Abstract
Ca2+ and nitric oxide (NO) are essential components involved in plant senescence signaling cascades. In other signaling pathways, NO generation can be dependent on cytosolic Ca2+. The Arabidopsis (Arabidopsis thaliana) mutant dnd1 lacks a plasma membrane-localized cation channel (CNGC2). We recently demonstrated that this channel affects plant response to pathogens through a signaling cascade involving Ca2+ modulation of NO generation; the pathogen response phenotype of dnd1 can be complemented by application of a NO donor. At present, the interrelationship between Ca2+ and NO generation in plant cells during leaf senescence remains unclear. Here, we use dnd1 plants to present genetic evidence consistent with the hypothesis that Ca2+ uptake and NO production play pivotal roles in plant leaf senescence. Leaf Ca2+ accumulation is reduced in dnd1 leaves compared to the wild type. Early senescence-associated phenotypes (such as loss of chlorophyll, expression level of senescence-associated genes, H2O2 generation, lipid peroxidation, tissue necrosis, and increased salicylic acid levels) were more prominent in dnd1 leaves compared to the wild type. Application of a Ca2+ channel blocker hastened senescence of detached wild-type leaves maintained in the dark, increasing the rate of chlorophyll loss, expression of a senescence-associated gene, and lipid peroxidation. Pharmacological manipulation of Ca2+ signaling provides evidence consistent with genetic studies of the relationship between Ca2+ signaling and senescence with the dnd1 mutant. Basal levels of NO in dnd1 leaf tissue were lower than that in leaves of wild-type plants. Application of a NO donor effectively rescues many dnd1 senescence-related phenotypes. Our work demonstrates that the CNGC2 channel is involved in Ca2+ uptake during plant development beyond its role in pathogen defense response signaling. Work presented here suggests that this function of CNGC2 may impact downstream basal NO production in addition to its role (also linked to NO signaling) in pathogen defense responses and that this NO generation acts as a negative regulator during plant leaf senescence signaling.
Senescence can be considered as the final stage of a plant’s development. During this process, nutrients will be reallocated from older to younger parts of the plant, such as developing leaves and seeds. Leaf senescence has been characterized as a type of programmed cell death (PCD; Gan and Amasino, 1997; Quirino et al., 2000; Lim et al., 2003). During senescence, organelles such as chloroplasts will break down first. Biochemical changes will also occur in the peroxisome during this process. When the chloroplast disassembles, it is easily observed as a loss of chlorophyll. Mitochondria, the source of energy for cells, will be the last cell organelles to undergo changes during the senescence process (Quirino et al., 2000). At the same time, other catabolic events (e.g. protein and lipid breakdown, etc.) are occurring (Quirino et al., 2000). Hormones may also contribute to this process (Gepstein, 2004). From this information we can infer that leaf senescence is regulated by many signals.
Darkness treatment can induce senescence in detached leaves (Poovaiah and Leopold, 1973; Chou and Kao, 1992; Weaver and Amasino, 2001; Chrost et al., 2004; Guo and Crawford, 2005; Ülker et al., 2007). Ca2+ can delay the senescence of detached leaves (Poovaiah and Leopold, 1973) and leaf senescence induced by methyl jasmonate (Chou and Kao, 1992); the molecular events that mediate this effect of Ca2+ are not well characterized at present.
Nitric oxide (NO) is a critical signaling molecule involved in many plant physiological processes. Recently, published evidence supports NO acting as a negative regulator during leaf senescence (Guo and Crawford, 2005; Mishina et al., 2007). Abolishing NO generation in either loss-of-function mutants (Guo and Crawford, 2005) or transgenic Arabidopsis (Arabidopsis thaliana) plants expressing NO degrading dioxygenase (NOD; Mishina et al., 2007) leads to an early senescence phenotype in these plants compared to the wild type. Corpas et al. (2004) showed that endogenous NO is mainly accumulated in vascular tissues of pea (Pisum sativum) leaves. This accumulation is significantly reduced in senescing leaves (Corpas et al., 2004). Corpas et al. (2004) also provided evidence that NO synthase (NOS)-like activity (i.e. generation of NO from l-Arg) is greatly reduced in senescing leaves. Plant NOS activity is regulated by Ca2+/calmodulin (CaM; Delledonne et al., 1998; Corpas et al., 2004, 2009; del Río et al., 2004; Valderrama et al., 2007; Ma et al., 2008). These studies suggest a link between Ca2+ and NO that could be operating during senescence.
In animal cells, all three NOS isoforms require Ca2+/CaM as a cofactor (Nathan and Xie, 1994; Stuehr, 1999; Alderton et al., 2001). Notably, animal NOS contains a CaM binding domain (Stuehr, 1999). It is unclear whether Ca2+/CaM can directly modulate plant NOS or if Ca2+/CaM impacts plant leaf development/senescence through (either direct or indirect) effects on NO generation. However, recent studies from our lab suggest that Ca2+/CaM acts as an activator of NOS activity in plant innate immune response signaling (Ali et al., 2007; Ma et al., 2008).
Although Arabidopsis NO ASSOCIATED PROTEIN1 (AtNOA1; formerly named AtNOS1) was thought to encode a NOS enzyme, no NOS-encoding gene has yet been identified in plants (Guo et al., 2003; Crawford et al., 2006; Zemojtel et al., 2006). However, the AtNOA1 loss-of-function mutant does display reduced levels of NO generation, and several groups have used the NO donor sodium nitroprusside (SNP) to reverse some low-NO related phenotypes in Atnoa1 plants (Guo et al., 2003; Bright et al., 2006; Zhao et al., 2007). Importantly, plant endogenous NO deficiency (Guo and Crawford, 2005; Mishina et al., 2007) or abscisic acid/methyl jasmonate (Hung and Kao, 2003, 2004) induced early senescence can be successfully rescued by application of exogenous NO. Addition of NO donor can delay GA-elicited PCD in barley (Hordeum vulgare) aleurone layers as well (Beligni et al., 2002).
It has been suggested that salicylic acid (SA), a critical pathogen defense metabolite, can be increased in natural (Morris et al., 2000; Mishina et al., 2007) and transgenic NOD-induced senescent Arabidopsis leaves (Mishina et al., 2007). Pathogenesis related gene1 (PR1) expression is up-regulated in transgenic Arabidopsis expressing NOD (Mishina et al., 2007) and in leaves of an early senescence mutant (Ülker et al., 2007).
Plant cyclic nucleotide gated channels (CNGCs) have been proposed as candidates to conduct extracellular Ca2+ into the cytosol (Sunkar et al., 2000; Talke et al., 2003; Lemtiri-Chlieh and Berkowitz, 2004; Ali et al., 2007; Demidchik and Maathuis, 2007; Frietsch et al., 2007; Kaplan et al., 2007; Ma and Berkowitz, 2007; Urquhart et al., 2007; Ma et al., 2009a, 2009b). Arabidopsis “defense, no death” (dnd1) mutant plants have a null mutation in the gene encoding the plasma membrane-localized Ca2+-conducting CNGC2 channel. This mutant also displays no hypersensitive response to infection by some pathogens (Clough et al., 2000; Ali et al., 2007). In addition to involvement in pathogen-mediated Ca2+ signaling, CNGC2 has been suggested to participate in the process of leaf development/senescence (Köhler et al., 2001). dnd1 mutant plants have high levels of SA and expression of PR1 (Yu et al., 1998), and spontaneous necrotic lesions appear conditionally in dnd1 leaves (Clough et al., 2000; Jirage et al., 2001). Endogenous H2O2 levels in dnd1 mutants are increased from wild-type levels (Mateo et al., 2006). Reactive oxygen species molecules, such as H2O2, are critical to the PCD/senescence processes of plants (Navabpour et al., 2003; Overmyer et al., 2003; Hung and Kao, 2004; Guo and Crawford, 2005; Zimmermann et al., 2006). Here, we use the dnd1 mutant to evaluate the relationship between leaf Ca2+ uptake during plant growth and leaf senescence. Our results identify NO, as affected by leaf Ca2+ level, to be an important negative regulator of leaf senescence initiation. Ca2+-mediated NO production during leaf development could control senescence-associated gene (SAG) expression and the production of molecules (such as SA and H2O2) that act as signals during the initiation of leaf senescence programs.
