google Scholar]
- Borner GHH, Sherrier DJ, Weimar T, Michaelson LV, Hawkins ND, Macaskill A, Napier JA, Beale MH, Lilley KS, Dupree P. (2005) Analysis of detergent-resistant membranes in Arabidopsis: evidence for plasma membrane lipid rafts. Plant Physiol 137: 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs CA. (2010) Fluorescence microscopy: a concise guide to current imaging methods. Curr Protoc Neurosci 50: 2.1.1–2.1.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnert G, Keller J, Medda R, Andrei MA, Rizzoli SO, Lührmann R, Jahn R, Eggeling C, Hell SW. (2006) Macromolecular-scale resolution in biological fluorescence microscopy. Proc Natl Acad Sci USA 103: 11440–11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggeling C, Ringemann C, Medda R, Schwarzmann G, Sandhoff K, Polyakova S, Belov VN, Hein B, von Middendorff C, Schönle A, et al. (2009) Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 457: 1159–1162 [DOI] [PubMed] [Google Scholar]
- Fitzgibbon J, Bell K, King E, Oparka K. (2010) Super-resolution imaging of plasmodesmata using three-dimensional structured illumination microscopy. Plant Physiol 153: 1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z. (2005) Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700 [DOI] [PubMed] [Google Scholar]
- Giepmans BN. (2008) Bridging fluorescence microscopy and electron microscopy. Histochem Cell Biol 130: 211–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann G, Malinsky J, Stahlschmidt W, Loibl M, Weig-Meckl I, Frommer WB, Opekarová M, Tanner W. (2008) Plasma membrane microdomains regulate turnover of transport proteins in yeast. J Cell Biol 183: 1075–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann G, Opekarová M, Novakova L, Stolz J, Tanner W. (2006) Lipid raft-based membrane compartmentation of a plant transport protein expressed in Saccharomyces cerevisiae. Eukaryot Cell 5: 945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MGL. (2005) Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci USA 102: 13081–13086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney CH, Long SR. (2010) Plant flotillins are required for infection by nitrogen-fixing bacteria. Proc Natl Acad Sci USA 107: 478–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell SW, Wichmann J. (1994) Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett 19: 780–782 [DOI] [PubMed] [Google Scholar]
- Hess ST, Girirajan TPK, Mason MD. (2006) Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J 91: 4258–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann U, Meckel T, Hewing J, Hütt MT, Hurst AC. (2007) Distinct fluorescent pattern of KAT1:GFP in the plasma membrane of Vicia faba guard cells. Eur J Cell Biol 86: 489–500 [DOI] [PubMed] [Google Scholar]
- Huang B, Bates M, Zhuang X. (2009) Super-resolution fluorescence microscopy. Annu Rev Biochem 78: 993–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianoul A, Grant DD, Rouleau Y, Bani-Yaghoub M, Johnston LJ, Pezacki JP. (2005) Imaging nanometer domains of beta-adrenergic receptor complexes on the surface of cardiac myocytes. Nat Chem Biol 1: 196–202 [DOI] [PubMed] [Google Scholar]
- Kner P, Chhun BB, Griffis ER, Winoto L, Gustafsson MGL. (2009) Super-resolution video microscopy of live cells by structured illumination. Nat Methods 6: 339–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka CA, Bednarek SY. (2008) Variable-angle epifluorescence microscopy: a new way to look at protein dynamics in the plant cell cortex. Plant J 53: 186–196 [DOI] [PubMed] [Google Scholar]
- Lalonde S, Ehrhardt DW, Loqué D, Chen J, Rhee SY, Frommer WB. (2008) Molecular and cellular approaches for the detection of protein-protein interactions: latest techniques and current limitations. Plant J 53: 610–635 [DOI] [PubMed] [Google Scholar]
- Lingwood D, Simons K. (2010) Lipid rafts as a membrane-organizing principle. Science 327: 46–50 [DOI] [PubMed] [Google Scholar]
- Malínská K, Malinsky J, Opekarová M, Tanner W. (2003) Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol Biol Cell 14: 4427–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrand S, Morel J, Laroche J, Claverol S, Carde JP, Hartmann MA, Bonneu M, Simon-Plas F, Lessire R, Bessoule JJ. (2004) Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J Biol Chem 279: 36277–36286 [DOI] [PubMed] [Google Scholar]
- Munro S. (2003) Lipid rafts: elusive or illusive? Cell 115: 377–388 [DOI] [PubMed] [Google Scholar]
- Owen DM, Oddos S, Kumar S, Davis DM, Neil MA, French PM, Dustin ML, Magee AI, Cebecauer M. (2010) High plasma membrane lipid order imaged at the immunological synapse periphery in live T cells. Mol Membr Biol 27: 178–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW. (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Pavani SRP, Thompson MA, Biteen JS, Lord SJ, Liu N, Twieg RJ, Piestun R, Moerner WE. (2009) Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc Natl Acad Sci USA 106: 2995–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike LJ. (2006) Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J Lipid Res 47: 1597–1598 [DOI] [PubMed] [Google Scholar]
- Raffaele S, Bayer E, Lafarge D, Cluzet S, German Retana S, Boubekeur T, Leborgne-Castel N, Carde JP, Lherminier J, Noirot E, et al. (2009) Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. (2006) Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods 3: 793–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl SJ, Leutenegger M, Hilbert M, Hell SW, Eggeling C. (2010) Fast molecular tracking maps nanoscale dynamics of plasma membrane lipids. Proc Natl Acad Sci USA 107: 6829–6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y, Kusumi A. (1994) Compartmentalized structure of the plasma membrane for receptor movements as revealed by a nanometer-level motion analysis. J Cell Biol 125: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff H, Galbraith CG, Galbraith JA, Betzig E. (2008) Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat Methods 5: 417–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Ikonen E. (1997) Functional rafts in cell membranes. Nature 387: 569–572 [DOI] [PubMed] [Google Scholar]
- Singer SJ, Nicolson GL. (1972) The fluid mosaic model of the structure of cell membranes. Science 175: 720–731 [DOI] [PubMed] [Google Scholar]
- Staiger CJ, Sheahan MB, Khurana P, Wang X, McCurdy DW, Blanchoin L. (2009) Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. J Cell Biol 184: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter JU, Campanoni P, Tyrrell M, Blatt MR. (2006) Selective mobility and sensitivity to SNAREs is exhibited by the Arabidopsis KAT1 K+ channel at the plasma membrane. Plant Cell 18: 935–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titapiwatanakun B, Blakeslee JJ, Bandyopadhyay A, Yang H, Mravec J, Sauer M, Cheng Y, Adamec J, Nagashima A, Geisler M, et al. (2009) ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. Plant J 57: 27–44 [DOI] [PubMed] [Google Scholar]
- Zappel NF, Panstruga R. (2008) Heterogeneity and lateral compartmentalization of plant plasma membranes. Curr Opin Plant Biol 11: 632–640 [DOI] [PubMed] [Google Scholar]