RESULTS
Leaf Ca2+ Accumulation Is Reduced in the dnd1 Mutant
Ca2+-conducting ion channels facilitate transitory cytosolic Ca2+ spikes as components of numerous signaling pathways in plant cells. They are also thought to play a role in Ca2+ nutrition (i.e. uptake and translocation of Ca2+ within the plant; White and Broadley, 2003; Hetherington and Brownlee, 2004). Patch clamp studies of plant cells also indicate that nonselective, weakly voltage gated (i.e. potentially ligand gated) channels contribute to Ca2+ uptake in plants (White et al., 2002; White and Broadley, 2003; Demidchik and Maathuis, 2007). However, current reviews indicate no gene product has yet been associated with Ca2+ uptake into plants or accumulation in leaves (Maathuis, 2009). Therefore, it is currently unclear from prior work that cation channels contribute to Ca2+ uptake into plants and accumulation in leaves during plant growth and development (in contrast to temporary influx of Ca2+ associated with signaling).
Of the 57 known cation conducting channels in Arabidopsis, 20 members are CNGCs. CNGCs are candidates for specific gene products involved in the plant Ca2+ uptake pathway (White et al., 2002; White and Broadley, 2003; Demidchik and Maathuis, 2007). The cytosolic secondary messenger cAMP activates inward Ca2+current through the plasma membrane in wild-type leaf cells while this current is absent in leaves of the CNGC2 loss-of-function mutant dnd1 (Lemtiri-Chlieh and Berkowitz, 2004; Ali et al., 2007). Pathogen recognition results in activation of CNGC-dependent inward Ca2+ current and downstream NO production during plant innate immune response signaling cascades (Ali et al., 2007; Ma et al., 2009a). These studies link CNGC channels with inward Ca2+ flux associated with signaling.
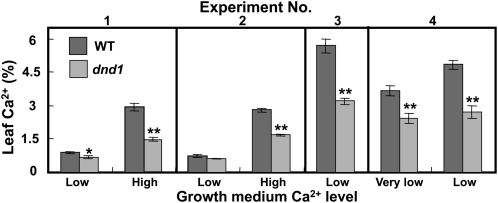
Here, we first investigated whether CNGC2 plays a role in Ca2+ uptake from a plant nutrition perspective by comparing leaf Ca2+content in wild-type and dnd1 plants grown with different Ca2+ levels in their growth medium. We found that the CNGC2 null mutation in dnd1 plants has effects on long-term Ca2+ acquisition (Fig. 1) at a range of external Ca2+, either when we measured total shoot Ca2+ levels in plants grown on solid (agar) medium (experiments 1 and 2) or grown hydroponically on liquid nutrient solution (experiments 3 and 4). In these experiments (Fig. 1), we observed a decrease in Ca2+ content of leaves of dnd1 plants compared with wild-type plants under a number of different growth conditions (solid and liquid media and varying external Ca2+). These results are consistent with a hypothesis that conductance through cation channels formed by CNGC2 contributes to Ca2+ nutrition of the plant. It appears that CNGC2 channels contribute to Ca2+ acquisition by leaves as part of the normal growth and development of the plant in addition to the Ca2+ conductance associated with innate immune signaling described by Ali et al. (2007) and Ma et al. (2009a).
Figure 1.
Leaf Ca2+ level is reduced in dnd1 plants compared to wild-type (WT) plants. Results from plants grown on solid medium (experiments 1 and 2) and hydroponic medium (experiments 3 and 4) are shown. The very low, low, and high Ca2+ treatments correspond to 0.75, 1.5, and 20 mm Ca2+ in the medium, respectively. Results for wild-type (dark bars) and dnd1 (light bars) plants are shown as means ± se (n 3). ANOVA analysis was used to evaluate means separation between paired wild-type and dnd1 measurements (for each experiment at a specific growth medium Ca2+ level). Significant differences (at P < 0.05 and P < 0.01, respectively) are indicated by * or ** above the light bars. The low Ca2+ treatment corresponds to the standard Ca2+ level in half-strength MS solid medium typically used for growth of Arabidopsis on agar plates. The high Ca2+ treatment corresponds to that used by Chan et al. (2003) for growth of wild-type and dnd1 plants. The very low Ca2+ treatment in experiment 4 (i.e. 0.75 mm in hydroponic nutrient solution) corresponds to a 50% reduction from that found in standard nutrient solutions. Plants grown on liquid hydroponic culture (experiments 3 and 4) had higher levels of leaf Ca2+ than that found in plants grown on solid medium (experiments 1 and 2). This could be due to either larger root systems in hydroponically grown plants or the increased availability of nutrients in a constantly mixed hydroponic solution compared to solid medium, where roots could deplete Ca2+ in localized zones. Leaves were harvested from plants grown for 3 (experiments 1 and 2), 4 (experiment 3), or 7 (experiment 4) weeks, respectively.
The work shown in Figure 1 is presented because the results are consistent with function of CNGC2 as a Ca2+ uptake pathway in leaves during growth and development (i.e. in addition to its role in immune responses). We do not assert here that the levels of leaf Ca2+ found in wild-type and dnd1 plants as shown in Figure 1 are specifically causal to differences in senescence programming in wild-type and mutant plants. Rather, we speculated that since CNGC2 is functioning as a Ca2+ uptake pathway during normal growth of wild-type plants, this channel may be responding to as yet unidentified signals during growth that impact the onset of senescence in leaves. CNGC2 loss of function in dnd1 plants could impact this signaling.
CNGC2 Is Involved in Plant Leaf Development/Senescence Signaling: The CNGC2 Loss-of-Function Mutant (dnd1) Displays Early Senescence Phenotypes
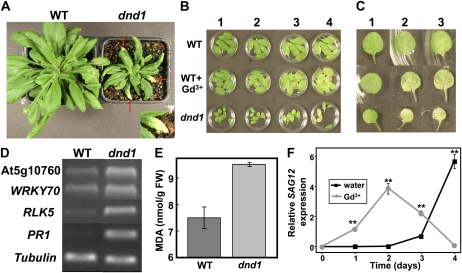
Results (Fig. 1) indicating that CNGC2 provides a pathway for Ca2+ uptake in plants beyond that related to immune signaling led us to investigate the link between CNGC2 and leaf senescence. We observed that dnd1 plants show early leaf senescence phenotypes compared to wild-type plants (Fig. 2). In Figure 2A, senescent yellowing leaves can be observed on dnd1 plants (highlighted by arrows) of the same age as wild-type plants, which do not have any leaves undergoing senescence at this point in plant development. The sporadic development of leaf tip yellowing shown here for dnd1 plants appears similar to that shown by Mishina et al. (2007) for plants with (apparently) low basal NO levels. Mishina et al. found that about a week after expression of a NOD, Arabidopsis plants had similar level and extent of leaf yellowing as shown here for dnd1 plants; this developmental change corresponds to senescence stage S2 (Mishina et al., 2007).
Figure 2.
Leaves of dnd1 plants display earlier senescence phenotypes compared to leaves of wild-type (WT) plants. A, dnd1 plants show earlier natural senescence (i.e. leaves begin turning yellow at the tip; highlighted by arrows). Inset shows an enlarged image of a portion from one leaf of the dnd1 plant. B, Leaves detached from wild-type plants (top and center rows) and dnd1 plants (bottom row) were incubated on water or water containing 100 μm Gd3+ (center row). From left to right, lanes 1 to 4 show leaves after 0, 3, 4, and 5 d in darkness, respectively. Counting from the first true leaf, leaves 3 to 8 of seedlings were used for this assay. C, Images are shown of individual detached leaf (in this case, leaves shown are either leaf 1 or 2) of the seedlings used for the experiment shown in B. Lanes 1 to 3 correspond to 0, 3, and 4 d in darkness treatment, respectively. D, RT-PCR products generated with primers corresponding to SAG genes and RNA prepared from leaves (the entire rosette) of wild-type or dnd1 plants as template were subjected to agarose gel electrophoresis. A band corresponding to tubulin is shown as a loading control. E, MDA levels in (rosette) leaves of wild-type and dnd1 plants (expressed per unit leaf fresh weight [FW]). Results are presented as mean (n = 3) ± se. ANOVA evaluation of means separation indicated differences between wild-type and dnd1 MDA levels were significant at P < 0.01. Similar results were found when the experiment was repeated three times. F, Quantitative real-time PCR analysis of SAG12 transcript accumulation (relative to tubulin transcript) in wild-type detached leaves (leaves 3–5) left in the dark (0–4 d) on water or Gd3+. Results are shown as means ± se (n = 3). ANOVA analysis was used to evaluate means separation at each time point. Significant differences (at P < 0.01) are indicated by ** above symbols.
Previous studies have reported that spontaneous necrotic lesions can appear in dnd1 leaves (Clough et al., 2000; Jirage et al., 2001). Here, our observation further expands our understanding of the dnd1 mutant phenotype. Subjecting detached young leaves to darkness also revealed that dnd1 leaves achieve senescence faster than wild-type leaves (Fig. 2, B and C). With leaves from wild-type plants, Gd3+, a Ca2+ channel blocker, hastens senescence, mimicking the early senescence phenotype of dnd1 leaves (Fig. 2, B and C). Results in Figure 2, A to C, are consistent with the hypothesis that the presence of a functional Ca2+ uptake pathway may act to defer senescence during development.
A recent microarray study from Chan et al. (2008) provides a wealth of information about differences in gene expression associated with the CNGC2 loss-of-function mutant. Among the genes whose expression was up-regulated in CNGC2 loss-of-function mutants as suggested by the global analysis undertaken by Chan et al. (2008), some are intimately associated with the senescence process (for information about those gene expression profiles in Arabidopsis tissues, refer to the Arabidopsis eFP browser at bar.utoronto.ca; Winter et al., 2007). Here, we monitored the expression of SAGs (WRKY70, RLK5, and At5g10760) in dnd1 and wild-type plants using semiquantitative reverse transcription (RT)-PCR, demonstrating up-regulation of these SAGs in leaves of dnd1 compared to wild-type plants (Fig. 2D). We also found increased PR1 transcript levels in dnd1 plants (Fig. 2D), similar to what was reported by Yu et al. (1998). Furthermore, measurements of lipid peroxidation (i.e. quantified by monitoring malondialdehyde [MDA] levels), which increases during senescence (Dhindsa et al., 1981; Buchanan-Wollaston, 1997; Berger et al., 2001; Wingler et al., 2004), indicate that dnd1 leaves have a higher level of lipid peroxidation than wild-type leaves (Fig. 2E). A number of different experimental approaches as delineated in Figure 2, then, indicate that senescence programs are activated in leaves of dnd1 plants compared to leaves of wild-type plants.
Results in Figure 2, B and C, indicate that with regard to the rapid senescence that occurs in detached leaves kept in the dark, application of Gd3+, a channel blocker that prevents inward currents through plasma membrane Ca2+-conducting channels (Tegg et al., 2005), mimics the phenotype shown by dnd1. Further studies were undertaken to determine if the effect of Gd3+ on senescence programming could be observed at the biochemical and gene expression level. Expression of SAG12 is increased during senescence of detached leaves kept in the dark as well as in planta in leaves undergoing natural senescence; not all SAG genes up-regulated in planta show a similar response to darkness in detached leaves (Weaver et al., 1998). Quantitative real-time PCR evaluation of SAG12 expression in detached wild-type leaves kept in the dark demonstrated that the hastening of senescence by Gd3+ could also be observed at the level of gene expression (Fig. 2F). SAG12 expression was greater at days 1 to 3 after detachment (Fig. 2F) in wild-type leaves when they were exposed to Gd3+. Further studies indicated that hastening of senescence by Gd3+ could be observed at the biochemical level as well. With detached leaves kept in the dark, MDA levels increased more quickly in the presence of Gd3+ (Supplemental Fig. S1). In other experiments, SAG12 expression was compared in leaves of wild-type and dnd1 plants (Supplemental Fig. S2). SAG12 expression was noted in leaves of dnd1 plants and not in leaves of wild-type plants (Supplemental Fig. S2, day 0). When leaves were detached from plants and kept in the dark, further SAG12 expression occurred in dnd1 leaves earlier than in wild-type leaves (compare expression on day 1 in Supplemental Fig. S2). Results from all of these aforementioned studies (Fig. 2; Supplemental Figs. S1 and S2) indicate that impairment of a leaf cell Ca2+ uptake pathway (either genetically or biochemically) hastens senescence programming.
dnd1 Has a Lower Endogenous NO Level in Leaves Compared to the Wild-Type Plant
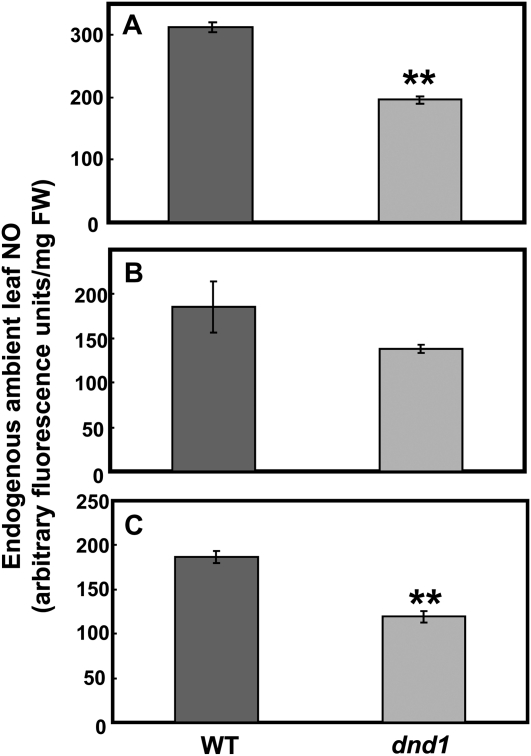
The hypothesis underlying the work presented in this report is that impairment of a Ca2+ uptake pathway functional during growth and development of dnd1 plants leads to loss of signal that represses senescence programming in leaves. We speculate that CNGC2-dependent Ca2+ uptake affects leaf senescence programming through an intermediary step of NO synthesis. Thus, in dnd1 plants with a null mutation in CNGC2, downstream Ca2+-dependent activation of NO generation may be impaired. Reduced NO generation could lead to a derepression of leaf senescence programming. The basis for this conjecture is as follows. (1) Prior studies (Poovaiah and Leopold, 1973; Chou and Kao, 1992; Corpas et al., 2004; Guo and Crawford, 2005; Mishina et al., 2007) discussed above suggest that both leaf Ca2+ and NO repress leaf senescence. (2) High levels of SA and expression of PR1 are associated with treatments that reduce NO production in leaves and induce early senescence (Morris et al., 2000; Mishina et al., 2007; Ülker, et al., 2007); SA level and PR1 expression are elevated in leaves of dnd1 plants (Yu et al., 1998). (3) Senescing leaves have reduced endogenous NO level (Corpas et al., 2004). (4) Recent studies from this lab (Ali et al., 2007) indicate that impairment of innate immune signaling in dnd1 plants occurs due to a lack of Ca2+-dependent NO generation. We measured the endogenous NO level in wild-type and dnd1 leaves. As shown in Figure 3, dnd1 plants have lower endogenous levels of leaf NO compared to the wild type.
Figure 3.
Endogenous leaf NO is reduced in dnd1 plants compared to wild-type (WT) plants. Genotype means of arbitrary fluorescence units (n = 3) ± se are shown for three independent experiments. A ** above a bar representing NO levels in dnd1 plants indicates the difference from the level found in wild-type leaves is significant at P < 0.01 for an individual experiment. ANOVA (paired t test) evaluation of means separation between wild-type and dnd1 genotypes for the pooled values from all three experiments indicated the genotype differences were significant at P < 0.01. FW, Fresh weight.
To our knowledge, endogenous basal NO (in contrast to NO generation in response to a specific signal) in leaves of any Arabidopsis genotype has not been previously reported. In prior studies from this lab (Ali et al., 2007), basal level of NO in guard cells was monitored in epidermal peels of wild-type and dnd1 plant leaves. No significant differences were noted, although the level in dnd1 guard cells was slightly higher. The difference between these prior results focusing on guard cells and the work reported here may be due to the measurement of total leaf NO in the work shown in Figure 3.
NO Donor Application Rescues dnd1 Senescence-Associated Phenotypes
With regard to impaired pathogen response signaling in dnd1 plants, application of the NO donor SNP to these mutant plants complemented the dnd1 pathogen response phenotype (Ali et al., 2007). We therefore tested the model that is the focus of the work presented here about involvement of NO in Ca2+ repression of senescence programming by examining the effect of SNP application on some senescence-related phenotypes in leaves of dnd1 plants. As is the case with other NO donors used with plants, NO release from SNP requires light (Floryszak-Wieczorek et al., 2006). Therefore, our evaluation of SNP effects on senescence in dnd1 plants focused on phenotypes occurring in the light.
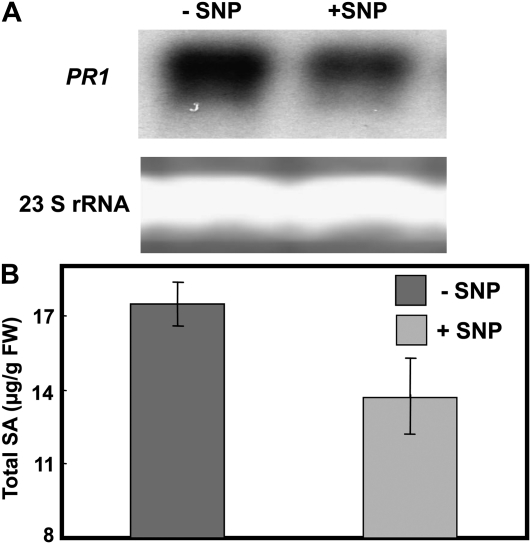
As mentioned above, prior studies have shown that PR1 expression and SA levels are elevated in dnd1 plants compared to wild-type plants (Yu et al., 1998). We confirmed these results (data not shown; also see Fig. 2D). We also find that SNP treatment reduced the high constitutive PR1 transcriptional level in dnd1 (Fig. 4A). In addition, SNP application to dnd1 plants also (modestly) lowered the constitutively high level of SA accumulation (Fig. 4B). SNP application had no significant effect on either PR1 expression or SA levels in leaves of wild-type plants (data not shown).
Figure 4.
PR1 expression (A) and total SA (B) in leaves of dnd1 plants treated with water (−SNP) or 100 μm SNP (+SNP). PR1 expression was monitored using northern-blot hybridization; loading controls show the 23S ribosomal RNA (rRNA) band from the corresponding agarose gel stained with ethidium bromide. The experiment shown in A was repeated two times. Results in B are presented as means (n = 3) ± se. FW, Fresh weight.
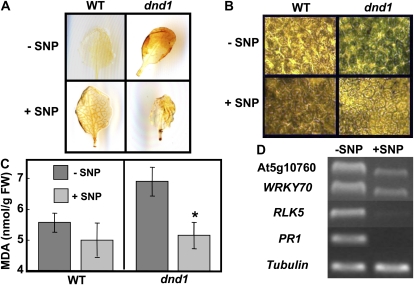
Effect of exogenous NO application was further tested on other senescence-related phenotypes in dnd1 plants. Increased endogenous H2O2 content (reported in Mateo et al., 2006) and extent of necrotic regions in leaves (reported in Jirage et al., 2001) of dnd1 plants decreased after the addition of SNP (Fig. 5, A and B, respectively). Although only qualitative effects of SNP treatment can be discerned from the images shown in Figure 5, A and B, it appears that the effect of exogenous NO supply on dnd1 plants, i.e. reduced H2O2 and necrosis, is not induced by SNP in leaves of wild-type plants. SNP application also was found to have an effect on the high level of lipid peroxidation in dnd1 plants (Fig. 2E). As shown in Figure 5C, application of the NO donor SNP reduced the high level of lipid peroxidation in dnd1 leaves, while SNP had no effect on the level of lipid peroxidation in leaves of wild-type plants.
Figure 5.
Application of NO donor SNP rescues senescence-associated phenotypes in dnd1 plants. A, Endogenous H2O2 levels in leaves of wild-type (WT; left panels) and dnd1 (right panels) plants were detected using 3,3′-diaminobenzidine staining. Leaves from plants treated with water or with SNP are shown in top and bottom panels, respectively. B, Necrosis, monitored using Trypan Blue staining, in leaf tissue of wild-type and dnd1 plants treated with water (−SNP) or 100 μm SNP (+SNP). Dead cells become blue after staining. This experiment was repeated three times. C, Lipid peroxidation (MDA level) in leaf tissue of wild-type and dnd1 plants treated with water (−SNP; dark bars) or 100 μm SNP (+SNP; light bars). Results are presented as means (n = 3) ± se. ANOVA evaluation of means separation between –SNP and +SNP treatments indicated a significant difference for dnd1 leaves (P 0.05; indicated with a * above bar representing the +SNP treatment) and no significant difference for wild-type leaves. FW, Fresh weight. D, Effect of (50 μm) SNP on SAG transcript accumulation in dnd1 leaves was analyzed by semiquantitative RT-PCR. This experiment was repeated three times.
The expression level of the SAGs At5g10760, WRKY70, RLK5, and PR1 is increased in (attached) leaves of dnd1 plants (Fig. 2D). Application of the NO donor SNP to dnd1 plants reduced the expression level (monitored using RT-PCR) of these SAGs (Fig. 5D). Analysis using quantitative PCR also demonstrated that exogenous NO application reduced expression of PR1 in leaves of dnd1 plants; SNP had no significant effect on PR1 expression in leaves of wild-type plants (Supplemental Fig. S3). Results shown in Figure 5D and Supplemental Figure S3 are from experiments performed on young seedlings grown on agar medium enclosed in sealed boxes. As discussed above, in an experiment performed on mature plants with fully expanded leaves, we also found that an SNP treatment (in this case, mature leaves of plants were sprayed with aqueous solutions containing SNP) reduced PR1 expression (monitored in this experiment using northern analysis) in leaves of dnd1 plants (Fig. 4A).
In this report, we have shown that a null mutation in CNGC2, a Ca2+-conducting plasma membrane cation channel, results in a reduction in leaf Ca2+ levels during growth and development of dnd1 plants. We associated the loss of function of this Ca2+ uptake pathway in dnd1 plants with a number of senescence-related phenotypes that are complemented by exogenous application of the NO donor SNP. In Figure 6, we identify another NO-related phenotype of dnd1 plants. Application of NO at high levels inhibits growth of, and is toxic to, wild-type Arabidopsis (He et al., 2004) and tobacco (Nicotiana tabacum; Morot-Gaudry-Talarmain et al., 2002) plants. In the experiment shown in Figure 6, wild-type and dnd1 seedlings were grown in sealed containers on growth medium containing varying concentrations of SNP. It is thought that in the light, SNP in aqueous solutions undergoes photochemical degradation to release gaseous NO (Floryszak-Wieczorek et al., 2006). Presumably, growth of seedlings in sealed, illuminated containers would allow for greater buildup of gaseous NO around (and in) plant tissue (compared with spraying SNP on leaves or adding the NO donor to irrigation solution). Under these conditions, we note that increasing SNP in growth medium (to 100 μm) is lethal to wild-type seedlings, while dnd1 seedlings are less affected (Fig. 6). At 50 μm SNP, there is a visible difference between wild-type (more wilted appearance) and dnd1 seedlings (less affected) as well (Fig. 6). This sensitivity of wild-type Arabidopsis seedlings to SNP addition to the growth medium we report here is similar to that shown by He et al. (2004). Current work notes the paucity of easily discerned plant phenotypes of CNGC loss-of-function mutants (Frietsch et al., 2007). Thus, our identification of a NO-related phenotype of the dnd1 mutant could provide the basis for further study of the effects of CNGC-dependent Ca2+ uptake on growth and development. One possible mechanism underlying the dnd1 phenotype shown in Figure 6 is as follows. Lower endogenous levels of NO present in dnd1 plants during growth (Fig. 3) could allow for tissue in this mutant to be less sensitive (i.e. in an additive sense) to addition of exogenous NO.
Figure 6.
Wild-type (WT) seedlings are more sensitive than dnd1 seedlings to exogenous NO. Wild-type and dnd1 seedlings were transplanted into (covered) Magenta boxes with half-strength MS medium containing 0, 50, or 100 μm SNP. Pictures were taken 4 d after transplanting. This experiment was repeated three times with similar results.
DISCUSSION
Evidence presented in this manuscript depicts a model linking the function of the Ca2+ conducting channel CNGC2 with downstream NO production and senescence programming. Prior work has shown that transitory Ca2+ uptake into plant cells through CNGC2 initiated during pathogen defense signaling cascades involves downstream NO production. Here, we show that the same channel contributes to Ca2+ uptake into the leaf during growth and development of the plant beyond this previously reported role in pathogen defense responses. The Ca2+ uptake capability provided by CNGC2 apparently impacts NO generation during growth and development as well; null mutation of CNGC2 results in reduced endogenous NO level in dnd1 plants compared to wild-type plants. We find that exogenous application of a NO donor to dnd1 plants reverses a number of senescence-related phenotypes. This link, between CNGC2-mediated uptake of Ca2+ into leaf tissue and NO, presumably mediates leaf senescence development as a negative regulator. Therefore, this work provides new information about the molecular mechanism of plant senescence signaling.
We provide new experimental evidence indicating that dnd1 mutants have reduced NO production during growth and development of the plant and associate this reduction in basal NO level with the absence of a Ca2+ uptake pathway that is operative during growth of the plant. Guo and Crawford (2005) also attributed the complementation of the early senescence phenotype of atnoa1 mutants by SNP to a reversal (by the NO donor) of depressed NO generation during leaf senescence in these plants; they did not monitor the basal level of NO in these mutants. Mishina et al. (2007) also attributed the induction of early senescence by a NO-degrading treatment (NOD expression) to presumed changes in the basal level of this signaling molecule in leaves. In this work, we observe that dnd1 plants have a reduced endogenous NO level in leaves compared with the wild type, which supports the early senescence phenotypes in dnd1 leaves that we found in this study.
Previous studies suggest that NO not only functions as a senescence signaling regulator (Guo and Crawford, 2005; Mishina et al., 2007) but also inhibits SA elevation during this process (Mishina et al., 2007). Work presented here is consistent with that model. Application of SNP to dnd1 plants reduces their high SA level as well as the transcript level of a marker gene (PR1) for SA generation.
In summary, our studies provide new genetic information linking Ca2+ uptake through the CNGC2 channel and accumulation in leaves during the course of plant growth and development as a component of leaf senescence signaling. NO is also proposed to be involved in this signaling cascade. Results presented here are consistent with NO action as a negative regulator during the developmental progression to leaf senescence.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) wild-type (Columbia ecotype) and dnd1 (Clough et al., 2000) plants were generally grown in a growth chamber on LP5 potting mix containing starter fertilizer (Sun Gro) at 16 h light (100 μmol m−2 s−1 illumination)/8 h dark (72% relative humidity) and 22°C. Seeds were vernalized at 4°C in the dark for 2 d prior to use. During growth, plants were irrigated with half-strength Murashige and Skoog (MS; Caisson) solution one to two times to provide supplementary fertilizer. Alternatively, seeds were surface sterilized and spread on petri dishes containing half-strength MS medium, 2.6 mm MES (adjusted to pH 5.7 with KOH), 1% Suc, and 0.8% agar (Ma et al., 2006). These seeds were germinated and grown in a growth chamber with a day (60–70 μmol m−2 s−1 illumination)/night cycle of 16/8 h at 25°C. Rosette leaves from plants were used for experiments unless noted otherwise in the figure legends. In summary, leaves obtained from plants grown on solid medium were used for experiments shown in Figures 1, A and B, 2D, 5D, and 6 and Supplemental Figure S3. Plants grown using hydroponic culture were used for experiments shown in Figure 1, C and D (for details, see following sections). For all other experiments, leaves were harvested from plants grown on soil.
As characterized by Clough et al. (2000), the dnd1 Arabidopsis genotype is homozygous for a null mutation in the gene (At5g15410) encoding CNGC2. The dnd1 allele contains a G-to-A point mutation that creates a stop codon in exon 3 at Trp-290, generating a severely truncated and nonfunctional CNGC2 coding sequence. All chemicals were purchased from Sigma-Aldrich unless noted otherwise.
Leaf Ca2+
Wild-type and dnd1 plants were grown either on solid agar medium or on liquid nutrient solution (hydroponic culture). For growth on solid medium (experiments 1 and 2 in Fig. 1), 15 seeds were germinated/petri plate on medium containing 0.8% (w/v) agar, 1% (w/v) Suc, 2.5 mm MES (pH 5.7), and half-strength MS medium with either 1.5 mm Ca2+ (low Ca2+ treatment) or 20 mm (high Ca2+ treatment) final concentration CaCl2.2H2O. Shoots (tissue growing above the solid medium) were harvested after 3 weeks. For hydroponic culture (experiments 3 and 4 in Fig. 1), seedlings (maximum of 30/tank) were grown on approximately 5-cm-long rock wool plugs extending into 18-L tanks containing 1.25 mm KNO3, 1.5 mm (low Ca2+ treatment) Ca(NO3)2.4H2O, 0.75 mm MgSO4, 0.5 mm KH2PO4, 72 μm EDTA (ferric sodium salt), 50 μm KCl, 50 μm H3BO3, 10 μm MnSO4, 2 μm ZnSO4, 1.5 μm CuSO4, and 0.525 μm Na2MoO4 with constant aeration. In the hydroponic culture for the very low Ca2+ treatment, the medium contained 0.75 mm Ca(NO3)2.4H2O, KNO3 was increased to 2.75 mm, and all the other components were added at the same concentration as stated above. Water was added every other day to maintain the level of solution in tanks, and leaves were harvested after 4 (experiment 3) or 7 (experiment 4) weeks of growth. Harvested leaves (hydroponics) or shoots (from agar plates) were dried at 70°C for 2 to 3 d prior to tissue ashing and analysis of Ca2+ (using an inductively coupled plasma mass optical emission spectrometer) by the National Science Foundation-funded Purdue Ionomics Facility as described (Salt, 2004). A minimum of 200 mg fresh weight of tissue was used for each tissue sample. Replicate tissue samples were taken from different plates in the case of the solid medium experiment and from different plants in the case of the hydroponic experiment.
Dark-Induced Leaf Senescence
The darkness-induced leaf senescence assay we used was a method adapted from Ülker et al. (2007). In brief, leaves were detached from 5-week-old plants and floated on water or water containing 100 μm Gd3+ in petri dishes as described in the figure legends. The first true leaf at the bottom of the shoot (arising after the cotyledons) was counted as the oldest leaf. Leaves with same age were used to ensure the accuracy of the experiments. Petri dishes were wrapped with aluminum foil and kept at 22°C. Pictures were taken after 3 to 5 d as indicated in the figure legends.
Lipid Peroxidation
The extent of lipid peroxidation in leaves was evaluated by measuring MDA formation as described by Guo and Crawford (2005). For these studies, 3-week-old plants were used except for the experiment shown in Supplemental Figure S1; in this case, plants were 5 weeks old. In brief, each leaf sample was either ground in chilled extraction buffer (containing 0.25% [w/v] thiobarbituric acid in 10% [w/v] trichloroacetic acid; Fisher Scientific) directly or first ground in liquid nitrogen followed by resuspension in extraction buffer. The homogenates were incubated in a water bath (90°C) for 20 min. Heated homogenates were then equilibrated to room temperature and centrifuged (12,000g for 15 min). Supernatants were decanted for optical density measurement at A532 and A600 for MDA determination using an extinction coefficient presented by Heath and Packer (1968). Effect of an exogenous NO donor on lipid peroxidation was evaluated. In this case, wild-type and dnd1 plants were treated with the NO donor by spraying leaves (once per day) with water containing 0 or 100 μm SNP for 2 d as described by Beligni and Lamattina (1999). When lipid peroxidation was monitored during dark-induced leaf senescence, plants were treated as noted above. Samples were harvested for the MDA measurement 0 to 3 d after initiating the dark treatment as indicated in the figure legends.
Gene Expression (RNA Extraction, Semiquantitative RT-PCR, Quantitative Real-Time PCR, and Northern Blots)
For experiments shown in Figures 2D, 5D, and Supplemental Figure S3, 3-week-old plants were used. Five-week-old plants were used for experiments shown in Figures 2F, 4B, and Supplemental Figure S2. For experiments involving application of an exogenous NO donor, 3-week-old seedlings were transplanted to clear polycarbonate boxes containing half-strength MS medium (with either 0 or 50 μm SNP). Five days after transplanting, leaves were harvested and frozen in liquid nitrogen for RNA extraction. The dark-induced leaf senescence experiment was performed as described in the above paragraph. Samples were harvested for the RNA extraction 0 to 4 d as indicated in the figure legends. Total RNA was isolated from leaf samples (ground in liquid nitrogen) using the RNA kit (Qiagen) following the supplier’s protocol. Genomic DNA contamination was removed using the DNA-free reagent (Ambion) following the manufacturer’s instructions. First-strand cDNA was synthesized in 20 μL using 1 μg RNA and reagents from the RETROscript kit (Ambion) following the manufacturers’ instructions. Subsequent PCR experiments were performed using 1.5 μL first-strand cDNA as template. Arabidopsis tubulin β-subunit (At5g44340) was used as the internal standard with the following gene-specific primers (forward [F], 5′-ACGTATCGATGTCTATTTCAACGA-3′, and reverse [R], 5′-ATATCGTAGAGAGCCTCATTGTCC-3′) as described by Ülker et al. (2007). Primers for PR1 (At2g14610; F, 5′-TCGTCTTTGTAGCTCTTGTAGGTG-3′, and R, 5′-TTCATTAGTATGGCTTCTCGTTCA-3′) and WRKY70 (At3g56400; F, 5′-TAATGGATACTAATAAAGCAAAAAAGC-3′, and R, 5′-CAGATAGATTCGAACATGAACTGAAG-3′) were described by Ülker et al. (2007). Primers for RLK5 (At4g23140; F, 5′-TAAAGCAAGTAACATTCTCCTAGATGC-3′, and R, 5′-AGCATGAAGTTTAAGGTCATATTTAGC-3′) were described by Galon et al. (2008). Primers for gene At5g10760 were designed as follows: F, 5′-CTTGTTGTGAGAGTAAGAGGGAGGT-3′, and R, 5′-CCGCAACCAAAGTAAACATCCTCCA-3′. No DNA contamination was found in the RNA sample after DNA-free reagent treatment.
Quantitative real-time PCR was performed using the LightCycler FastStart DNA MasterPLUS SYBR Green I kit (Roche Applied Science) or SYBR Green PCR Core Reagents mix (Applied Biosystems) according to the manufacturer’s manual. The level of gene transcript accumulation was normalized to an internal standard tubulin β-subunit (At5g44340). Primers used for real-time PCR were either the same as described above or as follows: SAG12 (At5G45890), F, 5′-GCGGATGTGAAGGAGGAAAA-3′, and R, 5′-CAATGCGTTCGACGTTGTTT-3′; tubulin β-subunit (At5g44340), F, 5′-TGTTTCGTTCATGTGTGTTCGTT-3′, and R, 5′-ACACGCAAAAGTTTAACAAATCCA-3′.
For northern-blot analysis, RNA was isolated from leaves of plants sprayed with water containing 0 or 100 μm SNP once daily for 2 d as described by Beligni and Lamattina (1999) using TRI reagent (Molecular Research Center) according to the manufacturer’s instructions. RNA blotting and probe labeling protocols followed the procedures described by Mercier et al. (2004). Primers for the PR1 probes were amplified using primers 5′-GTAGGTGCTCTTGTTCTTCCC-3′ and 5′-CACATAATTCCCACGAGGATC-3′ as described by Jurkowski et al. (2004).
H2O2 and Cell Death in Leaves
Rosette leaves of 4- to 6-week-old wild-type and dnd1 plants were irrigated with half-strength MS solution containing 100 μm SNP or half-strength MS solution alone (control) for 7 to 10 d as described by Ali et al. (2007). After treatment, the leaves were detached from the plant and used for either in vivo H2O2 detection or assay of cell death using Trypan Blue staining. The H2O2 detection assay was performed as previously described by Kwak et al. (2003). A 3,3′-diaminobenzidine (Sigma-Aldrich) stain was vacuum infiltrated into detached leaves for 1 to 2 min; leaves were then placed into a humid plastic box and left overnight. An ethanol:glycerol:85% lactic acid (3:1:1) fixation solution was added to the leaves, which were then shaken at 50 rpm for 24 to 48 h until pigments were completely removed from the leaf tissue. Trypan Blue staining was performed as previously described by Jirage et al. (2001). In brief, detached leaves were boiled in a staining solution containing Trypan Blue (Sigma-Aldrich) at a final working concentration of 250 μg/mL in 1:1:1:1 (v/v/v/v) water:1 m phenol:glycerol:85% lactic acid. Images were captured with an EPSON 1660 Perfection Photo Scanner using Adobe Photoshop Imaging software. All microscopy photographs were taken with an inverted Olympus IX70 microscope.
Endogenous NO in Leaves
The method used to measure endogenous NO in leaves was adapted from that described by Lamattina and colleagues (Graziano and Lamattina, 2007; Martin et al., 2009). NO was monitored using the NO-specific fluorescent dye 3-amino, 4-aminomethyl-2′,7′-difluorofluorescein diacetate (DAF-FM DA; Calbiochem). The use of DAF dye to measure NO in plant leaves has been used in conjunction with alternative methods for NO quantification in prior studies. In this prior work, effects of treatments on leaf NO as monitored with DAF fluorescence were comparable to NO analyses using alternative methods. For example, NO elevations due to exposure of leaves to lipopolysaccharide were detected similarly using either DAF fluorescence or electron paramagnetic resonance (Zeidler et al., 2004).
Leaves were detached from 4- to 5-week-old wild-type and dnd1 plants and weighed (each sample is around 100 mg). Each individual sample was then vacuum infiltrated with or without 10 μm DAF-FM DA (NO detection dye) in 20 mm HEPES/NaOH (pH 7.5) buffer. Leaves were kept in this solution for 10 min after vacuum infiltration. The samples were then washed three times (shaking 1 min each time) in petri dishes containing 20 mm HEPES/NaOH (pH 7.5) buffer. After washing, samples were blotted to remove excess buffer and then homogenized in 0.5 mL of 20 mm HEPES/NaOH (pH 7.5) buffer. The extracts were then centrifuged at 15,000g at 4°C for 10 min. One hundred microliters of collected supernatant was added to the well of a 96-well microplate (black walls and clear bottom), and fluorescence signals were quantified using a FLUOstar Optima microplate reader (BMG Labtech) at excitation and emission wavelengths of 485 and 520 nm, respectively. Endogenous NO content was ascertained by subtracting a background signal value from the individual sample. Background signal was typically obtained by measuring fluorescence signals in wild-type and dnd1 samples infiltrated with 20 mm HEPES/NaOH (pH 7.5) buffer alone.
Leaf SA
Eight-week-old plants were treated with a NO donor by spraying leaves with water containing 0 or 100 μm SNP daily for 2 d as described by Beligni and Lamattina (1999). SA and its glucoside were quantified using gas chromatography-mass spectrometry. One hundred milligrams of frozen leaf tissue were extracted twice with 800 μL acetone:50 mm citric acid (7:3, v/v) in 2-mL Fast Prep tubes containing ceramic beads using a FastPrep FP 120 tissue homogenizer (Qbiogene). Radiolabeled [2H6] SA (CDN Isotopes) was added to leaf samples as an internal standard. After evaporation of the acetone under vacuum, the aqueous solutions were extracted twice with 750 μL of diethyl ether. SA-glucoside was extracted from the remaining aqueous solution after acidification with 5 μL of HCl and hydrolysis at 90°C for 1 h by diethyl-ether extraction. All samples were then loaded on 1-mL Supelclean LC-NH2 SPE columns (Supelco). After washing with 1.2 mL chloroform:2-propanol (2:1, v/v), compounds were eluted with 1.5 mL diethyl-ether:formic acid (98:2, v/v). The eluates were then evaporated to dryness under a stream of N2, dissolved in 100 μL dichloromethane:methanol (8:2, v/v), and derivatized with 2 μL trimethylsilyl-diazomethane (Aldrich) for 20 min. The reaction was stopped by adding 2 μL of 2 m acetic acid in hexane (Schmelz et al., 2004). The resulting methyl esters of SA were analyzed by gas chromatography-mass spectrometry (6890 n GC connected to a 5975 mass selective detector; Agilent Technologies) in isobutene chemical ionization mode following the specifications of Schmelz et al. (2004). The methyl esters were measured using selected-ion monitoring with mass-to-charge ratio of 153 (SA) and 157 ([2H6] SA). Results are presented as total (conjugated and free) SA.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. MDA levels in leaves detached from wild-type plants and floated in the dark on water or 100 μm Gd3+ for 0 to 3 d.
Supplemental Figure S2. SAG12 transcript accumulation in wild-type (WT) and dnd1 detached leaves (kept in the dark on water for 0–2 d) was analyzed by semiquantitative RT-PCR.
Supplemental Figure S3. Quantitative real-time PCR analysis of PR1 transcript accumulation (relative to tubulin transcript) in leaves of dnd1 and wild-type (WT) plants treated with 0 or 50 μm SNP as described in Figure 5D.
Supplementary Material
Acknowledgments
We thank David Salt, Laura Loder, and Brett Lahner for technical assistance with the shoot Ca2+ analysis. We thank John Farley and his lab members for their efforts in SA measurement. We thank Shafiuddin Siddiqui and Zhi Qi for technical assistance with quantitative real-time PCR experiment.
References
- Alderton WK, Cooper CE, Knowles RG. (2001) Nitric oxide synthases: structure, function and inhibition. Biochem J 357: 593–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali R, Ma W, Lemtiri-Chlieh F, Tsaltas D, Leng Q, von Bodman S, Berkowitz GA. (2007) Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beligni MV, Fath A, Bethke PC, Lamattina L, Jones RL. (2002) Nitric oxide acts as an antioxidant and delays programmed cell death in barley aleurone layers. Plant Physiol 129: 1642–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beligni MV, Lamattina L. (1999) Nitric oxide protects against cellular damage produced by methylviologen herbicides in potato plants. Nitric Oxide 3: 199–208 [DOI] [PubMed] [Google Scholar]
- Berger S, Weichert H, Porzel A, Wasternack C, Kühn H, Feussner I. (2001) Enzymatic and non-enzymatic lipid peroxidation in leaf development. Biochim Biophys Acta 1533: 266–276 [DOI] [PubMed] [Google Scholar]
- Bright J, Radhika D, Hancock JT, Weir IS, Neill JS. (2006) ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J 45: 113–122 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V. (1997) The molecular biology of leaf senescence. J Exp Bot 48: 181–199 [Google Scholar]
- Chan CW, Schorrak LM, Smith RK, Jr, Bent AF, Sussman MR. (2003) A cyclic nucleotide-gated ion channel, CNGC2, is crucial for plant development and adaptation to calcium stress. Plant Physiol 132: 728–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CW, Wohlbach DJ, Rodesch MJ, Sussman MR. (2008) Transcriptional changes in response to growth of Arabidopsis in high external calcium. FEBS Lett 582: 967–976 [DOI] [PubMed] [Google Scholar]
- Chou CM, Kao CH. (1992) Methyl jasmonate, calcium, and leaf senescence in rice. Plant Physiol 99: 1693–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrost B, Daniel A, Krupinska K. (2004) Regulation of alpha-galactosidase gene expression in primary foliage leaves of barley (Hordeum vulgare L.) during dark-induced senescence. Planta 218: 886–889 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Fengler KA, Yu IC, Lippok B, Smith RK, Jr, Bent AF. (2000) The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci USA 97: 9323–9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, Carreras A, Quirós M, León AM, Romero-Puertas MC, Esteban FJ, Valderrama R, Palma JM, Sandalio LM, et al. (2004) Cellular and subcellular localization of endogenous NO in young and senescent pea plants. Plant Physiol 136: 2722–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Palma JM, del Río LA, Barroso JB. (2009) Evidence supporting the existence of l-arginine-dependent nitric oxide synthase activity in plants. New Phytol 184: 9–14 [DOI] [PubMed] [Google Scholar]
- Crawford NM, Galli M, Tischner R, Heimer YM, Okamoto M, Mack A. (2006) Response to Zemojtel et al.: Plant nitric oxide synthase: back to square one. Trends Plant Sci 11: 526–527 [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C. (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394: 585–588 [DOI] [PubMed] [Google Scholar]
- del Río LA, Corpas FJ, Barroso JB. (2004) Nitric oxide and nitric oxide synthase activity in plants. Phytochemistry 65: 783–792 [DOI] [PubMed] [Google Scholar]
- Demidchik V, Maathuis FJ. (2007) Physiological roles of nonselective cation channels in plants: from salt stress to signalling and development. New Phytol 175: 387–404 [DOI] [PubMed] [Google Scholar]
- Dhindsa RS, Plumb-Dhindsa P, Thorpe TA. (1981) Leaf senescence: correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J Exp Bot 32: 93–101 [Google Scholar]
- Floryszak-Wieczorek J, Milczarek G, Arasimowicz M, Ciszewski A. (2006) Do nitric oxide donors mimic endogenous NO-related response in plants? Planta 224: 1363–1372 [DOI] [PubMed] [Google Scholar]
- Frietsch S, Wang YF, Sladek C, Poulsen LR, Romanowsky SM, Schroeder JI, Harper JF. (2007) A cyclic nucleotide-gated channel is essential for polarized tip growth of pollen. Proc Natl Acad Sci USA 104: 14531–14536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon Y, Nave R, Boyce JM, Nachmias D, Knight MR, Fromm H. (2008) Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett 582: 943–948 [DOI] [PubMed] [Google Scholar]
- Gan S, Amasino RM. (1997) Making sense of senescence (Molecular genetic regulation and manipulation of leaf senescence). Plant Physiol 113: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepstein S. (2004) Leaf senescence-not just a ‘wear and tear’ phenomenon. Genome Biol 5: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano M, Lamattina L. (2007) Nitric oxide accumulation is required for molecular and physiological responses to iron deficiency in tomato roots. Plant J 52: 949–960 [DOI] [PubMed] [Google Scholar]
- Guo FQ, Crawford NM. (2005) Arabidopsis NO synthase1 is targeted to mitochondria and protects against oxidative damage and dark-induced senescence. Plant Cell 17: 3436–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FQ, Okamoto M, Crawford NM. (2003) Identification of a plant NO synthase gene involved in hormonal signaling. Science 302: 100–103 [DOI] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, et al. (2004) Nitric oxide represses the Arabidopsis floral transition. Science 305: 1968–1971 [DOI] [PubMed] [Google Scholar]
- Heath RL, Packer L. (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125: 180–198 [DOI] [PubMed] [Google Scholar]
- Hetherington AM, Brownlee C. (2004) The generation of Ca2+ signals in plants. Annu Rev Plant Biol 55: 401–427 [DOI] [PubMed] [Google Scholar]
- Hung KT, Kao CH. (2003) Nitric oxide counteracts the senescence of rice leaves induced by abscisic acid. J Plant Physiol 160: 871–879 [DOI] [PubMed] [Google Scholar]
- Hung KT, Kao CH. (2004) Nitric oxide acts as an antioxidant and delays methyl jasmonate-induced senescence of rice leaves. J Plant Physiol 161: 43–52 [DOI] [PubMed] [Google Scholar]
- Jirage D, Zhou N, Cooper B, Clarke JD, Dong X, Glazebrook J. (2001) Constitutive salicylic acid-dependent signaling in cpr1 and cpr6 mutants requires PAD4. Plant J 26: 395–407 [DOI] [PubMed] [Google Scholar]
- Jurkowski GI, Smith RK, Jr, Yu IC, Ham JH, Sharma SB, Klessig DF, Fengler KA, Bent AF. (2004) Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol Plant Microbe Interact 17: 511–520 [DOI] [PubMed] [Google Scholar]
- Kaplan B, Sherman T, Fromm H. (2007) Cyclic nucleotide-gated channels in plants. FEBS Lett 581: 2237–2246 [DOI] [PubMed] [Google Scholar]
- Köhler C, Merkle T, Roby D, Neuhaus G. (2001) Developmentally regulated expression of a cyclic nucleotide-gated channel from Arabidopsis indicates its involvement in programmed cell death. Planta 213: 327–332 [DOI] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI. (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F, Berkowitz GA. (2004) Cyclic adenosine monophosphate regulates calcium channels in the plasma membrane of Arabidopsis leaf guard and mesophyll cells. J Biol Chem 279: 35306–35312 [DOI] [PubMed] [Google Scholar]
- Lim PO, Woo HR, Nam HG. (2003) Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci 8: 272–278 [DOI] [PubMed] [Google Scholar]
- Ma W, Ali R, Berkowitz GA. (2006) Characterization of plant phenotypes associated with loss-of-function of AtCNGC1, a plant cyclic nucleotide gated cation channel. Plant Physiol Biochem 44: 494–505 [DOI] [PubMed] [Google Scholar]
- Ma W, Berkowitz GA. (2007) The grateful dead: calcium and cell death in plant innate immunity. Cell Microbiol 9: 2571–2585 [DOI] [PubMed] [Google Scholar]
- Ma W, Qi Z, Smigel A, Walker RK, Verma R, Berkowitz GA. (2009a) Ca2+, cAMP, and transduction of non-self perception during plant immune responses. Proc Natl Acad Sci USA 106: 20995–21000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Smigel A, Tsai YC, Braam J, Berkowitz GA. (2008) Innate immunity signaling: Cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiol 148: 818–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Smigel A, Verma R, Berkowitz GA. (2009b) Cyclic nucleotide gated channels and related signaling components in plant innate immunity. Plant Signal Behav 4: 277–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJ. (2009) Physiological functions of mineral macronutrients. Curr Opin Plant Biol 12: 250–258 [DOI] [PubMed] [Google Scholar]
- Martin M, Colman MJ, Gómez-Casati DF, Lamattina L, Zabaleta EJ. (2009) Nitric oxide accumulation is required to protect against iron-mediated oxidative stress in frataxin-deficient Arabidopsis plants. FEBS Lett 583: 542–548 [DOI] [PubMed] [Google Scholar]
- Mateo A, Funck D, Mühlenbock P, Kular B, Mullineaux PM, Karpinski S. (2006) Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J Exp Bot 57: 1795–1807 [DOI] [PubMed] [Google Scholar]
- Mercier RW, Rabinowitz NM, Ali R, Gaxiola RA, Berkowitz GA. (2004) Yeast hygromycin sensitivity as a functional assay of cyclic nucleotide gated cation channels. Plant Physiol Biochem 42: 529–536 [DOI] [PubMed] [Google Scholar]
- Mishina TE, Lamb C, Zeier J. (2007) Expression of a NO degrading enzyme induces a senescence programme in Arabidopsis. Plant Cell Environ 30: 39–52 [DOI] [PubMed] [Google Scholar]
- Morot-Gaudry-Talarmain Y, Rockel P, Moureaux T, Quilleré I, Leydecker MT, Kaiser WM, Morot-Gaudry JF. (2002) Nitrite accumulation and nitric oxide emission in relation to cellular signaling in nitrite reductase antisense tobacco. Planta 215: 708–715 [DOI] [PubMed] [Google Scholar]
- Morris K, MacKerness SA, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V. (2000) Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J 23: 677–685 [DOI] [PubMed] [Google Scholar]
- Nathan C, Xie QW. (1994) Nitric oxide synthases: roles, tolls, and controls. Cell 78: 915–918 [DOI] [PubMed] [Google Scholar]
- Navabpour S, Morris K, Allen R, Harrison E, A-H-Mackerness S, Buchanan-Wollaston V. (2003) Expression of senescence-enhanced genes in response to oxidative stress. J Exp Bot 54: 2285–2292 [DOI] [PubMed] [Google Scholar]
- Overmyer K, Brosché M, Kangasjärvi J. (2003) Reactive oxygen species and hormonal control of cell death. Trends Plant Sci 8: 335–342 [DOI] [PubMed] [Google Scholar]
- Poovaiah BW, Leopold AC. (1973) Deferral of leaf senescence with calcium. Plant Physiol 52: 236–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino BF, Noh YS, Himelblau E, Amasino RM. (2000) Molecular aspects of leaf senescence. Trends Plant Sci 5: 278–282 [DOI] [PubMed] [Google Scholar]
- Salt DE. (2004) Update on plant ionomics. Plant Physiol 136: 2451–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Engelberth J, Tumlinson JH, Block A, Alborn HT. (2004) The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J 39: 790–808 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Kaplan B, Bouche N, Arazi T, Dolev D, Talke IN, Maathuis FJ, Sanders D, Bouchez D, Fromm H. (2000) Expression of a truncated tobacco NtCBP4 channel in transgenic plants and disruption of the homologous Arabidopsis CNGC1 gene confer Pb2+ tolerance. Plant J 24: 533–542 [DOI] [PubMed] [Google Scholar]
- Stuehr DJ. (1999) Mammalian nitric oxide synthases. Biochim Biophys Acta 1411: 217–230 [DOI] [PubMed] [Google Scholar]
- Talke IN, Blaudez D, Maathuis FJM, Sanders D. (2003) CNGCs: prime targets of plant cyclic nucleotide signalling? Trends Plant Sci 8: 286–293 [DOI] [PubMed] [Google Scholar]
- Tegg RS, Melian L, Wilson CR, Shabala S. (2005) Plant cell growth and ion flux responses to the Streptomycete phytotoxin Thaxtomin A: calcium and hydrogen flux patterns revealed by the non-invasive MIFE technique. Plant Cell Physiol 46: 638–648 [DOI] [PubMed] [Google Scholar]
- Ülker B, Shahid Mukhtar M, Somssich IE. (2007) The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226: 125–137 [DOI] [PubMed] [Google Scholar]
- Urquhart W, Gunawardena AH, Moeder W, Ali R, Berkowitz GA, Yoshioka K. (2007) The chimeric cyclic nucleotide-gated ion channel ATCNGC11/12 constitutively induces programmed cell death in a Ca2+ dependent manner. Plant Mol Biol 65: 747–761 [DOI] [PubMed] [Google Scholar]
- Valderrama R, Corpas FJ, Carreras A, Fernández-Ocaña A, Chaki M, Luque F, Gómez-Rodríguez MV, Colmenero-Varea P, Del Río LA, Barroso JB. (2007) Nitrosative stress in plants. FEBS Lett 581: 453–461 [DOI] [PubMed] [Google Scholar]
- Weaver LM, Amasino RM. (2001) Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol 127: 876–886 [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM. (1998) A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol 37: 455–469 [DOI] [PubMed] [Google Scholar]
- White PJ, Bowen HC, Demidchik V, Nichols C, Davies JM. (2002) Genes for calcium-permeable channels in the plasma membrane of plant root cells. Biochim Biophys Acta 1564: 299–309 [DOI] [PubMed] [Google Scholar]
- White PJ, Broadley MR. (2003) Calcium in plants. Ann Bot (Lond) 92: 487–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Marès M, Pourtau N. (2004) Spatial patterns and metabolic regulation of photosynthetic parameters during leaf senescence. New Phytol 161: 781–789 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu IC, Parker J, Bent AF. (1998) Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci USA 95: 7819–7824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler D, Zähringer U, Gerber I, Dubery I, Hartung T, Bors W, Hutzler P, Durner J. (2004) Innate immunity in Arabidopsis thaliana: Lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc Natl Acad Sci USA 101: 15811–15816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemojtel T, Frohlich A, Palmieri MC, Kolanczyk M, Mikula I, Wyrwicz LS, Wanker EE, Mundlos S, Vingron M, Martasek P, et al. (2006) Plant nitric oxide synthase: a never-ending story? Trends Plant Sci 11: 524–525 [DOI] [PubMed] [Google Scholar]
- Zhao M, Zhao X, Wu Y, Zhang L. (2007) Enhanced sensitivity to oxidative stress in an Arabidopsis NO synthase mutant. J Plant Physiol 164: 737–745 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Heinlein C, Orendi G, Zentgraf U. (2006) Senescence-specific regulation of catalases in Arabidopsis thaliana (L.) Heynh. Plant Cell Environ 29: 1049–1060 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.